Abstract
Sustainable management of exploited and endangered species is facilitated by knowledge of their geographic genetic structure. Lake sturgeon (Acipenser fulvescens) epitomizes both categories, but genetic information has largely been limited to the Laurentian Great Lakes basin. We assessed the hierarchical geographic genetic structure of lake sturgeon across their Canadian range using a variation at 14 microsatellite loci. Observed patterns showed evidence of two ancestral groups which originated from Mississippian and Missourian glacial refugia. Coalescent analysis indicates the two lineages most recently shared common ancestry during the late Pleistocene and were likely isolated by the late Wisconsinan ice advance, with subsequent interpopulation divergences within each lineage reflecting their reciprocal isolation as glacial meltwaters receded. Hierarchical patterns of genetic relationships among contemporary populations largely reflect colonization histories and connections within primary and secondary watersheds. Populations in western Canada showed strong similarities based on their shared Missourian origins and colonization from glacial Lake Agassiz. By contrast, populations in the Great Lakes–St. Lawrence River drainage were largely founded from a Mississippian source. Sturgeon populations in northern parts of Ontario and Quebec showed evidence of mixed ancestry from secondary contact between the two refugial groups through Holocene meltwater lakes. Within major watersheds, the strong similarity among geographically separate populations reflects their shared ancestry during postglacial colonization. The general lack of structure within major river systems highlights historically continuous habitat (connectivity) and gene flow rather than contemporary barriers (dams). These data highlight the importance of Quaternary and prehistoric events on patterns of genetic diversity and divergence within and among contemporary populations, as well as the importance of these populations for conserving the species’ evolutionary legacy.
1. Introduction
Identifying and conserving the genetic diversity and structure of endangered species and populations is a central pillar of conservation biology. For many species, spatial genetic structure reflects phylogeographic patterns caused by glacial and postglacial processes [1], as well as contemporary influences from anthropogenic activities. Genetic criteria are often used to identify and prioritize populations for conservation, and maintaining genetic diversity within and among populations is paramount for many recovery and rehabilitation programs [2]. Many genetic investigations have been conducted on endangered species, characterizing genetic structure, and identifying appropriate units for conservation [2]. In many cases, species recovery plans incorporate recognition of genetic and adaptive resources within and among geographic elements of the species of concern, and accurate delineation of management units maximizes opportunities for the recovery of threatened species [2].
The lake sturgeon, Acipenser fulvescens, is one of the largest freshwater fishes in North America, reaching lengths of 2 m or more [3]. Like most species of sturgeon globally, the lake sturgeon is of significant conservation concern across most of its range [3,4,5,6]. Among the North American sturgeons, it is considered to have diverged from its congeners between 52.7 and 22.4 million years ago [7,8], has the largest range, and is unusual in having an entirely freshwater life cycle [4]. Lake sturgeon has a long generation time (20–30 years), longevity of 80–150 years, high larval mortality and low natural recruitment [3,4,5,6,9]. As a large-bodied, highly migratory species, lake sturgeon require hundreds of kilometers of continuous habitat to support viable populations [4]. Riverine habitat is particularly important for completing its life cycle, as adults depend on rapids for spawning habitat and larval sturgeon require substantial reaches with downstream flow before settling out of the current to begin foraging as juveniles [3,4]. Lake sturgeon historically had a broad geographic distribution, and formerly occurred through large portions of the Mississippi, Great Lakes, and Hudson Bay drainage basins [3,4]. Despite this broad distribution, the species is of conservation concern across its entire range [4,5,6,10], and is highly vulnerable to exploitation pressures, habitat alteration, and other anthropogenic disturbances [3,4,5,6].
These combined stressors and historical overharvest drastically reduced sturgeon numbers in the 19th and early 20th centuries [3], and many remaining populations are considered to be less than 10% of their historical abundances [10]. Commercial fisheries peaked in the late 19th century, only to collapse within a decade [3,6]. Industrial development and dam construction for hydroelectricity and water control through the 20th century severely impacted remaining lake sturgeon populations through blocked access for migration, alteration and loss of spawning and juvenile habitats, and disrupted connectivity between formerly continuous riverine habitats [3,4]. As a result of these combined historical impacts, lake sturgeon populations are greatly reduced through most of their range, and are listed as endangered, threatened, and of special concern in Canada [5], vulnerable, imperiled, or critically imperiled in 20 American jurisdictions and extirpated in two others [10] as well as endangered globally [6].
The zoogeography of lake sturgeon is poorly understood. Based on distributional data, lake sturgeon were considered to have colonized their current range from a Mississippian refugium at the end of the Pleistocene [11,12,13,14]. There was some speculation that lake sturgeon might also have dispersed from a western Missourian refugium [13], although lake sturgeon are not present in the upper Missouri drainage [11,12]. The distribution of lake sturgeon in Canada suggested that the species accessed Lake Agassiz from a Mississippian refugium approximately 11,000 years ago [12], and subsequently dispersed eastward into northern Ontario and central Quebec via Lake Ojibway-Barlow between 8300 and 7900 years ago [13,14].
Evidence of a second refugial source for lake sturgeon (Missourian or western Mississippian drainage) was first detected with genetic data. Analysis of mitochondrial DNA [15,16,17] showed evidence of two lineages with limited divergence in secondary contact in northeastern Ontario and northwestern Quebec. Based on the geographic distributions of the observed haplotypes, it was suggested these represented ancestral lineages originated from separate glacial refugia [17]. In contrast to other species with similar distributions, however, lake sturgeon show remarkably little mitochondrial variation [1], limiting the phylogeographic information value of mitochondrial data. Subsequent analysis of primarily Great Lakes populations showed marked differences from the Mattagami River population in northeastern Ontario, suggesting that Great Lakes and northern inland populations might differ in their ancestry [18,19,20]. This was confirmed by broader geographic studies of Canadian populations beyond the Great Lakes, which conclusively showed the presence of a second genetic group of lake sturgeon in western Canada and tributaries of Hudson Bay and James Bay in northern Ontario and Quebec [21,22].
Most recent genetic investigations of lake sturgeon population structure have focused largely but not exclusively on genetic patterns within and among remnant populations in the Great Lakes, due to historical losses and multijurisdictional conservation concerns [18,19,20,23,24,25,26]. Inclusion of samples from outside the Great Lakes basin was initially limited to lake sturgeon from one or two rivers draining into Hudson Bay or James Bay for comparison purposes as geographic outgroups [18,20], although more recent studies have addressed more populations outside the Great Lakes basin [21,22,25]. At a watershed scale, lake sturgeon generally demonstrate little genetic differentiation within rivers [23,24,25,26,27,28,29], but significant genetic differentiation among rivers and major drainages [17,21].
To expand our geographic knowledge of phylogeographic and genetic structure of lake sturgeon, we conducted a broad-scale genetic survey of populations across Canada. Using a suite of 14 microsatellite markers, we assessed the hierarchical genetic structure and diversity among native populations at several geographic scales in order to (1) identify the geographic extent of phylogeographic lineages and zones of secondary contact, and (2) assess hierarchical patterns of genetic diversity and differentiation within and among primary, secondary, and tertiary drainages. Our genetic results provide valuable information for delineating conservation and management units of sturgeon populations across Canada, as well as baseline information on levels of diversity for assessing potential changes in response to anthropogenic activities or changes in habitat quality.
2. Materials and Methods
2.1. Sample Collection
Fourteen microsatellite loci were used to examine the population structure of more than 4000 lake sturgeon from close to 50 locations across Canada and Wisconsin (Table 1 and Figure 1). Sampling was largely conducted by management agencies and consulting companies (listed in Acknowledgements). Many of the sampled rivers are fragmented by dams, primarily for water reservoirs in the prairie provinces and for hydroelectricity in rivers on and north of the Canadian Shield [3,4,5].

Table 1.
Summary of sampling locations by secondary watershed basins, showing sampled populations/waterbodies, sample sizes, geographic coordinates (latitude and longitude as decimal degrees), expected and observed heterozygosity (HE and HO, respectively), absolute and standardized allelic richness (AR and AS30, respectively), FIS, and estimated genetic effective population sizes (Ne) using a minimum allele frequency of 0.05. Ne superscripts indicate published Ne estimates from 1 [22], 2 [24].
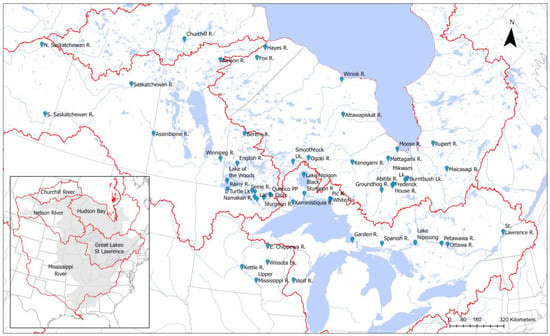
Figure 1.
Locations of 49 sampled lake sturgeon populations, showing sampling locations within major watersheds. Inset map shows the study’s extent within the historical range of lake sturgeon (shaded area) across North America.
Collection methods varied according to site-specific requirements, agency and/or company capture protocols, and preferences of field personnel. Most commonly, bottom-set gillnets (mesh size ranging from 12.7–30.48 cm) were set at depths ranging from 3 m to 15 m. A small portion of a pectoral fin (1–2 cm2) was clipped from captured adult fish, generally at known spawning sites during the spring spawning season (May to June) or standardized index netting projects (August to September), between 1991 and 2021 (Table 1). For large waterbodies such as the Saskatchewan and Ottawa Rivers, samples were collected in different locations and/or years. Individual fish were measured, often tagged, and then released. Tissue clips for genetic analysis were placed in either 95% ethanol or dried in a fin clip envelope. Larger individuals (>600 mm) were targeted in an attempt to minimize potential biases introduced by sampling across generations. It was beyond the scope of this study to incorporate a temporal component, but it is unlikely that significant differences would be seen within sites between two overlapping generations in such a long-lived species. Existing comparisons among temporal collections at the same site have shown no significant differences [20].
The sampling locations spanned the Canadian distribution of lake sturgeon, as well as five populations from Wisconsin (Table 1 and Figure 1). Sample collections represented all of the major drainage basins which still support extant populations of lake sturgeon (Table 1 and Figure 1). Several river basins and waterbodies were represented by multiple sampling locations (Saskatchewan River, Ottawa River, Winnipeg River, Lake of the Woods/Rainy River) and samples were provided from several tributaries of Lake Huron and Lake Superior (Table 1).
2.2. DNA Isolation and Genotyping
Whole genomic DNA was extracted using a simple isopropanol precipitation. Tissues were homogenized in 250 μL of 1X TNES buffer (50 mM Tris pH 8, 1000 mM NaCl, 1 mM EDTA, 1% sodium dodecyl sulphate (weight per volume)) with the addition of 1 mg Proteinase K (Bioshop, Burlington, ON, Canada) and incubated at 37 °C overnight (>16 h). DNA was precipitated using 500 μL of 80% isopropanol and centrifugation at 2000 gravities for 30 min, followed by removing the supernatant and rinsing the pellets with 1 mL of 70% ethanol and then air drying. DNA pellets were resuspended in 150 μL 1X TE (10 mM Tris pH8, 1 mM EDTA). Extraction yields and quantity were tested using electrophoresis in 1.5% agarose gel stained with SybrGreen (Cedar Lane Laboratories, Burlington, ON, Canada) alongside a molecular mass ladder (Bioshop, Burlington, ON, Canada). Stock DNA was diluted 1:30 to make a working solution for PCR amplification.
Samples were amplified at 14 microsatellite loci that have been previously described [18,30,31,32,33] and shown to be polymorphic and exhibit Mendelian inheritance [18,33]. Loci were amplified in four 15 µL multiplex PCR containing 5 µL of genomic DNA, 1X PCR buffer (containing 1.5 mM MgCl2), 0.5 mM additional MgCl2, 0.025 units µL−1 of Taq DNA polymerase (Qiagen, Germantown, MD, USA), and 0.2 mM of dNTPs (Bioshop, Burlington, ON, Canada), 0.3 mg mL−1 of Bovine Serum Albumin (BSA) (Bioshop, Burlington, ON, Canada). Multiplex reactions contained the following primer concentrations Multiplex 1- Afu68 (0.25 μM), AfuG63 (0.22 μM), AfuG122 (0.25 μM), AfuG195 (0.25 μM), AfuG74 (0.28 μM), and AfuG67 (0.25 μM); Multiplex 2- AfuG160 (0.25 μM), AfuG204 (0.23 μM), Afu68b (0.3 μM), AfuG61 (0.25 μM), and AfuG71 (0.25 μM); Multiplex 3- AfuG9 (0.35 μM) and AfuG112 (0.38 μM). The locus AfuG56 was amplified singly with a primer concentration of 0.3 μM due to problems with nonspecific amplification. All reactions used cycling parameters of 95 °C for 11 min, followed by 36 cycles of 1 min each of 94 °C, 55 °C and 72 °C, and a final extension phase of 60 °C for 45 min [29]. Amplified products were run on an AB3730 automated sequencer and standardized genotypes were determined using GeneMapper v. 3.1 (Applied Biosystems, Inc., Mississauga, ON, Canada). Samples that failed to amplify for more than four of the 14 loci were removed from all analyses.
2.3. Data Analysis
Genetic analyses were conducted in R (R Core Team 2014) using the adegenet [34] hierfstat, and mmod [35] packages unless otherwise specified. All loci were tested for Hardy–Weinberg equilibrium and linkage disequilibrium within sampling sites, using a table-wide significance level of p < 0.01 instead of Bonferroni correction for multiple simultaneous tests to guard against false significant results. A Monte Carlo simulation with 1000 iterations was conducted to simulate p-values instead of using a χ2 approximation when testing for Hardy–Weinberg equilibrium. The basic.stats function in hierfstat was used to calculate observed and expected heterozygosity (HO and HE), FIS, FST, Dest, and absolute allelic richness (AR). Observed and expected heterozygosity were determined for each locus and averaged across loci within each sampling site. Standardized allelic richness (AS) was calculated for a sample size of 30 individuals (60 allele copies) using the function rgenotypes.arich from the package R StandArich [36]. Potential differences in HO, HE, and AS among drainages were assessed using a Kruskal–Wallis test.
Several methods were employed to test for hierarchical genetic structuring among lake sturgeon populations and geographic localities or groups of localities, using both individual- and population-based analyses. The number of genetic clusters (K) within the dataset independent of sampling locations was assessed using Structure 2.3.4 [37], using a range of potential K values from 1 to 30 as competing hypotheses to identify proportional membership of individuals to each group at hierarchical levels of organization. Runs were conducted under the assumption of admixture and not using population information (i.e., drainage basins or waterbodies from which individuals were sampled) as prior information, and with the further assumption that allele frequencies were independent across loci. These model conditions minimize the potential bias for detecting structure by not assuming that capture locations are either informative or genetically distinct [37], as well as assuming that some potential for admixture was possible during secondary contact between separate glacial lineages as well as the potential for contemporary gene flow among connected watersheds or sites within major waterbodies such as Lake Superior and Lake Huron Analytical runs in structure which included 50,000 ‘burn-in’ iterations, followed by 100,000 re-sampling iterations, using 10 replicate trials for each estimate of K and altering the random generator seed at the start of each replicate. The optimal number of groups for lake sturgeon was determined by estimating the probability of the number of genetic groups (ln[P|K] [37]) and calculating ΔK, the change in the log probability of data solutions between successive K values [38]. The ideal K was identified by examining the biological feasibility of the K preferred by both methods. After the ideal K was chosen, the output from each of the 10 independent replicates was imported to CLUMPP [39] to identify the optimal alignment among replicate trials using the Greedy algorithm and visualized using the program DISTRUCT [40].
Independently, principal component analysis (PCA) was used to assess genetic diversity among individuals identified in putative groups (i.e., major drainage basins and sampled waterbodies). Allele frequencies were transformed by centering using the scaleGen function within adegenet, and missing values were replaced with 0. Similar populations would group based on their PCA scores. Discriminant analysis of principal components (DAPC) was conducted to provide descriptions of the genetic clusters and membership probabilities of each individual for the different groups based on the retained discriminant functions.
Pairwise genetic distances among populations were estimated using Nei et al.’s DA genetic distance [41] and plotted as a neighbor-joining (NJ) dendrogram using PopTreeW [42]. Bootstrap values were based on 1000 replicates. Pairwise genetic differences among populations were also quantified with FST and Dest [43] using the mmod package.
Effective population size (Ne) for each population was estimated using the linkage disequilibrium (LDNe) method of Waples and Do [44] as implemented in NeEstimator v2 [45]. Due to variable sample sizes among populations, Ne was estimated for each population using a minimum allele frequency of 0.05 and parametric 95% confidence intervals.
Divergence times within and among lineages and populations, or time since most recent common ancestry (TMRCA) was estimated using BEAST2, a Bayesian phylogenetic inference software (http://beast2.org; accessed on 17 October 2022) that can incorporate microsatellite genotypes to estimate TMRCA among populations [46]. Microsatellite data for BEAST2 were expressed as the number of motif repeats [47]. The number of motif repeats was determined by subtracting the smallest allele of that locus from the allele of interest, dividing by the bp of the motif (dinucleotide = 2) and adding one. The average motif lengths were computed for each locus within each population for the input file. The source code package ‘BEASTvntr’ was used for the microsatellite data [46]. The program BEAUti (included in the BEAST package) was used to set up and parameterize the model. The gamma category count was set at 6 with a shape of 1. The Sainudin model was selected using default values of bias magnitude, focal point, g, and one on A1 was selected using empirical frequencies. These parameters were ‘tuned’ based on BEAST recommendations after initial runs to improve model convergence. A strict clock was selected for model runs [47], and the coalescence constant population was selected as the tree prior. The timing of separation between the Pic River and Ottawa River was used as a prior for calibrating the tree: the two populations could have been connected as early as 10.8 Kya through the Fossmill Outlet of glacial Lake Algonquin [14] and as recently as 9.8–8.6 Kya through the North Bay (Mattawa) outlet [14]. The prior was set at 10.0 K years using a lognormal distribution with large variation which provided a central probability range covering 15.4–6.6 Kya (M = 2.3, S = 0.22). BEAST2 was used to run the scenario using 10,000,000 chain length. The resultant trace log file was imported into Tracer (http://tree.bio.ed.ac.uk/software/tracer/; accessed on 24 October 2022) and examined for model convergence. The tree log file was imported into TreeAnnotator (included in the BEAST package) to create the best supported phylogenetic tree. The resulting dendrogram was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/; accessed on 24 October 2022) to illustrate the variation (95% HPD) in node estimates.
3. Results
In total, 4639 lake sturgeon from 49 localities across North America were genotyped for 14 microsatellite loci (Table 1 and Figure 1). Of 237 samples (5%) that were re-genotyped, only two genotyping errors were detected and corrected through reamplification and rescoring. Mean expected and observed heterozygosity was broadly similar across sampling locations, averaging 0.48 and 0.47, respectively (Table 1). Several populations showed significant departures from Hardy–Weinberg equilibrium (HWE) expectations, with both heterozygote excesses and deficits observed (Table 1 and Supplemental Table S1). For the majority of these, deviations of mean HE from HWE expectations were within sampling locations with small sample sizes, although several sites with sample sizes greater than 25 individuals also showed significant deviations, suggesting the potential for local inbreeding or substructure (Table 1).
Population by locus testing showed potential evidence for null alleles at locus AfuG122 for populations outside of the Great Lakes and Mississippi drainages (Supplemental Table S1), so this locus was excluded from subsequent analyses. Single locus deviations from HWE expectations were detected at multiple loci in a number of populations, suggesting potential substructuring or Wahlund effects (Supplemental Table S1). These deviations were observed within multiple waterbodies from each of the northern drainages (Nelson River drainage: (Saskatchewan River, Berens River, Namakan River, and Winnipeg River); Hudson Bay drainage: (Attawapiskat River, Burntbush Lake, Kenogami River, Maicasagi River, and Moose River); Great Lakes drainage: (Lake Nipigon, Black Sturgeon River, Kaministiquia River, White River, Spanish River, and Ottawa River)).
Linkage disequilibrium (LD) was observed among some pairs of loci within populations, but was not consistent across populations (Supplemental Table S2). The majority of populations showed linkage disequilibrium at 0–2 locus pairs among 91 pairwise locus combinations, consistent with expectations for finite populations and multiple simultaneous comparisons. Several populations showed significant linkage disequilibrium for multiple locus pairs, indicating potential admixture or Wahlund effects (Supplemental Table S2): samples from the Rainy and Winnipeg rivers in the Nelson Bay drainage (29 and 13 LD pairs, respectively), the Ogoki River (11 LD pairs) in the Hudson Bay drainage, and Lake Nipigon (9 LD pairs), the Black Sturgeon and Kaministiquia rivers in Lake Superior (14 and 44 LD pairs), and the Spanish River (Lake Huron; 16 LD pairs) in the Great Lakes–St Lawrence drainage were most noticeable.
Single locus estimates of FIS varied within and among populations, with no consistent pattern (Supplemental Table S3). Mean FIS values within populations also varied among populations, with notable positive and negative differences from what would be expected under random mating (Table 1). This was evident in several waterbodies with small sample sizes, but was also observed in waterbodies with sample sizes ≥30 individuals (Table 1).
Mean expected and observed heterozygosity were lower in southern prairie populations in the Churchill and Nelson River drainages than Great Lakes populations and those in Hudson Bay tributaries independent of sample size, based on Kruskal–Wallis tests (H = 22.44, p < 0.01 and H = 21.20, p < 0.01, respectively). Mean allelic richness across loci was also broadly similar among sampling locations (Table 1). Standardized allelic richness for a minimum sample size of 30 individuals (=60 allele copies) differed among drainages, with populations in the Churchill and Nelson River drainages having lower AS values (median = 3.12) than those in the Hudson Bay and Mississippi River drainages (median = 3.68; H = 18.31, p < 0.01). For the most part, standardized allelic richness was similar among sampled populations within drainages, although several sites in the Nelson River basin (Berens River, English River, Quetico Provincial Park, and the Sturgeon River) showed lower AS values than other sites.
Individual-based analysis of the lake sturgeon genotypes confirmed hierarchical patterns of genetic structure among the sampled populations (Figure 2). Evaluating the greatest gain in probability between successive values of K (ΔK) [38] suggested an optimal solution of K = 2 (ln [P(X|K) = −126,806]; Supplementary Figures S1 and S2), reflecting differing ancestry for populations draining into Hudson Bay versus those in and south of the Great Lakes basin, with lake sturgeon in Lake Nipigon clustering within both groups (Figure 2). While there was minor evidence for putative admixture between these two groups, most localities assigned strongly to a single cluster only. Assessment of the probability values for varying values of K [37] suggested further local optima at K = 5 (ln [P(X|K) = −124,017]), and K = 8 (ln [P(X|K) = −120,616]), with decreasing probability for values of K greater than 16 (Supplemental Figure S1).
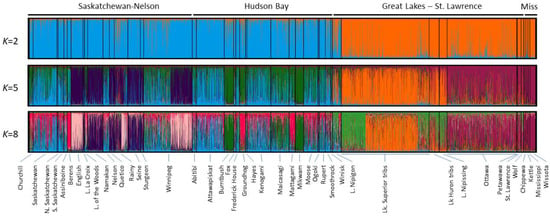
Figure 2.
Structure assignment plots for individual-based analyses of lake sturgeon across the species range for different solutions of K (number of genetic groups present), showing individual membership assignments to multiple groups for values of K = 2, 5, and 8 without using sampling locations as a priori information to identify putative genetic groups.
Population groupings at K = 5 identified substructure within the Nelson River and Hudson Bay basins, with three recognizable subgroups (Figure 2). In northwestern Ontario, samples from the English River, Namakan River, Rainy River, Lake of the Woods, Seine and Sturgeon rivers, and some Winnipeg River locations grouped together, with lake sturgeon from the other northwestern populations forming a separate group. Samples from some southern Hudson Bay (James Bay) tributaries assigned to the latter group as well, with others forming a third distinct subgroup. The additional substructure also clustered lake sturgeon from the Saskatchewan and Attawapiskat rivers, as well as some samples from the Winnipeg River watershed, with rivers in the Hudson Bay drainage, particularly the Attawapiskat and Kenogami rivers (Figure 2). Several waterbodies such as the Rainy River, Winnipeg River, and Lake of the Woods showed marked evidence of substructures (Figure 2). Samples from a number of locations within the Great Lakes basin showed potential minor ancestry from these latter two groups (Figure 2). The Great Lakes and Mississippi River drainage populations showed clear differentiation at K = 5 with lake sturgeon in the upper Great Lakes and Lake Nipissing forming one group, and populations in eastern Ontario outflows and the Mississippi River drainage forming a second group (Figure 2).
Increasing the resolution to K = 8 maintained the groupings observed for K = 5 with some additional substructure. Within the Nelson River and Hudson Bay drainages, samples from the Berens, Groundhog, and Mattagami rivers formed a distinct cluster, as did lake sturgeon from the English River and subsets from the Rainy and Winnipeg rivers (Figure 2). Additional differences were apparent among Great Lakes basin populations, with lakes Nipigon and Nipissing recognizably distinct as well as substructuring among Lake Superior and Lake Huron tributaries (Figure 2).
The relationships within and among drainages and populations was further clarified with PCA and DAPC (Figure 3). Ordination of the full dataset gave clear resolution among the major drainage basins, with populations in the Mississippi watershed (Wisconsin populations) clearly distinct from all Canadian populations. Further analysis among populations within the Nelson, Great Lakes, and Hudson Bay drainage basins also detected significant variation, with varying amounts of substructuring within each basin (Figure 3).
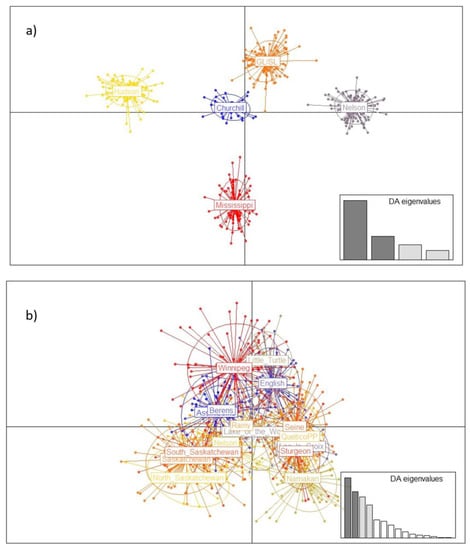
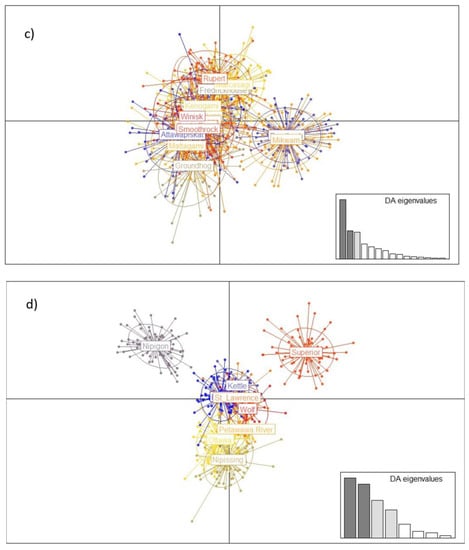
Figure 3.
Discriminant Analysis of Principal Components (DAPC) of lake sturgeon across the species range, (a) grouped by major watershed (“GL/SL” = Great Lakes/St. Lawrence River), and (b–d) for geographically distinct subgroups. (b) Saskatchewan–Nelson River watershed; (c) Hudson Bay watershed; (d) Great Lakes/St. Lawrence watershed. For drainage basins with large numbers of samples (groups in (a) and large populations in (b–d)), a random subsample of 100 genotypes was used to avoid biasing ordination outcomes from unequal sample sizes. Inset plots show significant axes and weighting of discriminant analysis eigenvalues.
Within the Nelson River drainage basin, comparatively little substructure was detected using DAPC despite spanning three provinces, although populations from the eastern and western portions of the drainage basin showed some degree of differentiation (Figure 3b). Similarly, populations within the Hudson Bay watershed had largely similar genetic composition, with only the Mikwam and Burntbush populations showing some divergence from the main grouping (Figure 3c). Within the Great Lakes–St. Lawrence River basin, lake sturgeon from Lake Nipigon and Lake Superior were recognizably distinct from all other populations and each other, and samples from Lake Nipissing showed some separation from the main cluster (Figure 3d).
The genetic relationships and divergences among the sampled populations were confirmed with the population-based analyses. Neighbor-joining clustering of genetic distance estimates among the population confirmed the presence of two distinct lineages with significant (97%) bootstrap support (Figure 4). Among the Churchill River, Nelson River, and Hudson Bay drainages, strong bootstrap support (>80%) was only observed for lake sturgeon from the north, south, and mainstem Saskatchewan River (84%), populations from the southeastern portion of the Nelson River basin (Seine, Namakan, Sturgeon rivers, Quetico Provincial Park, and Lac La Croix; 93%), and the Mikwam and Burntbush rivers which drain into James Bay in southern Hudson Bay (100%). Lake sturgeon from Lake Nipigon appeared to be somewhat intermediate between the two major groups but more closely related to populations in the Hudson Bay cluster; lake sturgeon from Lake Superior tributaries and the Ottawa River were similarly intermediate but more closely affiliated with Great Lakes populations (Figure 4).
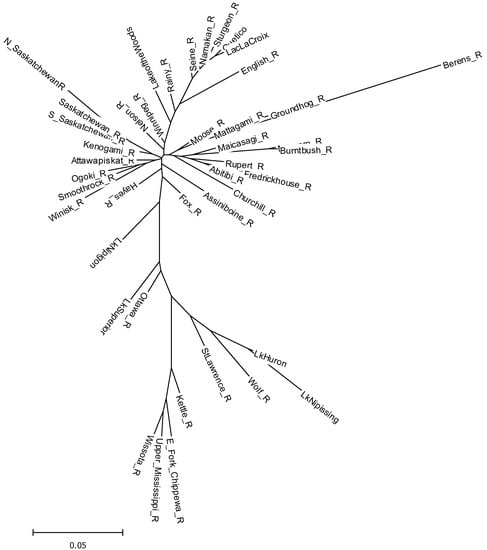
Figure 4.
Unrooted neighbor-joining dendrogram of genetic relationships among sampled populations of lake sturgeon based on Nei et al.’s (1983) genetic distance (Da). Populations with fewer than 10 samples were excluded to avoid sample-size artefacts in estimating genetic distance.
Pairwise assessment of differences among populations based on FST estimates confirmed significant divergence among the majority of populations, particularly between populations in the two major lineages (Supplemental Table S4). For the most part, pairwise FST values between populations in the two lineages were greater than 0.1, whereas pairwise divergences among populations within each lineage were typically less than 0.1. Notable exceptions to this were FST values for the Berens River versus all other populations (mean FST = 0.35, range 0.17 to 0.45), Little Turtle Lake in the Rainy River catchment versus all populations except the Rainy, Seine, and Winnipeg rivers (mean FST = 0.24, range 0.08 to 0.39), and divergences between Mississippi River drainage populations from the Kaministiquia and Black Sturgeon River populations in northern Lake Superior (Supplemental Table S4). Pairwise estimates of Jost’s D [43] showed a similar pattern, with pairwise values ranging from 0 to 0.45 (Supplemental Table S4).
Populations varied considerably for effective population size estimates (Table 1). In general, larger and more northern rivers had higher Ne estimates, although the majority of populations had Ne estimates of 100 individuals or less (Table 1). Population Ne estimates were not correlated with sample size (Supplemental Figure S3). Several populations had very low Ne estimates—for sites with low sample sizes (N ≤ 20), Ne estimates were very low or unresolved, and should be interpreted with caution. Several populations with sample sizes greater than 20 individuals also yielded low Ne values, including sizeable waterbodies such as the Rainy River and Lake Nipigon (Table 1). The large confidence intervals for many Ne estimates underscores the need for caution in interpreting these results, as sampling error effects from finite sample sizes can inflate estimated confidence intervals including values of infinity [44,45].
Coalescent analysis of time to most recent common ancestry among populations using BEAST indicated that the two major lineages diverged approximately 29.6 kya, with a substantial confidence interval in divergence time (95% CI 56.7–7.1 kya) (Figure 5).
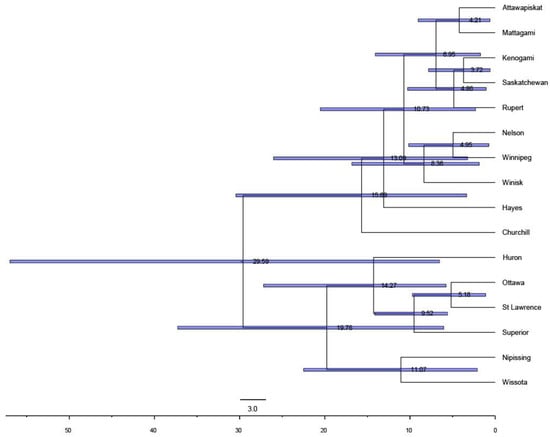
Figure 5.
Estimated phylogenetic relationships and divergence times in thousands of years between the major intraspecific lineages of lake sturgeon and geographically representative populations within each lineage based on coalescent analysis, with the horizontal axis showing estimated time for most recent common ancestry (TMRCA) in thousands of years. Shaded horizontal bars on each branch node represent 95% confidence intervals for estimated divergence times.
Estimates of among-population divergence times within each lineage similarly resulted in large confidence intervals, but mean values of divergence times largely coincide with glacial events. Among populations in the Churchill River, Nelson River, and Hudson Bay drainages, estimated mean times of most recent common ancestry ranged from 15.7 kya to as recently as 3.7 kya, with no clear separation in divergence times among the three drainage basins (Figure 5). Similarly, estimated divergence times among populations in the Mississippi and Great Lakes–St. Lawrence River basins ranged from 19.8 to 5.2 kya, with sizeable confidence intervals (Figure 5).
4. Discussion
The hierarchical genetic structure within and among lake sturgeon populations shows evidence of postglacial, contemporary, and anthropogenic influences. Our results corroborate and expand on previous studies documenting the presence of two ancestral lineages among contemporary lake sturgeon populations, resolving the phylogeographic structure and history of lake sturgeon across the majority of the species’ extant native range, with secondary contact between the two groups in northern Ontario [15,17,20,21]. The observed genetic structure and diversity was primarily influenced by glacial and postglacial colonization events, with comparatively little divergence due to differences among tertiary watersheds or substructuring within rivers and major waterbodies. Within-river fragmentation has had little effect on contemporary phylogeographic substructuring within watersheds, although basic diversity metrics and estimates of local effective population sizes suggest that some genetic fragmentation and genetic loss due to anthropogenic activity may have occurred.
Coalescent analysis indicates that the two intraspecific lineages likely diverged late in the Pleistocene, during the Wisconsinan glaciation approximately 90–10 kya [48]. The estimated mean time to most recent common ancestry between the two lineages of 29.6 kya fits closely with the onset of the late glacial maximum of the Laurentide Ice Sheet during the Wisconsinan glaciation approximately 26.5–19 kya [49,50], suggesting that the ice advance caused their vicariant divergence. The recent (in evolutionary time) divergence between the two lineages is surprising, considering that lake sturgeon purportedly diverged from its congeners more than 20 million years ago [7,8]. Given the antiquity of lake sturgeon as a species, it seems somewhat remarkable that the two intraspecific lineages shared common ancestry so recently and that no genetic evidence of older vicariant events from previous glaciations was detected. As glacial cycles of advance and retreat occurred repeatedly throughout the Pleistocene, however, lake sturgeon would have been directly affected by ice sheet movements as a strictly potamodromous species [4] and would have been able to utilize proglacial lakes at ice margins and newly formed rivers both during and between glacial cycles [13]. As the periodicity of glacial cycles shifted from 40 ky to 100 ky intervals in the late Pleistocene and glacial extends increased accordingly [48], it may be that smaller glacial advances more than 1MYA did not have the same genetic impact on lake sturgeon, or that secondary contact of divergent lineages displaced during previous glacial expansions resulted in admixture and loss of older vicariant signatures. It may be that historical populations in the lower Mississippi River watershed possessed different phylogeographic ancestry, as has been reported for the broadly co-distributed walleye (Sander vitreus) [51], but this can only be resolved if historical samples from extirpated populations are analyzed. The considerable variation in estimated time to most recent shared ancestry (TMRCA) between the two lineages as well as among populations within each lineage reflect the combined effects of coalescent properties, large ancestral populations over prehistoric as well as more recent pre-exploitation timeframes, and localized genetic drift due to finite populations in reciprocally isolated watersheds after periglacial meltwater connections receded. As lake sturgeon populations have retained substantial amounts of genetic diversity despite documented population declines from historical overharvesting [3,19,20,21] and habitat fragmentation [27,29], it seems unlikely that anthropogenic factors have significantly affected TMRCA estimation.
The analyses support the hypothesis that lake sturgeon colonized the Great Lakes primarily through a Mississippian refugium, with subsequent eastward colonization via outlets of Glacial Lake Algonquin. By contrast, populations in western Canada, northern Ontario, and Quebec were colonized from a western, presumably Missourian refugium, as previously hypothesized [17]. The two lineages appear to have largely separate distributions, although some evidence of mixing was apparent. Individual-based analyses for higher values of K showed evidence of limited Missourian ancestry in some Great Lakes and eastern populations, and the Mississippian lineage appears to have dispersed beyond the Great Lakes basin, albeit to a lesser extent. Secondary contact between the two lineages was apparent in northern Lake Superior, outflows of Glacial Lake Algonquin (Lake Nipissing and the Ottawa River), and to some extent in northeastern Ontario. Phylogeographic evidence from previous mitochondrial analyses detected the Mississippian haplotype at low frequency in the Nelson River, Manitoba [17] and in north-central Quebec [16], indicating that Mississippian-origin lake sturgeon were able to colonize Lake Agassiz at least several centuries before it and the connected Lake Ojibway-Barlow drained approximately 8 kya [52]. The stronger Missourian signature in areas of secondary contact likely reflects their order of colonization with early colonists having a priority effect, versus persistence of mitochondrial haplotypes due to their strictly maternal inheritance [53]. The presence of both lineages in Lake Nipigon may be due to recent as well as ancestral secondary contact, due to the 20th century diversion of the Ogoki River to flow into eastern Lake Nipigon [54]. The complex substructure in northwestern Ontario populations may also reflect secondary contact, but could also be due to a separate colonization event during a later phase of Lake Agassiz [21]. The pronounced differences among lake sturgeon populations in northern tributaries of Lake Superior ([20] and this study) suggest that lake sturgeon likely colonized these habitats in separate connection events with Lake Agassiz [14,55]. Genetic differences from contemporary populations in eastern Lake Superior and other parts of the Great Lakes [19,20] suggest both that Mississippian-ancestry lake sturgeon were well established in the other Great Lakes by then and that western Lake Superior may have a complex colonization history.
The two lineages differed in their timing for colonizing Lake Agassiz and subsequent dispersal. The Missourian lineage appears to have accessed Lake Agassiz relatively early, potentially before or during its Moorhead phase 10.9–10.2 kya [56], enabling it to colonize waterbodies across the prairie provinces and the Hudson Bay drainage basin as the ice sheet retreated. By contrast, the Mississippian lineage appears to have not gained access to Lake Agassiz until a later phase. Lake Agassiz’s connection with Lake Ojibway-Barlow enabled both lineages to disperse eastward [13,14,17], although the much stronger genetic signature of the Missourian lineage suggests it preceded the Mississippian lineage in colonizing Lake Agassiz or had greater representation in the founding population. The apparently differing interpretations of the two lineages’ contributions to contemporary populations with mixed phylogeographic ancestry based on mitochondrial DNA versus microsatellite data may also reflect the different modes of inheritance of these two marker systems (strictly maternal versus biparental inheritance) [53].
Genetic structure was largely partitioned among rather than within major drainage basins, with populations draining into Hudson Bay showing shared ancestry, as do populations in the Great Lakes and Mississippi basins. The observed substructure among populations within the Nelson River drainage likely reflects both historical and contemporary drainage patterns, as well as differences in topography between the western low-elevation prairie rivers (Saskatchewan and Assiniboine) versus eastern tributaries on the Canadian Shield. These differences would have been absent while submerged during different stages of Lake Agassiz [52,55,56]. Connectivity among sites would have ended with the abrupt draining of Lakes Agassiz and Ojibway-Barlow 7.9 kya [52,56] and subsequent isostatic rebound that established contemporary watershed boundaries.
The coalescent analysis indicated that the time to most recent common ancestry within each of these groups corresponds with early stages of deglaciation following the late Wisconsinan maximum. The estimated time to most recent common ancestry (TMRCA) for Mississippian and Great Lakes populations corresponds closely with the end of the late glacial maximum [48,49,50]. Although this timing appears to pre-date expansion from a Mississippian refugium and colonization events during postglacial retreat [50,52], the TMRCA estimate may reflect population collapses and losses in the Great Lakes and US portions of the species range [3,4,5,6] with concomitant loss of ancestral genetic diversity [19]. The estimated divergence times among Missourian-ancestry populations are largely congruent with early melting of the western portion of the Laurentide Ice Sheet, although some TMRCA estimates among populations in separate watersheds and drainage basins gave mean divergence estimates for well after glacial meltwaters had receded [50]. Given the large confidence intervals for each estimate, interpretations of the TMRCA values must remain somewhat speculative.
The combined coalescent, genetic distance, and individual-based analyses also showed that remarkably little divergence has occurred among populations within and among drainages with shared postglacial history. As the ice sheet and proglacial meltwaters receded, isostatic rebound separated the contemporary drainage basins, and local lake sturgeon populations became isolated from others in other basins as well as other watersheds within each basin. Prior to 19th century exploitation, divergence among populations would largely reflect genetic drift associated with local population sizes. With the lake sturgeon’s longevity, long generation time, overlapping generations, and large pre-exploitation populations [4], genetic drift would likely have been much less pronounced than for shorter-lived species.
The general lack of detectable substructuring within most river basins likely reflects both their historical connectivity and the lake sturgeon’s capacity to migrate hundreds of kilometers in unfragmented systems [4,57]. In cases where genetic discontinuities have been detected, these have coincided with natural impassable barriers [23,58]. Many of these rivers were historically unimpeded, and the dams that now exist on many of these rivers have been constructed within the last two to three generations of lake sturgeon. As a result, despite spanning three provinces and now dammed in several locations [59], no substructure was detected among the multiple sampling locations in the Saskatchewan River using individual-based analyses. Similarly, no genetic substructure was detected among fragmented segments of the Ottawa River despite being separated by multiple hydroelectric and control dams [29]. In this study, the extensively fragmented Winnipeg River basin [60] was a notable exception, showing two subgroups, although the observed structuring may also reflect the basin’s complex postglacial history or natural substructure from barrier waterfalls [58]. Ironically, the presence of lake sturgeon from both lineages in Lake Nipigon likely reflects the anthropogenic diversion of the Ogoki River into eastern Lake Nipigon [54] rather than fragmentation. Although the individual-based analyses failed to detect spatial substructuring within most sampled waterbodies, pooling of distinct genetic groups, or spatial Wahlund effects [61], was suggested in several waterbodies by heterozygote deficits and linkage disequilibria at multiple loci. This was further evidenced in waterbodies where the structure analysis detected multiple genetic groups, suggesting natural or anthropogenic fragmentation. Genetic effects of fragmentation may be more readily detectable with high-resolution markers [26] and as recruitment levels [62,63] are reflected by genetic diversity and Ne within dammed river segments [22,24,27].
The observed FIS values and deviations from Hardy–Weinberg expectations are consistent with both theoretical predictions and lake sturgeon life history. As the samples within many capture locations were obtained across several years and were likely comprised of individuals of varying ages (multiple cohorts), it is not surprising that many populations showed deviations from Hardy–Weinberg equilibrium expectations and FIS values that varied substantially [61,64]. Strongly positive and negative FIS values at individual loci were most likely due to pooling samples from multiple cohorts with differing sets of parents, resulting in temporal Wahlund effects and violating multiple assumptions for Hardy–Weinberg expectations [61,64]. This would be exacerbated by the life history and reproductive biology of lake sturgeon, where males mature at 12 to 20 years of age and females between ages 15 to 30, with males spawning every 1 to 3 years versus 2 to 7 years for females [3,5]. Deviations from Hardy–Weinberg equilibrium expectations in some locations may also be due to recent population declines and multiple depleted cohorts which would be unlikely to conform to Hardy–Weinberg expectations when pooled.
The observed genetic diversity was remarkably similar among populations despite the substantial variation in sample sizes and being sampled from multiple watersheds. Many of the populations showed higher levels of genetic diversity than might be expected given their endangered or threatened status and documented historical collapses [5]. With few exceptions, the observed and expected heterozygosity and allelic richness are comparable to those reported for Great Lakes populations, many of which have also undergone historical reductions [19,20]. As shown elsewhere, it may be the case that declines in population size are still too recent (too few generations) for genetic losses from anthropogenic activities to be readily apparent [19,27,29]. Heterozygosity and allelic richness estimates have been shown to be relatively insensitive for detecting declines in lake sturgeon populations despite demographic [19,27,62,63] and genetic evidence [20] of historical declines or bottlenecks.
The long generation time of lake sturgeon may have buffered local genetic diversity against the effects of population declines and habitat fragmentation or degradation, but also makes populations vulnerable to rapidly changing conditions [4,5,65]. Estimates of effective population size have been shown to be a sensitive indicator of fragmentation or recent population declines, in some cases after a single generation [66,67,68], including in lake sturgeon [9,24]. The linkage disequilibrium method has been shown to accurately estimate Ne in species with overlapping generations, based on both simulations and empirical data [69,70], and generational Ne can be effectively estimated using randomly sampled adults from multiple year classes [9,70]. These same studies noted that using linkage disequilibrium Ne estimates from multiple adult year classes are downward-biased [69,70], so the numbers reported here likely under-represent true Ne values of the populations. Although generational Ne can also be estimated by calculating the effective number of breeders (Nb) per year class for a generation [9], this was not possible in our study due to the absence of age data and sampling only adult-sized fish. The large confidence intervals for Ne in our study may have been exacerbated by temporal Wahlund effects within some populations as described earlier, and confidence limits of infinity with the linkage disequilibrium method can result from sampling error for finite sample sizes [45].
Despite these caveats, our results show clear evidence of low Ne in many lake sturgeon populations despite retention of substantial genetic diversity. Although the generally higher Ne values among northern Hudson Bay tributaries reflects previous assessment of the relative robustness of these populations [5,6,17], it should be stressed that Ne estimates reflect the genetic resources of populations, rather than actual numbers or census estimates of population size [59,60], and the values here should be considered minimum estimates with substantial uncertainty [44]. Genetic bottleneck effects from recent population losses or declines can persist for several generations [44], and temporal Wahlund-like effects from overlapping generations may also bias Ne estimates downwards [69]. Although the genetic estimates of Ne in this study are markedly lower than population and demographic estimates for the same waterbodies [5,59], comparison of evolutionary and contemporary Ne likewise shows significant declines, particularly in fragmented versus unfragmented river systems [27]. These changes are even more pronounced when temporal estimation of Ne is possible: analysis of samples from different time periods showed substantially greater loss of Ne in a fragmented versus unfragmented river [27]. Given the sizeable confidence intervals for most of the Ne estimates, however, the values reported here should be interpreted with caution.
The estimates of Ne obtained in this study also raise concerns relating to long-term sustainability, as all estimates are well below those recommended for maintaining short-term resilience and adaptive potential over ecological and evolutionary time frames [6,71]. The evolutionary effective population size (Nef) of lake sturgeon is among the lowest reported among North American freshwater fishes [1], and multiple Ne estimators show that local as well as ancestral values are quite limited despite historically large populations [22]. As Ne reflects both demographic and genetic parameters of populations [65,66], the observed low values underscore the importance of guarding against future erosion or loss of existing genetic diversity, as is predicted for fragmented rivers in particular [21,27,28]. Accordingly, the genetic data should not be considered in isolation: both genetic and demographic parameters should be factored in when assessing persistence and recovery probabilities of populations [5,59,71]. As well as minimizing harm to populations and their supporting habitats, this will also require protecting and ensuring connectivity among habitats utilized at different life stages in sufficient quantity and quality to maintain or improve local populations or help them recover [4].
5. Conclusions
Contemporary populations of lake sturgeon face an uncertain future. Although both intraspecific lineages are well represented by multiple populations across extensive portions of the species range, most lake sturgeon populations in Canada are listed as threatened or endangered, habitat fragmentation by dams remains as an obstacle for population recovery [5,6]. Despite this, recovery prospects in parts of the species range are improving with habitat rehabilitation efforts and fishery closures [6]. It is our hope that recognizing the genetic structure and diversity within and among contemporary populations of lake sturgeon will help inform management efforts to help ensure the long-term sustainability of this charismatic, long-lived species.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d15030385/s1, Figure S1: Probability values (ln [P(X|K)]) for the dataset for K = 1–30; Figure S2: Change in log probability of data solutions between successive K values (ΔK) for K = 1–30 [36]; Figure S3: Plot of waterbody sample size (N) versus estimated effective population size (Ne). Table S1: Locus x population testing for Hardy–Weinberg equilibrium; Table S2: Linkage disequilibrium tests for locus pairs within and among populations; Table S3: FIS values for individual loci within populations; Table S4: Pairwise divergence estimates among populations, showing FST estimates below diagonal and Jost’s D above diagonal.
Author Contributions
Conceptualization, S.L.K., N.R.L. and C.C.W.; methodology, S.L.K. and K.W.; data analysis, S.L.K., T.H. and C.C.W.; validation, K.W.; investigation, S.L.K., T.H. and K.W.; resources, T.H., N.R.L. and C.C.W.; data curation, S.L.K., K.W. and T.H.; writing—original draft preparation, S.L.K.; writing—review and editing, T.H., N.R.L., K.W. and C.C.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the University of Toronto (Graduate Fellowships and the Frederick P. Ide Award), NSERC (Alexander Graham Bell Canadian Graduate Scholarship and Discovery Grant to N. Lovejoy), the Ontario Ministry of Natural Resources and Forestry, and the Canada-Ontario Agreement Respecting the Great Lakes Ecosystems.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
Many individuals who generously provided tissues and support for this work: Terry Clayton (Alberta Sustainable Resource Development); Rob Wallace (Saskatchewan Department of Environment and Resource Management); Grant McVittie, Don MacDonald, Ken Kansas, and Ron Campbell (Manitoba Water Stewardship); Cam Barth (University of Manitoba); Patrick Nelson (North/South Consultants Inc.); Chris Chenier, Darryl MacLeod, Charles Hendry, and Nadine Thebeau (Ontario Ministry of Natural Resources and Forestry); Caroline Deary, Andrew Ecclestone, Kim Tremblay, and David Young (Anishinabek/Ontario Fisheries Resource Centre); Henri Fournier, Daniel Nadeau, Pierre Dumont, Pascal Ouellet, and Guy Trencia (Ministère des Ressources Naturelles et de la Faune, Quebec); Amy Welsh (West Virginia University), and Bill Franzin, Bill Gardiner, and Douglas Watkinson (Department of Fisheries and Oceans).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bernatchez, L.; Wilson, C.C. Comparative phylogeography of Nearctic and Palearctic fishes. Mol. Ecol. 1998, 7, 431–452. [Google Scholar] [CrossRef]
- Palsbøll, P.J.; Berube, M.; Allendorf, F.W. Identification of management units using population genetic data. Trends Ecol. Evol. 2007, 22, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Harkness, W.J.K.; Dymond, J.R. The Lake Sturgeon: The History of Its Fishery and Problems of Conservation; Ontario Department of Lands and Forests, Fish and Wildlife Branch: Toronto, ON, Canada, 1961; p. 121. [Google Scholar]
- Bruch, R.M.; Haxton, T.J.; Koenigs, R.; Welsh, A.; Kerr, S.J. Status of Lake Sturgeon (Acipenser fulvescens Rafinesque 1817) in North America. J. Appl. Ichthyol. 2016, 32, 162–190. [Google Scholar] [CrossRef]
- Committee on the Status of Endangered Wildlife in Canada [COSEWIC]. COSEWIC Assessment and Status Report on the Lake Sturgeon Acipenser fulvescens, Western Hudson Bay Populations, Saskatchewan-Nelson River Populations, Southern Hudson Bay James Bay Populations and Great Lakes-Upper St. Lawrence Populations in Canada. Ottawa. 2017, pp. xxx + 53. Available online: https://publications.gc.ca/collections/collection_2018/eccc/CW69-14-484-2017-eng.pdf (accessed on 17 November 2022).
- Haxton, T.; Bruch, R. Acipenser fulvescens. The IUCN Red List of Threatened Species. E.T223A58134229. 2022. Available online: https://www.iucnredlist.org/ja/species/223/58134229 (accessed on 17 November 2022).
- Peng, Z.; Ludwig, A.; Wang, D.; Diogo, R.; Wei, Q.; He, S. Age and biogeography of major clades in sturgeons and paddlefishes (Pisces: Acipenseriformes). Mol. Phylo. Evol. 2007, 42, 854–862. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Yang, N.; Liu, Z.; Chen, Q.; Li, Y. Phylogenetic perspective on the relationships and evolutionary history of the Acipenseriformes. Genomics 2020, 112, 3511–3517. [Google Scholar] [CrossRef] [PubMed]
- Duong, T.Y.; Scribner, K.T.; Forsythe, P.S.; Crossman, J.A.; Baker, E.A. Interannual variation in effective number of breeders and estimation of effective population size in long-lived iteroparous lake sturgeon (Acipenser fulvescens). Mol. Ecol. 2013, 22, 1282–1294. [Google Scholar] [CrossRef]
- NatureServe. NatureServe Explorer: An Online Encyclopedia of Life [Web Application]. Version 7.1. 2022. Available online: http://www.natureserve.org/taxon/ELEMENT_GLOBAL.@104232/Acipenser_fulvescens (accessed on 1 March 2023).
- Lee, D.S.; Gilbert, C.R.; Hocutt, C.H.; Jenkins, R.E.; McAllister, D.E.; Stauffer, J.R., Jr. Atlas of North American Freshwater Fishes; N.C. State Museum of Natural History: Raleigh, NC, USA, 1980; p. 854. [Google Scholar]
- Stewart, K.W.; Lindsey, C.C. Postglacial dispersal of lower vertebrates in the Lake Agassiz region. In Glacial Lake Agassiz; Geological Association of Canada Special Paper 26; Teller, J.T., Clayton, L., Eds.; Geological Association of Canada: St. John’s, NL, Canada, 1983; pp. 391–419. [Google Scholar]
- Crossman, E.J.; McAllister, D.E. Zoogeography of freshwater fishes of the Hudson Bay drainage, Ungava Bay and the Arctic Archipelago. In The Zoogeography of North American Freshwater Fishes; Hocutt, C.H., Wiley, E.O., Eds.; John Wiley and Sons: New York, NY, USA, 1986; pp. 53–104. [Google Scholar]
- Mandrak, N.E.; Crossman, E.J. Postglacial dispersal of freshwater fishes into Ontario. Can. J. Zool. 1992, 70, 2247–2259. [Google Scholar] [CrossRef]
- Ferguson, M.M.; Bernatchez, L.; Gatt, M.; Konkle, B.R.; Lee, S.; Malott, M.L.; McKinley, R.S. Distribution of mitochondrial DNA variation in lake sturgeon (Acipenser fulvescens) from the Moose River Basin, Ontario, Canada. J. Fish Biol. 1993, 43, 91–101. [Google Scholar] [CrossRef]
- Guénette, S.; Fortin, R.; Rassart, E. Mitochondrial DNA variation in lake sturgeon (Acipenser fulvescens) from the St. Lawrence River and James Bay drainage basins in Quebec, Canada. Can. J. Fish. Aquat. Sci. 1993, 50, 659–664. [Google Scholar] [CrossRef]
- Ferguson, M.M.; Duckworth, G.A. The status and distribution of lake sturgeon, Acipenser fulvescens, in the Canadian provinces of Manitoba, Ontario and Quebec: A genetic perspective. Env. Biol. Fish. 1997, 48, 299–310. [Google Scholar] [CrossRef]
- McQuown, E.; Gall, G.A.E.; May, B. Characterization and inheritance of six microsatellite loci in lake sturgeon. Trans. Amer. Fish. Soc. 2002, 131, 299–307. [Google Scholar] [CrossRef]
- DeHaan, P.W.; Libants, S.V.; Elliott, R.F.; Scribner, K.T. Genetic population structure of remnant lake sturgeon populations in the Upper Great Lakes Basin. Trans. Amer. Fish. Soc. 2006, 135, 1478–1492. [Google Scholar] [CrossRef]
- Welsh, A.; Hill, T.; Quinlan, H.; Robinson, C.; May, B. Genetic assessment of Lake Sturgeon population structure in the Laurentian Great Lakes. N. Am. J. Fish. Man. 2008, 28, 572–591. [Google Scholar] [CrossRef]
- McDermid, J.; Wozney, K.; Kjartanson, S.; Wilson, C.C. Quantifying historical, contemporary, and anthropogenic influences on the geographic genetic structure and diversity of lake sturgeon in northern Ontario. J. Appl. Ichthyol. 2011, 27 (Suppl. 2), 12–23. [Google Scholar] [CrossRef]
- Wilson, C.C.; McDermid, J.L.; Wozney, K.M.; Kjartanson, S.L.; Haxton, T. Genetic estimation of evolutionary and contemporary effective population size in lake sturgeon (Acipenser fulvescens) populations. J. Appl. Ichthyol. 2014, 30, 1290–1299. [Google Scholar] [CrossRef]
- Welsh, A.; McLeod, D.T. Detection of natural barriers to movement of lake sturgeon (Acipenser fulvescens) within the Namakan River, Ontario. Can. J. Zool. 2010, 88, 390–397. [Google Scholar] [CrossRef]
- Wilson, C.C.; Haxton, T.J.; Wozney, K.M.; Friday, M. Historical watershed-level connectivity of lake sturgeon in a dammed Great Lakes tributary. J. Great Lakes Res. 2022, 48, 798–805. [Google Scholar] [CrossRef]
- Drauch, A.M.; Fish, B.E.; Latch, E.K.; Fike, J.A.; Rhodes, O.E., Jr. Evaluation of a remnant lake sturgeon population’s utility as a source for reintroductions in the Ohio River system. Cons. Genet. 2008, 9, 1195–1209. [Google Scholar] [CrossRef]
- Whitaker, J.M.; Price, L.E.; Boase, J.C.; Bernatchez, L.; Welsh, A.B. Detecting fine-scale population structure in the age of genomics: A case study of lake sturgeon in the Great Lakes. Fish. Res. 2020, 230, 105646. [Google Scholar] [CrossRef]
- McDermid, J.L.; Nienhuis, S.; Alshamlih, M.; Haxton, T.J.; Wilson, C.C. Evaluating the genetic consequences of river fragmentation on lake sturgeon populations. J. Appl. Ichthyol. 2014, 30, 1514–1523. [Google Scholar] [CrossRef]
- Miliot, E.; Côté, G.; Papillon, L.; Nelson, P.A.; Bernatchez, L. Preliminary Investigation of Population Genetics of Lake Sturgeon from the Lower Nelson River; Manitoba Hydro: Winnipeg, MB, Canada, 2007; p. 60. [Google Scholar]
- Wozney, K.; Haxton, T.; Kjartanson, S.L.; Wilson, C.C. Genetic assessment of Lake Sturgeon (Acipenser fulvescens) population structure in the Ottawa River. Env. Biol. Fishes 2011, 90, 183–195. [Google Scholar] [CrossRef]
- May, B.; Krueger, C.C.; Kincaid, H.L. Genetic variation at microsatellite loci in sturgeon: Primer sequence homology in Acipenser and Scaphirhynchus. Can. J. Fish. Aquat. Sci. 1997, 54, 1542–1547. [Google Scholar] [CrossRef]
- Pyatskowit, J.D.; Krueger, C.C.; Kincaid, H.L.; May, B. Inheritance of microsatellite loci in the polyploid lake sturgeon (Acipenser fulvescens). Genome 2001, 44, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Welsh, A.; Blumberg, M.; May, B. Identification of microsatellite loci in lake sturgeon, Acipenser fulvescens, and their variability in green sturgeon, A. medirostris. Mol. Ecol. Notes 2003, 3, 47–55. [Google Scholar] [CrossRef]
- Welsh, A.; May, B. Development and standardization of disomic microsatellite markers for lake sturgeon genetic studies. J. Appl. Ichthyol. 2006, 22, 337–344. [Google Scholar] [CrossRef]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Winter, D.J. NMOD: An R library for the calculation of population differentiation statistics. Mol. Ecol. Resour. 2012, 12, 1158–1160. [Google Scholar] [CrossRef]
- Alberto, F. standArich_v1.00: An R Package to Estimate Population Allelic Richness Using Standardized Sample Size [Internet]. 2006. Available online: http://www.ccmar.ualg.pt/maree/software.php?soft=sarich (accessed on 25 November 2022).
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREEW: Web version of POPTREE for constructing population trees from allele frequency data and computing other population statistics. Mol. Biol. Evol. 2014, 31, 1622–1624. [Google Scholar] [CrossRef]
- Jost, L. GST and its relatives do not measure differentiation. Mol. Ecol. 2008, 17, 4015–4026. [Google Scholar] [CrossRef]
- Waples, R.S.; Do, C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: A largely untapped resource for applied conservation and evolution. Evol. Appl. 2010, 3, 244–262. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator V2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Res. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchêne, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kühnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Wu, C.H.; Drummond, A.J. Joint inference of microsatellite mutation models, population history and genealogies using transdimensional Markov Chain Monte Carlo. Genetics 2011, 188, 151–164. [Google Scholar] [CrossRef]
- Batchelor, C.L.; Margold, M.; Krapp, M.; Murton, D.K.; Dalton, A.S.; Gibbard, P.L.; Stokes, C.R.; Murton, J.B.; Manica, A. The configuration of Northern Hemisphere ice sheets through the Quaternary. Nat. Comm. 2019, 10, 3713. [Google Scholar]
- Clark, P.U.; Dyke, A.S.; Shakun, J.D.; Carlson, A.E.; Clark, J.; Wohlfarth, B.; Mitrovica, J.X.; Hostetler, S.W.; McCabe, A.M. The last glacial maximum. Science 2009, 325, 710–714. [Google Scholar]
- Dyke, A.S.; Andrews, J.T.; Clark, P.U.; England, J.H.; Miller, G.H.; Shaw, J.; Veillette, J.J. The Laurentide and Innuitian ice sheets during the last glacial maximum. Quat. Sci. Rev. 2002, 21, 9–31. [Google Scholar] [CrossRef]
- Billington, N.; Strange, R.M. Mitochondrial DNA analysis confirms the existence of a genetically divergent walleye population in northeastern Mississippi. Trans. Am. Fish. Soc. 1995, 124, 770–776. [Google Scholar] [CrossRef]
- Dyke, A.S. An outline of North American deglaciation with emphasis on central and northern Canada. Dev. Quat. Sci. 2004, 2, 373–424. [Google Scholar]
- Billington, N.; Hebert, P.D.N. Mitochondrial DNA diversity in fishes and its implications for introductions. Can. J. Fish. Aquat. Sci. 1991, 48 (Suppl. 1), 80–94. [Google Scholar] [CrossRef]
- Wilson, C.C.; Haxton, T.J. Contemporary genetic structure of walleye (Sander vitreus) reflects a historical inter-basin river diversion. J. Great Lakes Res. 2021, 47, 884–891. [Google Scholar] [CrossRef]
- Teller, J.T.; Clayton, L. (Eds.) Glacial Lake Agassiz; Special Paper 26; Geological Association of Canada: St. John’s, NL, Canada, 1983. [Google Scholar]
- Leverington, D.W.; Teller, J.T. Paleotopographic reconstructions of the eastern outlets of glacial Lake Agassiz. Can. J. Earth Sci. 2003, 40, 1259–1278. [Google Scholar] [CrossRef]
- Haxton, T.; Friday, M.; Gillespie, M. Dynamics of lake sturgeon (Acipenser fulvescens Rafinesque, 1817) in a ‘pristine’ river. J. Appl. Ichthyol. 2018, 34, 290–301. [Google Scholar] [CrossRef]
- McDougall, C.A.; Welsh, A.B.; Gosselin, T.; Anderson, W.G.; Nelson, P.A. Rethinking the influence of hydroelectric development on gene flow in a long-lived fish, the lake sturgeon Acipenser fulvescens. PLoS ONE 2017, 12, e0174269. [Google Scholar] [CrossRef]
- McLeod, C.; Hildebrand, L.; Radford, D. A synopsis of lake sturgeon management in Alberta, Canada. J. Appl. Ichthyol. 1999, 15, 173–179. [Google Scholar] [CrossRef]
- George, S.S. Streamflow in the Winnipeg River basin, Canada: Trends, extremes and climate linkages. J. Hydrol. 2007, 332, 15–17. [Google Scholar]
- Hedrick, P.W. Genetics of Populations, 3rd ed.; Jones and Bartlett Publishers: Sudbury, MA, USA, 2005. [Google Scholar]
- Haxton, T.; Friday, M.; Cano, T.; Hendry, C. Assessing the magnitude of effect of hydroelectric production on lake sturgeon abundance in Ontario. N. Am. J. Fish. Man. 2015, 35, 930–941. [Google Scholar] [CrossRef]
- Haxton, T.J.; Findlay, C.S. Variation in lake sturgeon abundance and growth among river reaches in a large regulated river. Can. J. Fish. Aquat. Sci. 2008, 65, 645–657. [Google Scholar] [CrossRef]
- Waples, R.S. Testing for Hardy–Weinberg proportions: Have we lost the plot? J. Hered. 2015, 106, 1–19. [Google Scholar] [CrossRef]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar]
- Luikart, G.; Ryman, N.; Tallmon, D.A.; Schwartz, M.K.; Allendorf, F.W. Estimation of census and effective population sizes: The increasing usefulness of DNA-based approaches. Cons. Genet. 2010, 11, 355–373. [Google Scholar] [CrossRef]
- England, P.R.; Luikart, G.; Waples, R.S. Early detection of population fragmentation using linkage disequilibrium estimation of effective population size. Cons. Genet. 2010, 11, 2425–2430. [Google Scholar] [CrossRef]
- Antao, T.; Perez-Figueroa, A.; Luikart, G. Early detection of population declines: High power of genetic monitoring using effective population size estimators. Evol. Appl. 2011, 4, 144–154. [Google Scholar] [CrossRef]
- Waples, R.S.; Antao, T.; Luikart, G. Effects of overlapping generations on linkage disequilibrium estimates of effective population size. Genetics 2014, 197, 769–780. [Google Scholar] [CrossRef]
- Robinson, J.D.; Moyer, G.R. Linkage disequilibrium and effective population size when generations overlap. Evol. Appl. 2013, 6, 290–302. [Google Scholar] [CrossRef]
- Velez-Espino, L.A.; Koops, M.A. Recovery potential assessment for lake sturgeon in Canadian Designatable Units. N. Am. J. Fish. Man. 2009, 29, 1065–1090. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).