Abstract
Chickpeas are a very important legume due to their nutritional richness and high protein content and they are used as food for humans and as fodder for livestock. However, they are susceptible to fungal infections and mycotoxin contamination. The Alternaria genus was among the main fungi isolated from chickpea samples in Argentina. The species within this genus are able to produce several mycotoxins such as alternariol (AOH), alternariol monomethyl ether (AME), and tenuazonic acid (TA). So, the objectives of this study were to identify the Alternaria spp. found in the chickpea samples and to determine their toxigenic potential in vitro. A phylogenetic analysis of 32 Alternaria strains was carried out based on the combined sequences of the tef1, gpd, and Alt a1 genes. All Alternaria strains clustered into the section Alternaria and were identified as A. alternata and A. arborescens. Further, the toxigenic profile of each strain was determined in a ground rice–corn steep liquor medium and analysed by HPLC. Most strains were able to co-produce AOH, AME, and TA. These results indicate a potential risk for human health when consuming chickpeas since this legume could be contaminated with Alternaria and its mycotoxins, which are not yet regulated in food.
1. Introduction
Since ancient times, chickpeas (Cicer arietinum L.) have been a major legume crop, particularly in Asia. Chickpeas are the world’s most important crop, mainly used as food for humans and valued for their nutritious seeds with high protein content, which is of comparable or better quality than other legumes. Chickpeas can also be used as fodder for livestock [1]. Chickpeas are cultivated in a wide variety of agro-ecological conditions worldwide [2]. In Argentina, the crop is cultivated in fall–winter with low water requirements in comparison to other crops. Chickpea crops are susceptible to fungal contamination with both pathogens and saprophytes, some of them mycotoxin producers, so there is a potential risk of contamination with said agents. This can affect the vigor and longevity of the seeds [3]. The grain quality achieved by Argentina has enabled the country to be an important exporter and to be competitive in the international market. In 2021, in Argentina, chickpea cultivation spanned 81.236 ha, reaching 84.709 tons. The major portion of Argentina´s production is exported, although over the last few years, chickpea production has decreased due to unfavourable weather conditions, but a rise in output and export is expected for the next growing seasons [4].
The chickpea is susceptible to over 25 well-documented fungal pathogens and is often attacked—before and after harvest—by fungi, which greatly affects productivity. Some of these fungi have been identified as weak parasites as well as important contaminants and can damage the seeds during storage due to their nutritional richness and the texture of the seed coat. The most widespread fungi in chickpea plants are species belonging to the genera Aspergillus, Fusarium, Penicillium, Alternaria, and Rhizopus [5].
Recently (in the 2018 harvest season), we analysed 70 asymptomatic samples of chickpea from a local cereal storage company located in the northern region of the Córdoba province (the main chickpea production area) in Argentina. The analysis of the mycobiota present in the samples showed that species of the genus Aspergillus (54%) and Alternaria (22%) were isolated as the predominant ones. In addition, contamination by mycotoxins was determined in 10 chickpea samples using LC-MS/MS for preliminary screening. While Aspergillus was the predominant fungal genus isolated, with A. flavus being the most common species identified, we did not detect aflatoxins; instead, we detected Fusarium mycotoxins such as beauvericin, deoxynivalenol, and zearalenone. Among the Alternaria mycotoxin analysed, we detected alternariol monomethyl ether (AME) in all samples, at concentrations ranging from 0.7 to 14.5 ng/g, and alternariol (AOH) in 30% of samples at concentrations ranging from 1.4 to 2.3 ng/g [6].
Alternaria’s contamination of crops is particularly relevant due to their ability to produce a wide variety of mycotoxins that can have adverse effects on human and other animals’ health. The toxins produced by Alternaria species can be classified into host/non-host specific toxins [7] or grouped depending on their chemical structure [8], which is a preferred approach in matters of food security. The dibenzo-α-pyrone derivative group contains alternariol (AOH) and many modified forms, including alternariol mono methylether (AME), altenuene (ALT), isoaltenuene (isoALT), and altenuisol (ATL). Then comes the group derived from tetramic acid with tenuazonic acid (TA), followed by the perylene quinone derivative altertoxin I, II, and III (ATX-I, -II, -III); alterperylenol, also called alteichin (ALP); and stemphyltoxin III (STTX-III). The fourth group consists of various structures such as tentoxin (TEN), altenuic acid III (AA-III), and infectopyrone (INF), which was originally isolated from A. infectoria [3,9]. The toxins within these four groups belong to the group of non-host specific toxins and can be formed by different strains of Alternaria in a wide range of living host plants and agricultural products, such as harvested wheat grains, vegetables, cereal products, fruits, and oil seeds [10,11,12]. While there are currently no global regulations setting limits for such toxins in food and feed, the European Food Safety Authority (EFSA) expresses public health concerns regarding Alternaria mycotoxins [8,13].
The taxonomy of the genus Alternaria is an extremely controversial subject. The classification of the genus Alternaria has long been based solely on morphological traits, for example, colony morphology, mycelial growth, sporulation patterns, and conidia size and shape in accordance with Simmons’ proposed taxonomy key [14], who introduced the concept of species-groups to make identification easier. This approach is, however, not always efficient because of the strong influence of growing conditions on the morphological characteristics, the high level of similarity between certain species, and the presence of several strains with intermediate features [15]. As a consequence, many studies were devoted to Alternaria and related genera, often confirming a merged taxonomic background, which resulted in a continuous process of taxonomic revision. Conversely, multigenic phylogenetic analyses strongly supported the redefinition of the Alternaria genus. Several sequences from genetic regions such as glyceraldehyde-3-phosphate dehydrogenase (gpd), Alternaria major allergen (Alt a1), endopolygalacturonase (endoPG), translation elongation factor 1-alpha (tef1), calmodulin, plasma membrane ATPase (ATPase), the second largest subunit of RNA polymerase II (RPB2), and others have been applied to delimit the genus [16,17,18,19,20,21,22]. In recent studies, both morphological and molecular analyses have been used to delineate the genus Alternaria, which has been divided into 28 sections and eight monotypic lineages. The small-spored Alternaria species belongs to the section Alternaria under the species type A. alternata [20,23,24,25]. Alternaria species’ numbers have been continuously growing after redescriptions and new discoveries [26,27,28,29]. Coincidentally, several phylogenetic lineages have strongly supported morphology-based sections, while others have not [14,30].
For food-supplying countries, such as Argentina, contamination with mycotoxins can result in a negative impact or problems in trade in terms of rejection, restrictions, or unjustified demands. Thus, determining the distribution of the small-spored Alternaria in chickpea production areas is the initial step in determining the mycotoxigenic potential of crop contaminants and the risk in the final product.
As a result of the lack of information in our country on the contamination of this legume by species of the genus Alternaria, the objective of this study was to identify by phylogenetic analysis the Alternaria spp. found in samples of chickpea plants and to determine their toxigenic potential in vitro.
2. Materials and Methods
2.1. Isolates of Alternaria spp.
Out of 100 putative small-spored catenulate Alternaria strains previously isolated from chickpea grains (asymptomatic) in Argentina during 2018 harvest season [6], 32 single-spore strains were randomly selected for phylogenetic and toxicogenesis analyses. Isolates were morphologically characterized according to Simmons [14], mostly with respect to three-dimensional sporulation patterns. All the strains are deposited at the Universidad Nacional de Río Cuarto (UNRC) culture (RC) collection and preserved as spore suspensions in 15% glycerol frozen at −80 °C.
2.2. DNA Extraction, PCR Amplification, and Sequencing
Each strain was inoculated as a spore suspension in Erlenmeyer flasks containing 50 mL of Wikerman medium [31] and incubated at 25 °C under shaking (150 rpm) for three days. After this period, mycelia were obtained by filtration through non-gauze milk filters (Ken AG, Ashland, OH, USA) and, once dried, were stored frozen at −20 °C until ground with liquid nitrogen. The cetyltrimethylammonium bromide (CTAB) method [32] was used to obtain the fungal DNA. Its quality was analysed by electrophoresis and it was quantified using a spectrophotometer (model ND-1000; NanoDrop Technologies Inc, DE, USA).
The gpd, Alt a1, and tef1 gene regions were selected for a molecular characterisation. For PCR reactions, the following specific primer pairs were used: gdp1/gdp2 [33], alt-for/alt-rev [17], and Alt-tef1/Alt-tef2 [34].
The amplification of the three fragments was performed in a PTC-2000 Thermal Cycler (MJ Research Inc., Watertown, MA, USA) with the following parameters: 94 °C for 1 min, then 31 cycles at 94 °C for 30 s, 56 °C for 45 s, and 72 °C for 1 min, followed by 72 °C for 5 min and at 4 °C. After electrophoretic separation in 1X TAE buffer on 1.5% agarose gel, the PCR products were visualized with UV. Then, these PCR products were purified and sequenced by Macrogen, Inc., Seoul, South Korea, using the same primers previously used for the PCR amplification. Using BioEdit Sequence Alignment Editor 7.1.3.0 [35], sequences were edited and compared with reference sequences available on GenBank databases for identification of the field isolates.
2.3. Phylogenetic Analyses
According to BLAST searches, sequences of 32 field strains were aligned with 13 reference sequences downloaded from the National Center for Biotechnology Information (NCBI) and Somma et al. [34]. The present 32 strains and other 13 species were used during the phylogenetic analysis with A. infectoria CBS 210.86 as the outgroup (Table 1).

Table 1.
Source information of Alternaria strains used in the phylogenetic analyses.
Nucleotide sequences of amplicons of gpd, Alt a1, and tef1 were aligned with the MAFFT online version 7 [36]. Ends of alignments were cut in order to avoid regions with missing data. Each locus and the combined data set were subjected to Bayesian phylogenetic inference using MrBayes 3.2.6 [37]. The best substitution model was determined for each data set using JModelTest [38] and scored following the Akaike Information Criterion (AIC). The General Time-Reversible (GTR) substitution model was used for tef1, GTR + gamma-distributed rate variation across sites (G) was used for gpd, and GTR + proportion of invariable sites (I) was used for Alt a1. For the combined data set, each gene was treated as a separate partition with independent parameter estimations. Two runs with four chains each were run for ten million generations with a sampling frequency of every 100 generations. Trees after the initial 25% trees of each run were discarded as burn-in.
Tree topologies were adjusted using FigTree v1.4.3. DNA sequences generated in this study were deposited in GenBank under accession numbers (Table 1).
2.4. Mycotoxins Production and Extraction
A 4 mm diameter agar disk of each Alternaria strain grown on synthetic nutrient agar (SNA) [39] for 7 days was used to centrally inoculate Petri plates containing ground rice–corn steep liquor medium (GRCS; ground rice 50 g, corn steep liquor 5 g, agar 15 g, and 1000 mL distilled water). The plates were incubated for 14 d at 25 °C in darkness [40]. The extraction method consisted of a three-step extraction procedure based on a microscale extraction [41,42]. First, from the edge of each colony, 3 agar plugs (4 mm diameter) were cut from every Petri plate and were extracted using 1.5 mL chloroform/methanol (2:1 v/v) for 60 min in an ultrasonic bath. After that, the extract was transferred to clean 4 mL amber vials and evaporated to dryness (air, 50 °C). In a second step, the same plugs were extracted in 1.3 mL ethyl acetate with 1% formic acid for 60 min in an ultrasonic bath. This second extract was transferred to the amber vial with the first dried extract and evaporated. Finally, the plugs were extracted a third time with 1.5 mL of 2-propanol for 60 min using a ultrasonic bath, and then the extract was transferred to the amber vial with the two previous extracts and evaporated. Before the HPLC analysis, the pooled, dried extract was ultrasonically re-dissolved in 1 mL of methanol and 1 mL of acetonitrile:water (25:75 v/v) and filtered through a 0.45 µm filter.
2.5. HPLC Analysis
The HPLC system consisted of a Hewlett Packard model 1100 pump (Palo Alto, CA, USA) connected to a Hewlett Packard 1100 Series variable wavelength detector and a data module Hewlett Packard Kayak XA (HP Chem Station Rev. A.06.01, Palo Alto, CA, USA). Chromatographic separations were performed on a Symmetry C18 (100 × 4.6 mm i.d., 5 µm particle size) connected to a guard column Security Guard (20 × 4.6 mm i.d.) filled with the same phase. The mobile phase consisted of two consecutive isocratic mobile phase mixtures containing acetonitrile:0.027 M sodium dihydrogen phosphate solution (25:75, v/v Sn A) and acetonitrile:0.027 M sodium dihydrogen phosphate solution (50:50, v/v Sn B). Solvent A was pumped for 3.5 min at 1.0 mL/min followed by solvent B which was pumped for 16.5 min at 1.0 mL/min. The detector was set at 256 nm for AOH and AME and 279 nm for TA. Injection volume was 50 µL and the retention times were 11.8, 17.5, and 7.0 min for AOH, AME, and TA, respectively. Quantification was relative to external standards of 0.5, 1.0, 2.0, and 3 g/mL in acetonitrile:0.027 M sodium dihydrogen phosphate solution (25:75, v/v).
A recovery experiment was performed in GRCS medium at 0.1 to 10 µg/g levels of AOH, AME, and TA, respectively. Mean recovery and repeatability (relative standard deviation) ranged from 85 to 98% (0.2 to 1.4%), from 88 to 97% (0.1 to 2%), and from to 86% to 92% (0.5 to 2.5%) for AOH, AME, and TA, respectively. For the three toxins, the limit of detection (LOD; signal-to noise ratio 3) was 0.01 µg/g, and the limit of quantification (LOQ) was established as three times the detection limit.
3. Results
Thirty-two Alternaria strains isolated from chickpea plants were identified through a phylogenetic analysis. Sequences of the tef1, gpd, and Alt a1, genes were generated, yielding fragments of about 560 bp for tef1, 578 bp for gpd, and 497 bp for the Alt a1 gene. The combined tef1, gpd, and Alt a1 sequence data set consisted of 1635 base alignments. One Bayesian phylogenetic tree was generated for each sequenced gene and for the concatenated sequences of the three genes (Figure 1). The phylogenetic tree obtained showed three well-defined clades with high support values (> 90) corresponding to the sections Alternaria, Porri, and Brassicicola. All Alternaria strains isolated from the chickpeas clustered into the section Alternaria (100% support value) and were separated into three different groups. Seventeen strains (53%) clustered with the A. tenuissima and A. alternata reference strains (90% support), showing high homology among them, while four other strains (12.5%) clustered with the A. arborescens reference strain (62% support). In particular, the strain RC-CR639 was very close to the reference strain while the three other strains (RC-CR910, RC-CR908, and RC-CR88) were similar among themselves but distant from the A. arborescens reference strain. The remaining eleven strains (34.3%) formed a separated cluster, which included the A. citriarbusti reference strain (91% support). Within this group, a high variability was observed since most of the strains were closely related to the reference strain but did not cluster with it.
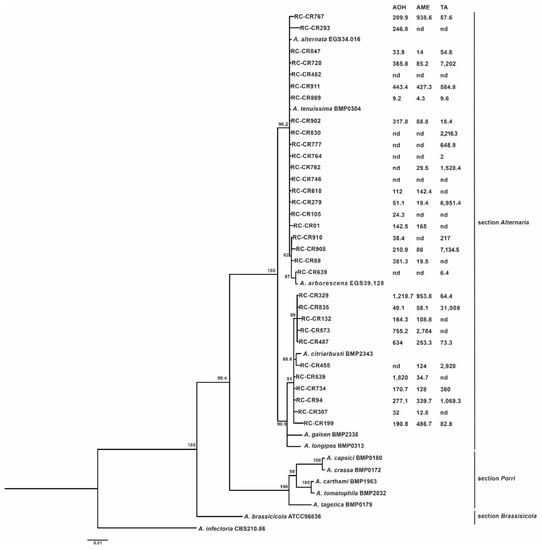
Figure 1.
Bayesian phylogenetic tree based on the combined gene sequences of tef, gpd, and Alt a1 of 32 Alternaria strains isolated from chickpea. Bayesian posterior probabilities (>50) are shown at nodes. Alternaria infectoria CBS 210.86 was used as the outgroup. Mycotoxin profile (µg/g) of Alternaria strains is indicated next to each strain. AOH: alternariol, AME: alternariol monomethylether, and TA: tenuazonic acid. nd: not detected (≤LOQ).
The mycotoxin production profile of the 32 Alternaria section Alternaria stains in the GRCS medium is reported in Figure 1. Among all the Alternaria strains tested, 94% of them were able to produce at least one of the mycotoxins evaluated. The most frequent profile found was the co-production of AOH, AME, and TA by 14 strains, followed by AOH and AME profiles found in 7 strains. The percentage of strains that did not produce any of the tested toxins was very low (6%), while TA yielded the highest concentration.
4. Discussion
During the present study, strains of Alternaria spp. isolated from chickpea plants in Argentina were phylogenetically identified as A. alternata and A. arborescens, which belong to the small-spored section Alternaria. The taxonomy of the small-spored Alternaria spp. has been based mostly on both morphological characteristics and molecular analysis. However, due to the absence of coherent morphological features and the limited variability of the molecular markers, it may be difficult to define a single species based only on the agreement between the morphological and phylogenetic lineages [15,30,43,44,45]. Our combined phylogenetic tree confirmed the difficulties in the identification of the different morphospecies in this section. In the present study, the phylogenetic analysis of the Alt a1, gpd, and tef1 genes demonstrated that all the isolates were grouped into three distinct clades: A. arborescens (n = 4), A. citriarbusti (n = 11), and a cluster containing A. alternata and A. tenuissima, which could not be clearly separated and, therefore, were grouped into a big cluster (n = 17). This close relation between A. alternata and A. tenuissima has also been reported using other molecular loci [15,43,46,47,48]. As in the phylogenetic analysis, there is no distinct separation between A. alternata and A. tenuissima; researchers recently suggested that these two species should be combined into a single species—A. alternata. Several studies that initially identified A. alternata and A. tenuissima using morphological features then found that the two species could not be separated by phylogenetic analysis using multiple molecular markers and, therefore, proposed that A. tenuissima be referred to as A. alternata [15,30,43,44,46,49]. Further, a study conducted by Dettman and Eggertson [50] compared—through a high-resolution, genome-wide study— the species within the section Alternaria, and they also proposed to merge A. alternata and A. tenuissima into one species, namely, A. alternata. Consequently, in the present study, the strains grouped into the alternaria/tenuissima clade were identified as A. alternata. Additionally, A. arborescens, defined by Woudenberg et al. [30] and Dettman and Eggertson [50] as a species complex, formed a separate clade from A. alternata in our analysis as well. These researchers also suggested that A. citriarbusti is synonymous with A. alternata species; however, in the present study, the A. citriarbusti isolates were not grouped together with A. alternata species. The incongruity between our results and the data produced by Woudenberg et al. [30] and Dettman and Eggertson [50] may be attributable to the different genomic regions used for the phylogenetic analysis. Based on our results, we agree with Dettman and Eggertson’s [50] conclusion that most existing loci may be able to place an unknown Alternaria strain into a section, although section-specific molecular markers are required to differentiate lineages in the section Alternaria. Nevertheless, until a common marker is not unanimously defined by the scientific community dealing with the taxonomy of the Alternaria genus, the phylogenetic relationships between species will remain confused.
All the strains in this study were characterized through the determination of their in vitro mycotoxin production. It is remarkable that most of the strains of the Alternaria section were able to produce at least one of the mycotoxins tested, and co-production was the prevalent manner by which this occurred. We failed to differentiate the A. alternata strains from the A. arboresces strains based on the mycotoxin profiles (types and concentrations); this result is in agreement with that reported by Zwickel et al. [51]. A previous study on the natural occurrence of Alternaria toxins in the same chickpea grains detected AME as the most frequent toxin contaminating the samples (100% positive) followed by AOH (30% positive). Thus, the degree of toxin contamination in the samples was in accordance with the metabolite profile of the predominant Alternaria species present.
In particular, A. alternata is the most important mycotoxin producer within the genus, since this mentioned species is widely reported to produce TA, AME, AOH, ALT, and ATX [52,53,54,55,56,57]. A recent study performed by Huybrechts et al. [58] has related the chronic exposure of a low-dose of Alternaria mycotoxins in food commodities to the onset of colorectal cancer in humans. In addition, consumer exposure and associated toxic effects are plausible due to the absence of regulation in food.
5. Conclusions
In conclusion, the present study contributed to attaining a successful classification of the Alternaria strains isolated from chickpea plants using a multi-locus analysis, and the toxigenic profile was also assessed. All the strains belong to the Alternaria section Alternaria, with A. alternata being the most common species found. Additionally, we were able to identify species within the A. arboresens species group. The analysis of the secondary metabolite production showed that most of the strains were able to produce at least one of the mycotoxins analysed. We were not able to find any differences between the mycotoxin profiles of the Alternaria species studied. The knowledge of the fungal populations infecting crops is a valuable tool for monitoring and establishing mycotoxin controls. Since Alternaria section Alternaria species are frequently found in chickpeas, and members of this section are well-known mycotoxin producers, food safety authorities should consider the wide diversity of toxic metabolites that may contaminate this pulse when reviewing the risks and safety aspects of Alternaria toxins in food and feed.
Author Contributions
M.J.N. and E.C. performed the whole experiment, J.F.H., C.J.R. and V.G.L.Z. performed the analysis of the metabolites, S.A.P., M.J.N. phylogenetic analysis, M.L.R. conceived and designed the experiments. M.L.R., M.J.N., S.A.P. analysed the data and wrote the paper. M.L.R. revised the manuscript. M.L.R. project administration and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants ANPCyT (Agencia Nacional de Promoción Científica y Tecnológica) PICT 2493/2017 and Consejo Nacional de Investigaciones Científicas y Técnicas: PUE 22920200100004.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Acknowledgments
C.J.R. and M.J.N. are fellow of CONICET and M.L.R., E.C., V.G.L.Z. and S.A.P. are members of the Research Career of CONICET. The authors are grateful to A. Moretti and M. Masiello for sharing sequences data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wood, J.A.; Grusak, M.A. Nutritional value of chickpea. In Chickpea Breeding and Management; Yadav, S., Redden, B., Chen, W., Sharma, B., Eds.; CAB International: Wallingford, UK, 2007; pp. 101–142. [Google Scholar]
- Berger, J.D.; Turner, N.C. The ecology of chickpea. In Chickpea Breeding and Management; Yadav, S., Redden, B., Chen, W., Sharma, B., Eds.; CAB International: Wallingford, UK, 2007; pp. 47–71. [Google Scholar]
- Chen, W.; Sharma, H.C.; Muehlbauer, F.J. Compendium of Chickpea and Lentil Diseases and Pests; American Phytopathological Society: St. Paul, MN, USA, 2011; p. 164. [Google Scholar]
- Agricultura, Ganadería y Pesca. Available online: http://www.minagri.gob.ar (accessed on 14 September 2022).
- Ramirez, M.L.; Cendoya, E.; Nichea, M.; Zachetti, V.; Chulze, S. Impact of toxigenic fungi and mycotoxins in chickpea: A review. Curr. Opin. Food Sci. 2018, 23, 32–37. [Google Scholar] [CrossRef]
- Romero, C.J.; Sulyok, M.; Chulze, S.; Ramirez, M.L. Mycobiota and mycotoxin occurrence in chickpea produced in Argentina. In Proceedings of the International Commission on Food Mycology Conference, Freising, Germany, 26 July 2019. [Google Scholar]
- Meena, M.; Gupta, S.K.; Swapnil, P.; Zehra, A.; Dubey, M.K.; Upadhyay, R.S. Alternaria toxins: Potential virulence factors and genes related to pathogenesis. Front. Microbiol. 2017, 8, 1451. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. Scientific opinion on the risks for animal and public health related to the presence of Alternaria toxins in feed and food: Alternaria toxins in feed and food. EFSA J. 2011, 9, 2407. [Google Scholar] [CrossRef]
- Zwickel, T.; Klaffke, H.; Richards, K.; Rychlik, M. Development of a high performance liquid chromatography tandem mass spectrometry based analysis for the simultaneous quantification of various Alternaria toxins in wine, vegetable juices and fruit juices. J. Chromatogr. A 2016, 1455, 74–85. [Google Scholar] [CrossRef]
- Pinto, V.E.; Patriarca, A. Alternaria species and their associated mycotoxins. Methods Mol. Biol. 2017, 1542, 13–32. [Google Scholar] [PubMed]
- Prendes, L.P.; Fontana, A.R.; Merín, M.G.; D´ Amario Fernández, A.; Bottini, R.; Ramirez, M.L.; Morata de Ambrosini, V.I. Natural occurrence and production of tenuazonic acid in wine grapes in Argentina. Food Sci. Nutr. 2018, 6, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Mao, X.; Sun, Q.; Wei, Z.; Li, J.; You, Y.; Zhao, J.; Jiang, G.; Wu, Y.; Wang, L. Alternaria mycotoxins: An overview of toxicity, metabolism, and analysis in food. J. Agric. Food Chem. 2021, 69, 7817–7830. [Google Scholar] [CrossRef] [PubMed]
- EFSA European Food Safety Authority Dietary exposure assessment to Alternaria toxins in the European population. EFSA J. 2016, 14, e04654. [CrossRef]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Andrew, M.; Peever, T.L.; Pryor, B.M. An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 2009, 101, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Pryor, B.M.; Gilbertson, R.L. Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mt SSU rDNA sequences. Mycol. Res. 2000, 104, 1312–1321. [Google Scholar] [CrossRef]
- Hong, S.G.; Cramer, R.A.; Lawrence, C.B.; Pryor, B.M. Alt a 1 allergen homologs from Alternaria and related taxa: Analysis of phylogenetic content and secondary structure. Fungal Genet. Biol. 2005, 42, 119–129. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Park, M.S.; Pryor, B.M. Nimbya and Embellisia revisited, with nov. comb. for Alternaria celosiae and A. perpunctulata. Mycol. Prog. 2012, 11, 799–815. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Gannibla, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.H.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Poursafar, A.; Ghosta, Y.; Orina, A.S.; Gannibal, P.B.; Javan-Nikkhah, M.; Lawrence, D.P. Taxonomic study on Alternaria sections Infectoriae and Pseudoalternaria associated with black (sooty) head mold of wheat and barley in Iran. Mycol Prog. 2018, 17, 343–356. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Rotondo, F.; Gannibal, P.B. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol Prog. 2015, 15, 3. [Google Scholar] [CrossRef]
- Ghafri, A.A.; Maharachchikumbura, S.S.N.; Al-Saady, N.A.; Al-Sadi, A.M. A new section and a new species of Alternaria encountered from Oman. Phytotaxa 2019, 405, 279–289. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Deng, J.K.; Li, M.J.; Paul, C.N.; Oo, M.M.; Lee, H.B.; Oh, S.K.; Yu, S. Alternaria brassicifolii sp. nov. isolated from Brassica rapa subsp. pekinensis in Korea. Mycobiology 2018, 46, 172–176. [Google Scholar] [CrossRef]
- Ahmadpour, A. Alternaria caricicola, a new species of Alternaria in the section Nimbya from Iran. Phytotaxa 2019, 405, 65–73. [Google Scholar] [CrossRef]
- Liu, H.F.; Liao, J.; Chen, X.Y.; Liu, Q.K.; Deng, J.K. A novel species and a new record of Alternaria isolated from two Solanaceae plants in China. Mycol. Prog. 2019, 18, 1005–1012. [Google Scholar] [CrossRef]
- Bessadat, N.; Hamon, B.; Bataille-Simoneau, N.; Mabrouk, K.; Simoneau, P. Alternaria telliensis sp. nov., a new species isolated from Solanaceae in Algeria. Phytotaxa 2020, 440, 89–100. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Mulè, G.; Susca, A.; Stea, G.; Moretti, A. Specific detection of the toxigenic species Fusarium proliferatum and F. oxysporum from asparagus plants using primers based on calmodulin gene sequences. FEMS Microbiol. Lett. 2004, 230, 235–240. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell Publishing: Hoboken, NJ, USA, 2006; p. 388. [Google Scholar]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Somma, S.; Amatulli, M.T.; Masiello, M.; Moretti, A.; Logrieco, A.F. Alternaria species associated to wheat black point identified through a multilocus sequence approach. Int. J. Food Microbiol. 2019, 293, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef]
- Gerlach, W.; Nirenberg, H.I. The genus Fusarium—A pictorial atlas; Dahlem: Berlin, Germany, 1982; p. 230. [Google Scholar]
- Chulze, S.; Torres, A.; Dalcero, A.; Combina, M. Production of alternarioi and alternariol monomethyl ether in natural substrates in comparison with semisynthetic culture medium. Mycotox. Res. 1994, 10, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Smedsgaard, J. Micro-scale extraction procedure for standardized screening of fungal metabolite production in cultures. J. Chromatogr. A 1997, 760, 264–270. [Google Scholar] [CrossRef]
- Andersen, B.; Krøger, E.; Roberts, R.G. Chemical and morphological segregation of Alternaria alternata, A. gaisen and A. longipes. Mycol. Res. 2001, 105, 291–299. [Google Scholar] [CrossRef]
- Armitage, A.D.; Barbara, D.J.; Harrison, R.J.; Lane, C.R.; Sreenivasaprasad, S.; Woodhall, J.W.; Clarkson, J.P. Discrete lineages within Alternaria alternata species group: Identification using new highly variable loci and support from morphological characters. Fungal Biol. 2015, 119, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Armitage, A.D.; Cockerton, H.M.; Sreenivasaprasad, S.; Woodhall, J.; Lane, C.R.; Harrison, R.J.; Clarkson, J.P. Genomics evolutionary history and diagnostics of the Alternaria alternata species group including apple and Asian pear pathotypes. Front. Microbiol. 2020, 10, 3124. [Google Scholar] [CrossRef] [PubMed]
- da Cruz Cabral, L.; Rodriguero, M.; Stenglein, S.; Nielsen, K.F.; Patriarca, A. Characterization of small-spored Alternaria from Argentinean crops through a polyphasic approach. Int. J. Food Microbiol. 2017, 257, 206–215. [Google Scholar] [CrossRef]
- Luo, Y.; Hou, L.; Förster, H.; Pryor, B.; Adaskaveg, J.E. Identification of Alternaria species causing heart rot of pomegranates in California. Plant Dis. 2017, 101, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, F.; Collina, M.; Brunelli, A.; Pryor, B.M. Comparison of Alternaria spp. collected in Italy from apple with A. mali and other AM-toxin producing strains. Phytopathology 2012, 102, 1130–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Q.; Xiao, C.L. Phylogenetic, morphological, and pathogenic characterization of Alternaria species associated with fruit rot of blueberry in California. Phytopathology 2015, 105, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Saito, S.; Michailides, T.J.; Xiao, C.-L. Phylogenetic, morphological, and pathogenic characterization of Alternaria species associated with fruit rot of mandarin in California. Plant Dis. 2021, 105, 2606–2617. [Google Scholar] [CrossRef] [PubMed]
- Dettman, J.R.; Eggertson, Q. Phylogenomic analyses of Alternaria section Alternaria: A high-resolution, genome-wide study of lineage sorting and gene tree discordance. Mycologia 2021, 113, 1218–1232. [Google Scholar] [CrossRef] [PubMed]
- Zwickel, T.; Kahl, S.M.; Rychlik, M.; Müller, M.E. Chemotaxonomy of mycotoxigenic small-spored Alternaria fungi–do multitoxin mixtures act as an indicator for species differentiation? Front. Microbiol. 2018, 9, 1368. [Google Scholar] [CrossRef] [PubMed]
- Logrieco, A.; Moretti, A.; Solfrizzo, M. Alternaria toxins and plant diseases: An overview of origin, occurrence and risks. World Mycotoxin J. 2009, 2, 129–140. [Google Scholar] [CrossRef]
- Patriarca, A.; da Cruz Cabral, L.; Pavicich, M.A.; Nielsen, K.F.; Andersen, B. Secondary metabolite profiles of small-spored Alternaria support the new phylogenetic organization of the genus. Int. J. Food Microbiol. 2019, 291, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Ramires, F.A.; Masiello, M.; Somma, S.; Villani, A.; Susca, A.; Logrieco, A.F.; Luz, C.; Meca, G.; Moretti, A. Phylogeny and mycotoxin characterization of Alternaria species isolated from wheat grown in Tuscany, Italy. Toxins 2018, 10, 472. [Google Scholar] [CrossRef]
- Siciliano, I.; Ortega, S.F.; Gilardi, G.; Bosio, P.; Garibaldi, A.; Gullino, M.L. Molecular phylogeny and characterization of secondary metabolite profile of plant pathogenic Alternaria species isolated from basil. Food Microbiol. 2018, 73, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Gobashy, E.; Fawzy, S.; Mikhail, W.Z.; Ismail, A.M.; Zekry, A.; Moretti, A.; Susca, A.; Soliman, A.S. Phylogenetic, toxigenic and virulence profiles of Alternaria species causing leaf blight of tomato in Egypt. Mycol. Prog. 2018, 17, 1269–1282. [Google Scholar] [CrossRef]
- Chalbi, A.; Sghaier-Hammami, B.; Giuseppe, M.; Quiles, J.M.; Abdelly, C.; Marangi, C.; Logrieco, A.F.; Moretti, A.; Masiello, M. Characterization of mycotoxigenic Alternaria species isolated from the Tunisian halophyte Cakile maritima. Phytopathol. Mediterr. 2020, 59, 107–108. [Google Scholar] [CrossRef]
- Huybrechts, I.; De Ruyck, K.; De Saeger, S.; De Boevre, M. Uniting large-scale databeses to unravel the impact of chronic multi-mycotoxins exposures on colorectal cancer incidence in Europe. In Proceedings of the Proceedings of the 2nd MycoKey International Conference, Wuhan, China, 16 September 2018. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).