Abstract
Sturgeons are ancient and endangered species whose populations have been greatly reduced over the past few centuries due to habitat destruction and overfishing for the production of expensive caviar. All sturgeon species are listed in CITES Appendix II to limit their trade; therefore, accurate species identification is crucial. In this study, we performed whole-genome resequencing of five sturgeon species, including Kaluga sturgeon (H. dauricus), Siberian sturgeon (A. baerii), Sterlet sturgeon (A. ruthenus), Russian sturgeon (A. gueldenstaedtii), and Amur sturgeon (A. schrenckii), to obtain species-specific InDel-based nucleotide sequences for Kaluga sturgeon. Through whole-genome screening within these five sturgeon species, we developed methods for the rapid identification of Kaluga sturgeon germplasm. Using dominant/co-dominant molecular markers designed for Huso dauricus, purebred or hybrid samples can be identified through a PCR reaction. These markers allow for the precise identification of Kaluga sturgeon lineages from at least seven sturgeon species (H. dauricus, A. schrenckii, A. ruthenus, A. baerii, A. gueldenstaedtii, A. stellatus, and H. huso) and their hybrids. This development is expected to have a positive effect on both the sturgeon trade and the conservation of sturgeon germplasm resources.
1. Introduction
Sturgeon are one of the most ancient fish whose origin can be traced back to the Jurassic period, two hundred million years ago (200 MYBP) [1]. They are a well-known long-lived fish that can live for over 100 years. Sturgeon also exhibit late sexual maturity, with some individuals taking more than 20 years to reach sexual maturity. Sturgeon have extremely high economic value. Caviar, the main product of sturgeon, is made from sturgeon eggs through a process of salting and is renowned for its delicious taste. However, due to its low production and exorbitant price, in the past few centuries, illegal poaching of wild sturgeon by humans, as well as human activity that has destroyed the habitat where sturgeon reside, have led to a sharp decline in the number of wild sturgeon. According to statistical data released by the IUCN in June 2022 [2], Chinese paddlefish (Psephurus gladius), the unique sturgeon species of China, have been declared extinct officially, and the remaining 26 sturgeon species in the world are all facing the risk of extinction, listed in CITES Appendix II to limit their trade [3].
Over the past few decades, the development of the sturgeon industry has gradually shifted from primarily relying on catching wild sturgeon to artificial breeding with the advancement of sturgeon introduction efforts and breakthroughs in sturgeon breeding technology. Currently, China has the highest sturgeon aquaculture output in the world, accounting for nearly 80% of the total global output. In China, the five primary sturgeon species for aquaculture are the Amur sturgeon (Acipencer schrenckii) and Kaluga sturgeon (Huso dauricus), which are distributed in local watersheds, as well as the Siberian sturgeon (Acipencer baerii), Russian sturgeon (Acipencer gueldenstaedtii), and Sterlet sturgeon (Acipencer ruthunus) [4]. Moreover, hybrid sturgeon species, such as the Huso dauricus × Acipenser schrenckii and Acipencer baerii × Acipencer schrenckii, are also bred due to their relatively superior characteristics, such as faster growth rate, compared to their parent species [5]. However, due to the high fertility of sturgeon hybrids and the mismanagement of some fisheries [6], the genetic purity of farmed sturgeon is often unclear, and hybrid sturgeon are difficult to distinguish from purebred sturgeon in early growth stages based on their external features, resulting in considerable risks for aquaculture companies in both breeding and sales processes. Additionally, the ecological issues caused by the escape of hybrid sturgeon into the natural environment cannot be ignored.
In recent decades, significant progress has been made in identifying sturgeon germplasm using molecular markers. Previous studies have utilized SSCP (single strand conformation polymorphism) [7], RFLP (Restriction Fragment Length Polymorphism) [8], and species-specific PCR [9] based on mitochondrial genomes to identify some sturgeon species. However, RFLP and other methods have exhibited poor reproducibility and are no longer preferred. Moreover, due to the maternal inheritance of the mitochondrion, mitochondrial-based markers have limitations in identifying hybrid sturgeon. In contrast, nuclear genomic molecular markers have demonstrated accurate identification of hybrid sturgeon germplasm. For instance, Milos et al. used ddRAD sequencing to identify di-nucleotide SNPs specific to Siberian and Russian sturgeons and designed molecular markers capable of distinguishing these two species from other sturgeon species [10]. Similarly, Havelka et al. utilized ddRAD to pinpoint specific dinucleotide SNPs of sterlet (Acipenser ruthenus) and beluga (Huso Huso) and designed molecular markers for identifying sterlet, beluga, and bester (sterlet × beluga) [11]. In addition, Boscari et al. amplified two intron sequences of the nuclear-coding gene Ribosomal Protein L8 (RPL8) from individuals of Kaluga and Amur sturgeon species [12] and compared the sequencing results to discover Kaluga and Amur sturgeon-specific mutations in the introns. Primers were designed for these sites, but no clear fixed allele sites were found. It was also found that there may be some degree of hybridization between Kaluga and Amur sturgeon, which poses new challenges for the accurate identification of these two species.
InDel-based molecular markers, in contrast to SNP markers, exhibit greater specificity in primer design due to longer base mutations and can be designed as co-dominant markers, making them more conducive to observation of identification outcomes. The major advantage of co-dominant molecular markers is that they are judged based on the size of the target PCR product rather than its presence or absence, which can prevent false- negative results in dominant markers from affecting the accuracy of the detection results. Additionally, the entire detection process can be confined to a single PCR reaction, reducing the complexity of the detection process. This approach has also been widely used in crop research such as rice subspecies identification [13].
Kaluga sturgeon (Huso dauricus) are distributed in the Amur River Basin of China. A large number of Amur sturgeon, Kaluga sturgeon, and their hybrids are bred in China, and natural hybrids of Amur and Kaluga sturgeon have also been detected in nature [14,15]. Therefore, the development of a molecular marker to accurately identify purebred and hybrid species of Kaluga sturgeon has a positive effect both on species identification in the sturgeon trade and on the conservation of this endangered sturgeon species. The purpose of this study was to establish a molecular marker for the identification of purebred and hybrid species of Kaluga sturgeon.
2. Results
2.1. Identification of Kaluga Sturgeon (H. dauricus) and Hybrids
To ensure the accuracy of both sequencing and PCR validation samples, all samples used for sequencing and PCR validation were screened with mitochondrial DNA species-specific markers [9]. Consistent with the results of previous studies, the maternal origin of all the sturgeon samples of five species matched their declared species (Figure 1), including Amur sturgeon, Kaluga sturgeon, Sterlet sturgeon, Siberian sturgeon, and Russian sturgeon. This allowed for a higher expected accuracy in the work of using purebred sturgeon samples for sequencing analysis, developing corresponding molecular markers, and conducting validation.
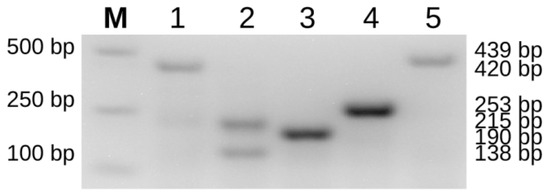
Figure 1.
PCR amplification results of sturgeon mitochondrial species-specific molecular markers. A schematic diagram of the five detected sturgeon species using the mitochondrial-specific molecular markers reported by Mugue et al. previously [9]; M = DL2000 DNA marker; 1 = A. gueldenstaedtii (AGF-AHR, 420 bp); 2 = A. baerii (ABF-AHR, 215 bp; ABF-ABRM, 138 bp); 3 = A. ruthenus (RutF-AHR, 190 bp); 4 = A. schrenckii (SchF-AHR, 253 bp); 5 = H. dauricus (DauF-AHR, 439 bp).
2.2. Specific Variant Identification of Kaluga Sturgeon
A 50 bp-length insertion mutation was found, which was specific to Kaluga sturgeon, located at an intergenic region (NC_048359.1:5385908) of sterlet genome (NCBI RefSeq sequence: GCF_010645085.2) [16] (Supplementary Information, Figure S1), where the inserted fragment was detected in all 69 reads of Kaluga sturgeon data, and the mutation was absent in the other sturgeons at the same loci. As this locus showed a clear homozygous pattern in the alignment results, we believe it may serve as a potential fixed allele specific to Kaluga sturgeon for the development of Kaluga sturgeon-specific molecular markers.
2.3. Dominant/Co-Dominant Primers Design for Kaluga Sturgeon and Validation
Primers according to this insertion site were designed. Two ways of primer design (dominant/co-dominant) for Kaluga sturgeon were adopted. One end of the dominant primer (Hdau216p) was designed within the specific insertion of Kaluga sturgeon, so the primer should lead to a specific amplification in Kaluga sturgeon and no amplification in other sturgeons, while the co-dominant primer Hdau195l was designed to span the 50 bp specific insertion in Kaluga sturgeon. The 195 bp and 145 bp PCR products should be amplified for Kaluga sturgeon and other sturgeon species, respectively (Table 1).

Table 1.
Dominant/co-dominant primers Hdau216p/Hdau195l developed for identification of Kaluga sturgeon (Huso dauricus).
Using the dominant primer Hdau216p to amplify Kaluga sturgeon from total seven species of purebred sturgeon, 100% samples of Kaluga sturgeon had a positive result of 216 bp, while other sturgeon species had negative amplification results (Figure 2a). Using the co-dominant primer to amplify seven species of sturgeon and five hybrids containing Kaluga sturgeon, the sample of Kaluga sturgeon showed a single band at 195 bp, while other sturgeon specimens showed a single band at 145 bp. Two bands at 145 bp and 195 bp appeared in the hybrid containing Kaluga sturgeon, as expected (Figure 2b) (Table 2 and Table 3).
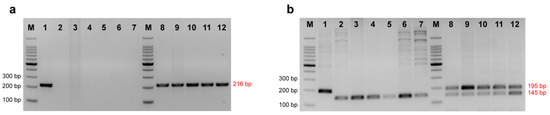
Figure 2.
Detection of the amplification efficiency of Kaluga sturgeon dominant primer Hdau216p (a) and co-dominant primer Hdau195l (b). 1 = H. dauricus; 2 = A. schrenckii; 3 = A. baerii; 4 = A. gueldenstaedtii; 5 = A. ruthenus; 6 = A. stellatus; 7 = H. huso; 8 = A. gueldenstaedtii (♀) × H. dauricus (♂); 9 = A. baerii (♀) × H. dauricus (♂); 10 = A. ruthenus (♀) × H. dauricus (♂); 11 = A. schrenckii (♀) × H. dauricus (♂); 12 = H. dauricus (♀) × A. schrenckii (♂); M = 100 bp DNA ladder.

Table 2.
Validation of Dominant/co-dominant primers Hdau216p/Hdau195l for seven purebred sturgeon specimens.

Table 3.
Validation of Dominant/co-dominant primers Hdau216p/Hdau195l for hybrid sturgeon specimens.
It proved that these two markers were accurately validated in purebred Kaluga sturgeon and other six purebred sturgeon species sampled from three different farms. Furthermore, various hybrid combinations containing the lineage of Kaluga (Amur sturgeon (♀) × Kaluga sturgeon (♂), Kaluga sturgeon (♀) × Amur sturgeon (♂), Siberian sturgeon (♀) × Kaluga sturgeon (♂), Sterlet sturgeon (♀) × Kaluga sturgeon (♂), and Russian sturgeon (♀) × Kaluga sturgeon (♂)) were also verified accurately.
Both the dominant marker Hdau216p and the co-dominant marker Hdau195l show correct specificity in the expected PCR results shown in Figure 3. For the dominant marker Hdau216p, positive bands at 216 bp were obtained for pure and hybrid Kaluga samples, while other sturgeon species had negative results; for the co-dominant marker Hdau195l, the sample of pure Kaluga sturgeon obtained only one positive band at 195 bp, while the sample of hybrid Kaluga sturgeon had positive bands at both 195 and 145 bp, and other sturgeon species had a band at 145 bp. The results match the conclusion we drew through resequencing analysis.
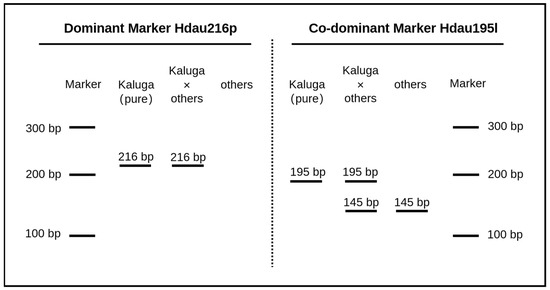
Figure 3.
Expected PCR results for Kaluga-specific dominant (Hdau216p) and co-dominant (Hdau195l) markers.
2.4. Validation and Comparison with Other Markers
We attempted to use the RP4L8 marker reported in previous studies [12] in order to ensure the correction of the new markers developed in this study, strictly following the PCR conditions. However, we found that the amplification results were unstable to some extent. As shown in the figure (Figure 4a), positive bands were detected not only in Kaluga sturgeon but also in other purebred and hybrid samples that did not contain Kaluga sturgeon, and a negative result was observed in a hybrid of Kaluga sturgeon. However, the expected results were obtained using the Hdau195l marker for detection (Figure 4b). Therefore, we sequenced the target band obtained by amplifying sample 12 (Amur sturgeon × Siberian sturgeon), and found that both the 3’ end of the forward and reverse primers of RP4L8 contained base pairs that did not match the specific allele in Kaluga sturgeon, indicating that the amplification product was not a Kaluga sturgeon-specific target band (Figure 5).

Figure 4.
Validation of the amplification efficiency for Kaluga-specific marker RP4L8 (a) and co-dominant primer Hdau195l (b) using PrimeSTAR® Max DNA Polymerase. (−) = negative control; (+) = positive control; 1, 2 = H. dauricus; 3, 4 = A. ruthenus; 5, 6 = A. gueldenstaedtii; 7, 8 = A. schrenckii; 9, 10 = A. baerii; 11 = A. schrenckii (♀) × H. dauricus (♂);12 = A. schrenckii (♀) × A. baerii (♂); M = 100 bp DNA ladder.

Figure 5.
Sequencing results of RP4L8 PCR product (sample 12: A. schrenckii (♀) × A. baerii (♂)). (a,b) show the binding regions of RP4L8_H.dau_F and RP4L8_H.dau_R primers, respectively. The template sequences bound by the primers are indicated by red underlines, and the bases at the 3’ end of the primers (non-Kaluga sturgeon specific) are marked by red arrows.
In order to identify the cause of the unexpected results, we expanded the sample size (including the previous 12 samples) and tried different PCR reagents (Figure 6a). We found that samples from non-Kaluga sturgeon lineages did not show false-positive results as previously observed, although one Kaluga sturgeon hybrid (sample 26) still showed negative results. The same samples were also amplified using Hdau195l, and the results were consistent with the expected results (Figure 6b). This suggests that different PCR systems may have some impact on the specificity, particularly for dominant primers.
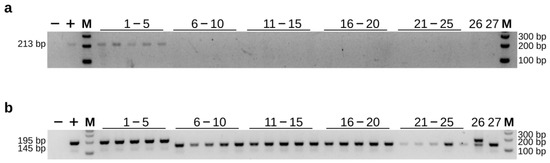
Figure 6.
Validation of the amplification efficiency for Kaluga-specific marker RP4L8 (a) and co-dominant primer Hdau195l (b) using TaKaRa Ex Taq®. (−) = negative control; (+) = positive control; 1–5 = H. dauricus; 6–10 = A. ruthenus; 11–15 = A. gueldenstaedtii; 16–20 = A. schrenckii; 21–25 = A. baerii; 26 = A. schrenckii (♀) × H. dauricus (♂); 27 = A. schrenckii (♀) × A. baerii (♂); M = 100 bp DNA ladder.
In addition to the experimental validation, we also analyzed the amplification sites of RP4L8 primers based on sequencing data. We found that the SNP site specific to Kaluga sturgeon in the terminal region of RP4L8 primers did show sturgeon-specific characteristics, but it seemed to be in a heterozygous state (Figure 7 and Figure 8). This explains why the marker showed low specificity in other sturgeon species.
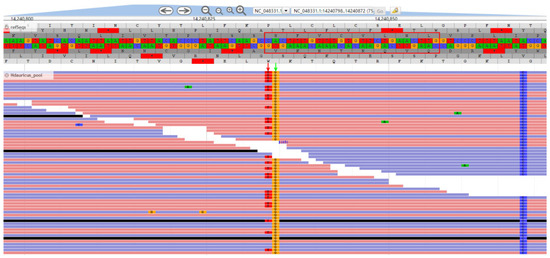
Figure 7.
Visualization of resequencing data alignment region of RP4L8_H.dau_F. The red box indicates the genomic region where the primer sequence matches, with the red arrows pointing to the Kaluga sturgeon-specific SNP site and the green arrows pointing to the artificially introduced mismatch position in the second base of the 3′ end of the primer.
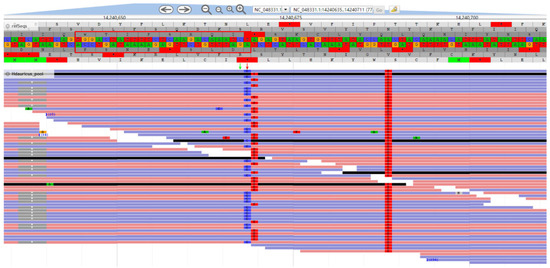
Figure 8.
Visualization of resequencing data alignment region of RP4L8_H.dau_R. The red box indicates the genomic region where the primer sequence matches, with the red arrows pointing to the Kaluga sturgeon-specific SNP site and the green arrows pointing to the artificially introduced mismatch position in the second base of the 3′ end of the primer.
3. Discussion
In this study, we aimed to develop new molecular markers for accurate identification of purebred and hybrid Kaluga sturgeon, addressing some of the limitations of existing methods for genetic identification of Kaluga sturgeon. We identified Kaluga-specific genetic variations using whole-genome sequencing data to develop these markers.
Previous studies have reported several methods for genetic identification of Kaluga sturgeon, among which the most accurate is mitochondrial DNA-specific PCR [9]. All samples used for sequencing and validation in this study were also tested for Kaluga sturgeon-specific mitochondrial markers. However, as mitochondrial DNA is maternally inherited, this marker can only determine the presence of Kaluga sturgeon maternal lineage in the specimen and cannot distinguish purebred from hybrid Kaluga sturgeon, which is its biggest limitation.
Microsatellite markers have also been used for Kaluga sturgeon identification [17]. For instance, the absence of amplified product of the microsatellite marker Afug41 in purebred Kaluga sturgeon can be used for its identification. However, this method may result in false negatives due to poor DNA quality, and positive results from other sturgeon species may interfere with the detection of Kaluga sturgeon lineage in hybrid specimens. Some studies have also shown that purebred Kaluga sturgeon can have amplified products of Afug41 [12]. The microsatellite marker An20 can produce a specific allele 169 in Kaluga sturgeon, but its detection efficiency is significantly reduced in hybrid individuals due to the reliance on allele frequencies in the population. Previous studies have also developed the RP4L8 marker based on the intron of the nuclear-coding gene Ribosomal Protein L8, which shows specificity to Kaluga sturgeon [12]. However, this specificity may be incomplete due to hidden hybridization events between Kaluga sturgeon and Amur sturgeon, and combination with other markers is still necessary for accurate identification of purebred and hybrid Kaluga sturgeon.
In our study, we first identified a 50 bp insertion mutation located in the intergenic region with Kaluga sturgeon specificity, based on the whole-genome resequencing data of five sturgeon species and the reference genome of Sterlet sturgeon. The dominant primer Hdau216p, which spans the insertion site, specifically amplified a 216 bp fragment from all purebred and hybrid Kaluga sturgeon samples. In contrast, the co-dominant primer Hdau195l, designed to flank the insertion site, amplified a 195 bp fragment from all purebred Kaluga sturgeon samples and a 145 bp fragment from other sturgeon species, including Siberian sturgeon, Amur sturgeon, Russian sturgeon, Sterlet sturgeon, Beluga sturgeon, and Stellate sturgeon. For Kaluga sturgeon hybrids, both 195 bp and 145 bp fragments were amplified. This marker exhibited expected amplification patterns in all seven claimed purebred sturgeon species and five Kaluga sturgeon hybrids.
We attempted to validate some purebred and hybrid sturgeon samples using the Kaluga-specific RP4L8 marker [12]. Through amplification and sequencing, we found that the specificity of this primer may be influenced by the PCR system, leading to false-positive results (Figure 4a and Figure 6a), though more accurate results may be obtained by adjusting the PCR system. As for the co-dominant marker Hdau195l developed in this study, it was designed within the conserved sequences shared by all species that we sequenced and achieved species identification by 50 bp difference of amplifying lengths between primers. Therefore, it may be less affected by the PCR system and is better at avoiding false- positive or -negative results.
We also attempted to validate previously reported markers RP4L8 [12] using our sequencing data. We found that the Kaluga sturgeon-specific SNP reported in RP4L8’s forward and reverse primers 3’ end was present in our data, but it appeared to be heterozygous, with both the specific and the other sturgeon species’ alleles present (Figure 7 and Figure 8). This may reduce its detection power in hybrids due to the parents only passing on half of the alleles. However, the insertion mutation targeted by our developed marker was a homozygous site in Kaluga sturgeon data, assuring us that the markers based on this site’s mutation would have an advantage in Kaluga sturgeon identification (especially in hybrids).
As the Kaluga sturgeon sample we used for sequencing was obtained from one fishery, there may be a risk of insufficient genetic diversity, which may result in the species-specific loci we found actually being population specific. However, they were proven to be species-specific markers from the validation results of the purebred and hybrid Kaluga sturgeon samples obtained from several other fisheries. In these samples, our markers, both the dominant and co-dominant markers, showed 100% accuracy, which means that the variant loci we found should be specific to Kaluga sturgeon rather than limited to individuals from certain populations. Although we have expanded the sampling range as much as possible to ensure the specificity of this marker for Kaluga sturgeon, it is still needed to further verify the specificity of Kaluga sturgeon using more sturgeon species samples from different sources.
We provide a co-dominant marker-based method that can also be used for other research in sturgeons, such as gynogenesis. Gynogenesis plays a crucial role in the study of sex determination in fish and in the production of sex-controlled individuals, such as in turbot [18]. As gynogenesis technology artificially induces the multiplication of haploid gametes into polyploid individuals, inactivated sperm from closely related species is often used to fertilize fish eggs. However, screening for true gynogenetic individuals among the surviving individuals can be challenging. In this case, our co-dominant marker can greatly reduce the difficulty of screening. On one hand, if fish eggs from purebred Kaluga sturgeon females and sperm from other sturgeon species are used to prepare gynogenetic individuals(F1), all surviving individuals’ DNA can be PCR-amplified with primer Hdau195l. If a single 195 bp band appears without a 145 bp band, the individual shows a purebred Kaluga sturgeon lineage at this locus and can be selected as a gynogenetic candidate for further screening. On the other hand, if gynogenetic individuals of other sturgeon species (except for Kaluga sturgeon) need to be prepared, fish eggs from other sturgeon species and Kaluga sturgeon sperm can be used for preparation, and all surviving individuals’ DNA can be PCR-amplified. If a single band of 145 bp appears without a 195 bp band, the individual shows no Kaluga sturgeon lineage at this locus and can be selected as a candidate for gynogenesis screening. Although it is possible to differentiate between gynogenetic and hybrid individuals for specific species of sturgeons based on their appearance, this co-dominant marker-based method can avoid the ambiguity and uncertainty of morphological features and can quickly and accurately identify individuals in juvenile fish after several months of growth, which is more practical than traditional methods such as microsatellite amplification we used before.
According to previous studies, hybridization between Amur sturgeon and Kaluga sturgeon may exist to some extent [12], which could be a reason for the contradictory results of different markers, as the standard individuals claimed to be purebred Amur sturgeon or Kaluga sturgeon may conceal hybrid ancestry. However, in the samples we tested, all purebred Amur sturgeon and Kaluga sturgeon samples showed different single bands of sizes (Kaluga sturgeon: 195 bp, Amur sturgeon: 145 bp) after the co-dominant marker Hdau195l detection, indicating that, at least at this locus, no hybridization events occurred between the tested Amur sturgeon and Kaluga sturgeon samples. This finding suggests that this locus may be a truly fixed allele specific to Kaluga sturgeon, although it is only located in the intergenic region.
In addition, we extracted DNA from a caviar sample claimed to be from a purebred Kaluga sturgeon and conducted PCR testing. Both of the markers we developed confirmed that the caviar contained Kaluga sturgeon germplasm and was indeed from a purebred Kaluga sturgeon (Figure 9). This demonstrates that our tool not only has good detection performance in sturgeon tissue samples but also has potential for identifying germplasm in sturgeon products such as caviar. However, not all the caviar samples have stable results during detection, and the quality of caviar DNA is thought to be responsible for the erratic results, which need to be further improved.

Figure 9.
Detection of the amplification efficiency of Kaluga sturgeon dominant primer Hdau216p (a) and co-dominant primer Hdau195l (b) with a caviar(purebred Kaluga sturgeon). M: 100 bp DNA ladder.
The purebred and hybrid sturgeon samples used in this study were F0 and F1 individuals, and the 100% accuracy of the developed marker also demonstrated its detection efficiency in the F0 and F1 generations. However, since there is currently a lack of clear F2 hybrid samples for validation, the detection efficiency of this marker after multiple generations of hybridization still needs further verification.
Kaluga sturgeon is an important aquaculture species in China distributed naturally in the Heilongjiang River Basin. Due to the general advantages of hybrid sturgeon in terms of growth, stress resistance, and reproductive traits [5], hybrids of Amur sturgeon and Kaluga sturgeon are widely bred in China, with the largest number of A. schrenckii × H. dauricus hybrids. As early as 2004, literature reports indicated that hybrid species of Amur and Kaluga sturgeon had been captured in the wild. The tools used in this study have been tested for accurate identification efficiency across various hybrids of Kaluga sturgeon. Therefore, they can play an important role in managing broodstock in breeding enterprises and protecting the sturgeon resources of Kaluga sturgeon in the wild.
4. Conclusions
Based on the InDels identified from whole-genome resequencing data, we have developed a series of tools for the identification of Kaluga sturgeon and related hybrids using both dominant and co-dominant markers. The identification of purebred and hybrid sturgeon can be accomplished using a single PCR reaction with co-dominant markers, which holds significant implications for the identification of Kaluga sturgeon, as well as the related hybrids, given their high commercial value and significant aquaculture production, particularly in China. These markers, unlike those reported in previous studies, have demonstrated unambiguous accuracy that we validated in both purebred and hybrid samples. Furthermore, they have shown potential in species identification of sturgeon-related products, such as caviar.
This technological advancement will contribute to the promotion of the sturgeon trade and the conservation of sturgeon germplasm resources.
5. Materials and Methods
5.1. Sampling and DNA Extraction
In this study, samples from fin tissues of five sturgeon species were used for sequencing and validation, including Amur sturgeon (Acipenser schrenckii), Kaluga sturgeon (Huso dauricus), Siberian sturgeon (Acipenser baerii), Sterlet sturgeon (Acipenser ruthenus), and Russian sturgeon (Acipenser gueldenstaedtii), among six sturgeon fish breeding farms in Beijing that cover Fangshan Shidu affiliated with the Beijing Academy of Agriculture and Forestry Sciences. In addition, some of the validation samples, both Beluga sturgeon (Huso huso) and Starry sturgeon (Acipenser stellatus), were obtained from fish farms owned by Hangzhou Qiandaohu Xunlong Sci-Tech Co. Ltd. (Hangzhou, China) and were not subjected to resequencing. To avoid low genetic diversity caused by close kinship among the sturgeon samples used for sequencing and PCR validation, we selected sturgeon samples from six fish farms around Beijing covering different batches and age groups (except for Kaluga sturgeon) to maximize genetic diversity within the samples. For Kaluga sturgeon, all samples used for sequencing were collected from the sturgeon fish breeding farm in Fangshan Shidu affiliated to Beijing Academy of Agriculture and Forestry Sciences; and 41 Kaluga sturgeon samples were collected from Zhongketianli Aquatic science and technology Co. Ltd. (Beijing, China) farm for PCR validation. To ensure population diversity for re-sequencing, we collected 30 samples from each of the five sturgeon species (Amur sturgeon; Sterlet sturgeon; Siberian sturgeon; Russian sturgeon; and Kaluga sturgeon) and pooled them for genome resequencing. All fin tissue samples for sequencing and validation were preserved in 95% ethanol and DNA was extracted using the DNeasy Blood & Tissue Kit (NO.69504) from Qiagen (Hilden, Germany) following the manufacturer’s instructions. All sturgeon DNA samples used for sequencing and PCR validation were subjected to Nanodrop detection to ensure their qualified DNA quality and concentration.
In order to ensure that the germplasm of the sequenced and verified sturgeon samples is consistent with their claimed germplasm, the species-specific mitochondrial markers [9] were prioritized for PCR detection of all samples, and the detection results (maternal germplasm) were consistent with their claimed species.
5.2. Whole-Genome Resequencing and Variant Identification
DNA was extracted from fin tissues of 30 samples collected from 5 species (A. schrenckii, H. dauricus, A. baerii, A. ruthenus, and A. gueldenstaedtii) and mixed in equal amounts for each species. Library preparation and whole-genome resequencing were perfomed by Novogene Co. Ltd. (Beijing; China) with an Illumina Hiseq 2000 platform. The low-quality reads and adapters in the raw data were filtered using Fastp software (v0.20.1) [19]. The clean data were mapped to the sterlet reference genome [16] (ASM1064508v1) (Du et al., 2020) using bwa (v0.7.17-r1188) [20] with default parameters, and then the MarkDuplicates module in the GATK software [21] was used to remove PCR duplicates. After reads mapping, the Haplotypecaller module in the GATK software (v4.1.8.1) was used for genomic variant sites calling, and then all GVCF files were merged to identify and filter InDel sites to obtain high-quality InDel sites among the whole genome. Then a custom Python script was used to screen the specific homozygous InDel sites (sequencing depth greater than 10, and InDel length greater than 20 bp and less than 60 bp) in the screening results of H. dauricus as candidate sites which might be a fixed allele in Kaluga sturgeon, and JBrowse [22] was used to manually verify the candidate sites visually.
5.3. Primer Design and Validation
A sequence of 600 bp (300 bp upstream and downstream) at the candidate InDel sites from sterlet genome was extracted for primer design. For species-specific insertion, the original sequence was substituted for the insertion sequence to construct the corresponding species-specific genome sequence. Use Primer Premier 5 [23] software was used to design primers for candidate sequences and avoid non-specific amplification.
For Kaluga sturgeon (Huso dauricus), in the design of dominant primers, most of the forward primer’s sequences (except for the 3’-terminal four bases) were designed within a 50 bp insertion mutation to ensure high specificity for Kaluga sturgeon during the amplification process (Figure 10a). For the design of co-dominant primers, the forward and reverse primers were designed on either side of the 50 bp insertion mutation (Figure 10b). Kaluga sturgeon samples exhibited a larger fragment size (195 bp) in the amplification result due to the presence of the specific insertion mutation in the amplification fragment, whereas other sturgeon species produced smaller fragments (145 bp) upon amplification. The difference in amplification fragment size can be rapidly distinguished by gel electrophoresis and a standard marker with 100 and 200 bp.

Figure 10.
Dominant/co-dominant primer design Hdau216p (a) and Hdau195l (b) for Kaluga sturgeon (H. dauricus).
Firstly, part of the sturgeon DNA specimens for sequencing was used for preliminary verification of the primers, and the primers were further verified by PCR using DNA specimens of seven species of sturgeon other than the sequencing samples and hybrid sturgeon samples. All reactions were performed using Takara PrimeSTAR® Max DNA Polymerase (2×) system, containing for each primer 0.5 µM, 50 ng template DNA, and 10 µL PrimeSTAR® Max DNA Polymerase (2×) in a reaction with a total volume of 20 µL. All PCR reactions were run using the following program: 94 °C for 5 min; 35 cycles of 94 °C for 30 s, 66 °C for 30 s, and 72 °C for 10 s; and 72 °C for 10 min for final extension. All PCR amplification products were inspected on 3% agarose gel using ImageLab software (v3.0).
5.4. Species-Specific Marker RP4L8 PCR Validation
For the previously reported species-specific molecular markers RP4L8 (RP4L8_H.dau_F: 5′-CAAGTTCAGAACACAAACAAAGGA-3′), RP4L8_H.dau_R: 5′-TGGACTATTTTCTCAAGACAAATGC-3′) designed based on the fourth (RP4) introns of nuclear-coding gene Ribosomal Protein L8 (RPL8) for Kaluga sturgeon, we amplified the markers using the PCR profile reported in the previous study (a first denaturation step at 95 °C for 3 min, 35 cycles at 95 °C for 30 s, 61 °C for 20 s, 72 °C for 30 s, and a final elongation step at 72 °C for 7 min), and used two different PCR reagents (PrimeSTAR® Max DNA Polymerase and TaKaRa Ex Taq®) simultaneously for amplification.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/d15050689/s1, Figure S1: Alignment of consensus sequence of H.dauricus and other sturgeon around the insertion.
Author Contributions
Conceptualization, H.H.; methodology, H.H. and X.Y.; software, X.Y. and H.S.; validation, X.Y., Y.D. and T.D.; sampling, H.H., Y.D. and W.W.; writing—original draft preparation, X.Y.; writing—review and editing, H.H.; project administration, H.H.; funding acquisition, H.H. and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by [Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding], Grant number [G20220628008] and [the Youth Foundation of Beijing Academy of Agriculture and Forestry Sciences] Grant number [QNJJ202207]. The APC was funded by [Beijing Joint Research Program for Germplasm Innovation and New Variety Breeding] Grant number: [G20220628008].
Data Availability Statement
All the publishable relevant data are within the manuscript and its Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bemis, W.E.; Findeis, E.K.; Grande, L. An overview of Acipenseriformes. Environ. Biol. Fishes 1997, 48, 25–71. [Google Scholar] [CrossRef]
- International Union for Conservation of Nature. The IUCN Red List of Threatened Species, Version 2022-1. 2022. Available online: www.iucnredlist.org (accessed on 21 July 2022).
- Convention on International Trade in Endangered Species of Wild Fauna and Flora. The tenth meeting of the Conference of the Parties to CITES (COP10), Harare, Zimbabwe. 9-20 June 1997. Available online: https://www.govinfo.gov/content/pkg/FR-1997-08-22/html/97-22402.htm (accessed on 21 July 2022).
- Bronzi, P.; Chebanov, M.; Michaels, J.T.; Wei, Q.; Rosenthal, H.; Gessner, J. Sturgeon meat and caviar production: Global update 2017. J. Appl. Ichthyol. 2019, 35, 257–266. [Google Scholar] [CrossRef]
- Shivaramu, S.; Vuong, D.T.; Havelka, M.; Šachlová, H.; Lebeda, I.; Kašpar, V.; Flajšhans, M. Influence of interspecific hybridization on fitness-related traits in Siberian sturgeon and Russian sturgeon. Czech J. Anim. Sci. 2019, 64, 78–88. [Google Scholar] [CrossRef]
- Havelka, M.; Arai, K. Hybridization and polyploidization in sturgeon. Sex Control. Aquac. 2018, Chapter 34, 669–687. [Google Scholar]
- Ludwig, A.; May, B.; Debus, L.; Jenneckens, I. Heteroplasmy in the mtDNA control region of sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 2000, 156, 1933–1947. [Google Scholar] [CrossRef]
- Ludwig, A.; Debus, L.; Jenneckens, I. A molecular approach to control the international trade in black caviar. Int. Rev. Hydrobiol. A J. Cover. All Asp. Limnol. Mar. Biol. 2002, 87, 661–674. [Google Scholar]
- Mugue, N.S.; Barmintseva, A.E.; Rastorguev, S.M.; Mugue, V.N.; Barmintsev, V.A. Polymorphism of the mitochondrial DNA control region in eight sturgeon species and development of a system for DNA-based species identification. Russ. J. Genet. 2008, 44, 793–798. [Google Scholar] [CrossRef]
- Havelka, M.; Arai, K.; Boscari, E.; Congiu, L.; Sergeev, A.; Mugue, N. A new marker, isolated by ddRAD sequencing, detects Siberian and Russian sturgeon in hybrids. Anim. Genet. 2019, 50, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Havelka, M.; Fujimoto, T.; Hagihara, S.; Adachi, S.; Arai, K. Nuclear DNA markers for identification of Beluga and Sterlet sturgeons and their interspecific Bester hybrid. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Boscari, E.; Barmintseva, A.; Zhang, S.; Yue, H.; Li, C.; Shedko, S.V.; Lieckfeldt, D.; Ludwig, A.; Wei, Q.W.; Mugue, N.S.; et al. Genetic identification of the caviar-producing Amur and Kaluga sturgeons revealed a high level of concealed hybridization. Food Control. 2017, 82, 243–250. [Google Scholar] [CrossRef]
- Yuan, H.; Yang, W.; Zou, J.; Cheng, M.; Fan, F.; Liang, T.; Yu, Y.; Qiu, R.; Li, S.; Hu, J. InDel Markers Based on 3K Whole-Genome Re-Sequencing Data Characterise the Subspecies of Rice (Oryza sativa L.). Agriculture 2021, 11, 655. [Google Scholar] [CrossRef]
- Omoto, N.; Maebayashi, M.; Hara, A.; Adachi, S.; Yamauchi, K. Gonadal maturity in wild sturgeons, Huso dauricus, Acipenser mikadoi and A. schrenckii caught near Hokkaido, Japan. Environ. Biol. Fishes 2004, 70, 381–391. [Google Scholar] [CrossRef]
- Shedko, S.V.; Miroshnichenko, I.L.; Nemkova, G.A. Asymmetric Hybridization of Kaluga Acipenser dauricus Georgi, 1775 and Amur Sturgeon A. schrenckii Brandt, 1869 (Acipenseridae) in Nature as Follows from Analysis of Mitochondrial and Nuclear DNA Markers. Russ. J. Genet. 2020, 56, 718–724. [Google Scholar] [CrossRef]
- Du, K.; Stöck, M.; Kneitz, S.; Klopp, C.; Woltering, J.M.; Adolfi, M.C.; Feron, R.; Prokopov, D.; Makunin, A.; Kichigin, I.; et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 2020, 4, 841–852. [Google Scholar] [CrossRef]
- Barmintseva, A.E.; Mugue, N.S. The use of microsatellite loci for identification of sturgeon species (Acipenseridae) and hybrid forms. Russ. J. Genet. 2013, 49, 950–961. [Google Scholar] [CrossRef]
- Taboada, X.; Robledo, D.; Bouza, C.; Piferrer, F.; Viñas, A.M.; Martínez, P. Reproduction and sex control in turbot. Sex Control Aquac. 2018, Chapter 28, 565–582. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).