Characterization of Okra (Abelmoschus esculentus L.) Accessions with Variable Drought Tolerance through Simple Sequence Repeat Markers and Phenotypic Traits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction, Purification, and Quantification
2.3. Polymerase Chain Reaction (PCR) and SSR Analysis
2.4. Marker Data Analysis
2.4.1. Computation of Principal Coordinate Analysis (PCoA) and Genetic Parameters
2.4.2. Cluster Analysis
2.5. Phenotyping Okra Accessions
2.5.1. Experimental Design and Crop Establishment
2.5.2. Phenotypic Data Collection
2.5.3. Phenotypic Data Analysis
3. Results and Discussion
3.1. Marker Characterization
3.2. Principal Coordinate Analysis (PCoA) of 26 Okra Accessions Genotyped Using 9 SSR Markers
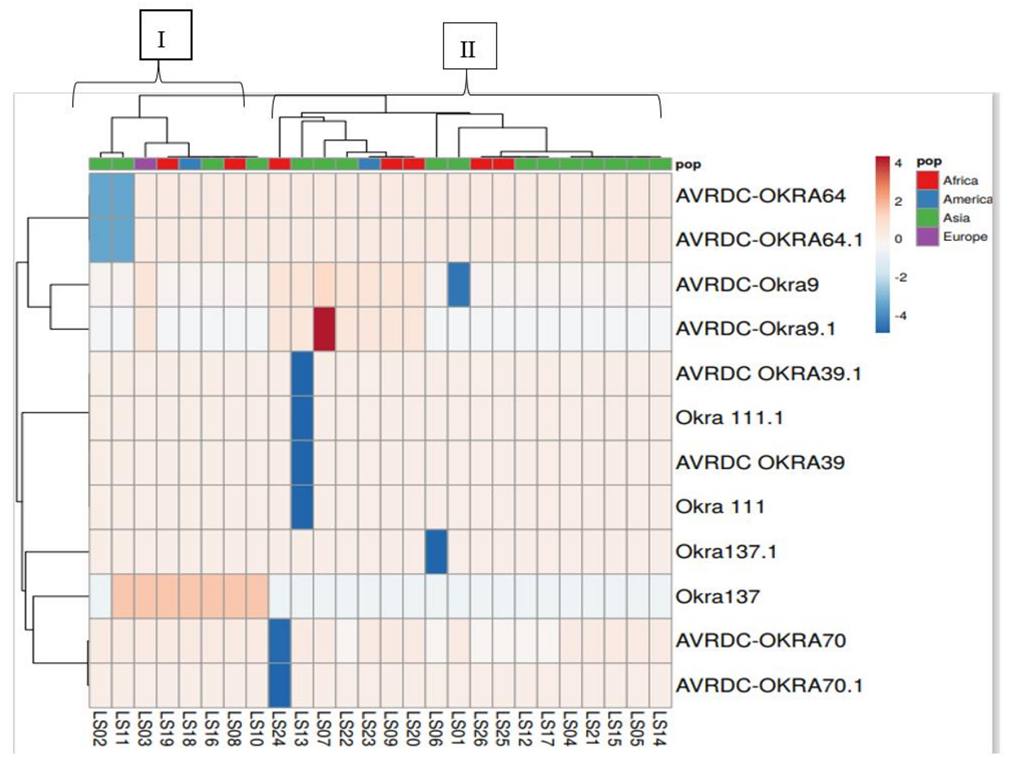
3.3. Heatmap Cluster
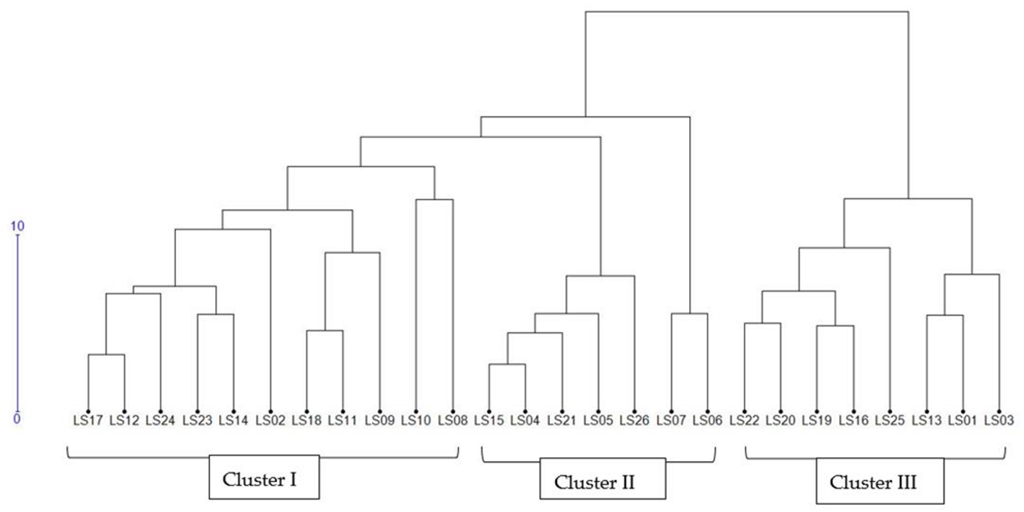
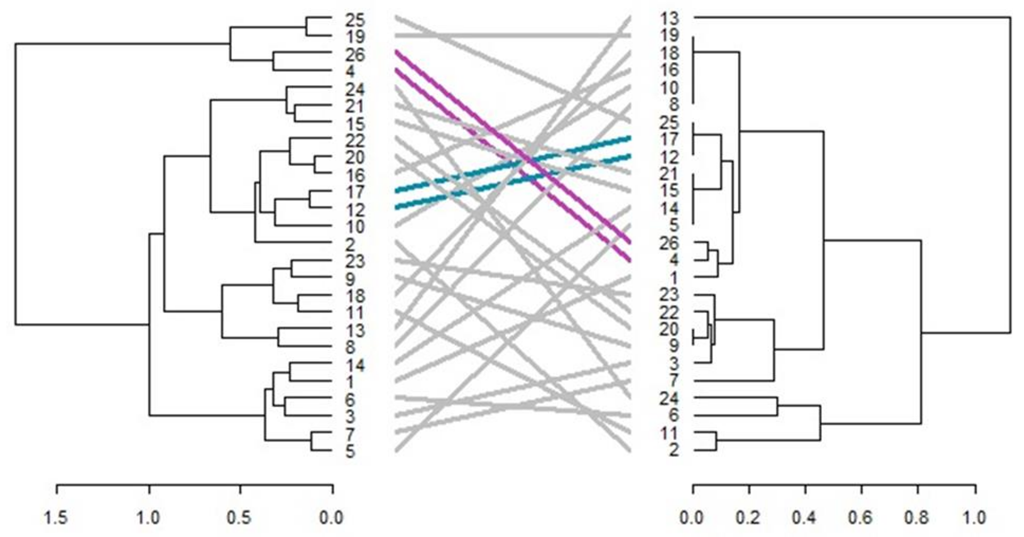
3.4. Cluster Analysis
3.5. Accession and Environmental Effects on Phenotypic Traits
3.5.1. Performance of Okra Accessions for Phenotypic Traits under Drought-Stressed and Non-Stressed Conditions
3.5.2. Associations among Phenotypic Traits under Drought-Stressed and Non-Stressed Conditions
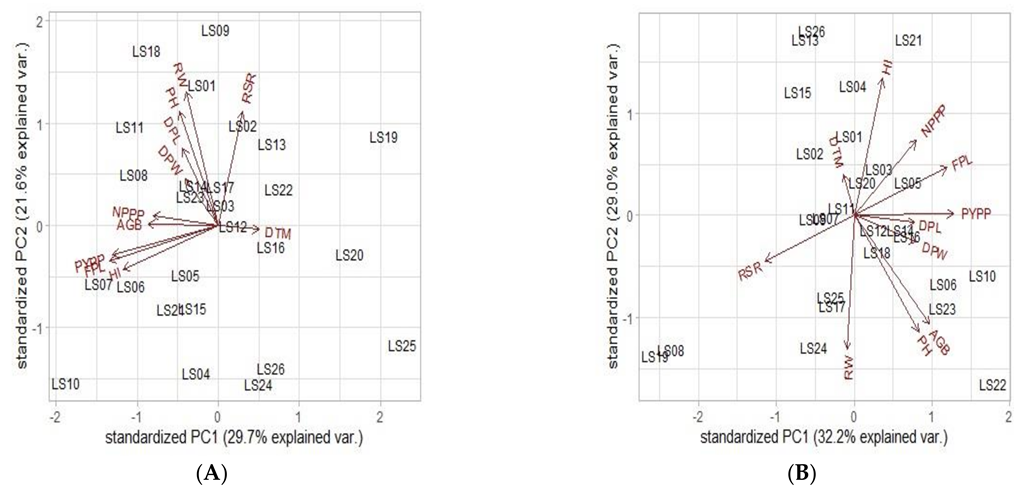
3.5.3. Principal Component Analysis (PCA)
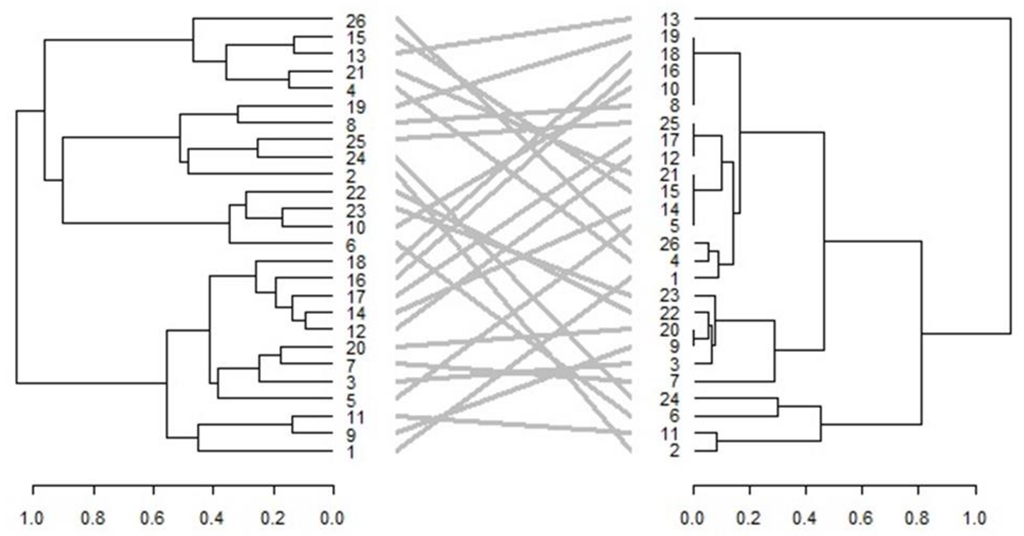
3.5.4. Phenotypic Hierarchical Clustering
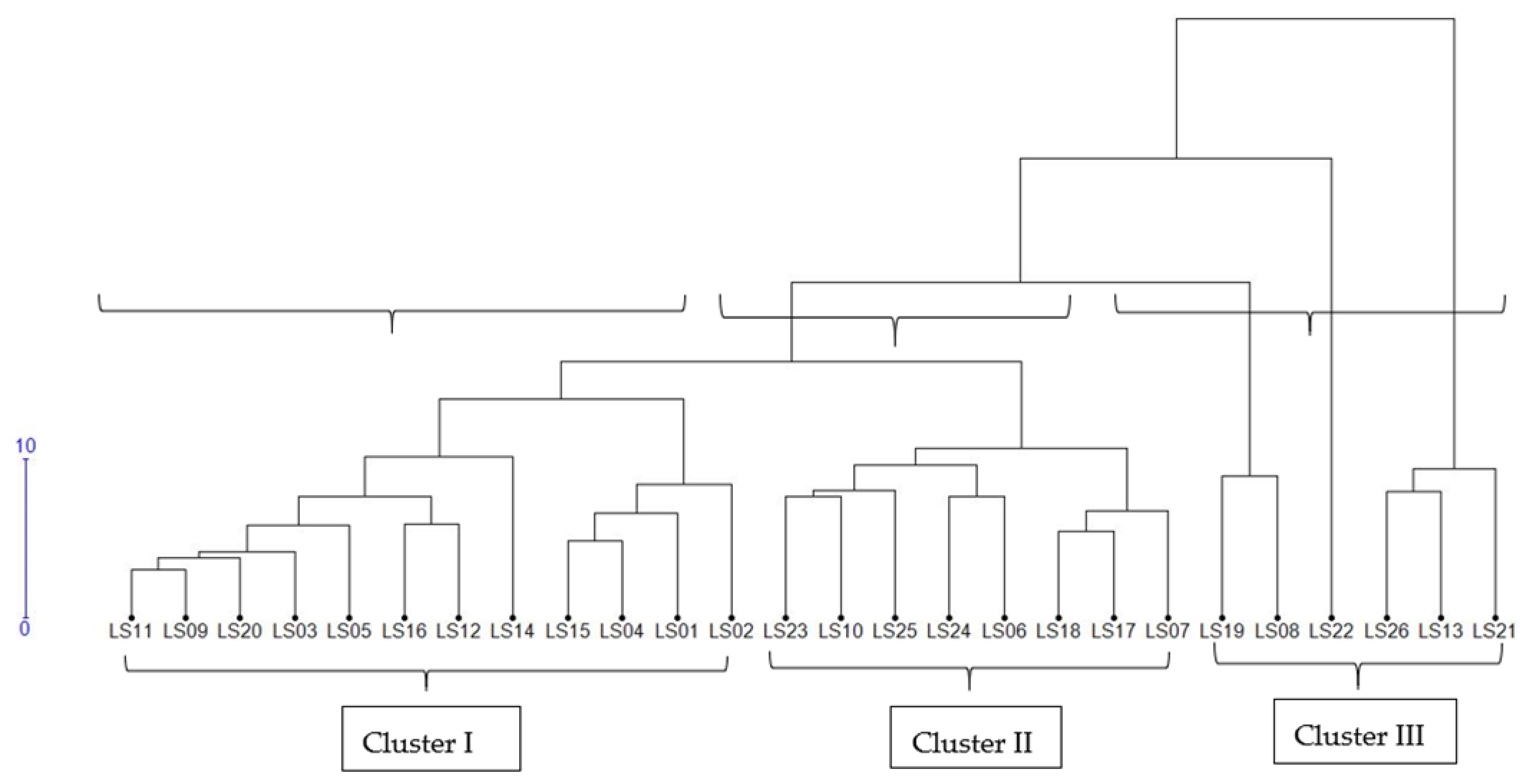
3.6. Comparison of Phenotypic and Genotypic Hierarchical Clusters
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Düzyaman, E. Phenotypic Diversity within a Collection of Distinct Okra (Abelmoschus esculentus) Cultivars Derived from Turkish Land Races. Genet. Resour. Crop Evol. 2005, 52, 1019–1030. [Google Scholar] [CrossRef]
- Kumar, M.; Sharma, V.R.; Kumar, N.; Sirohi, U.; Naresh, R.; Chaudhary, V. Screening of Microsatellite Markers for Genetic Diversity Assessment and Conservation of Germplasm in Okra (Abelmoschus esculentus L. Moench). Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 509–520. [Google Scholar] [CrossRef]
- Komolafe, R.J.; Ariyo, O.J.; Alake, C.O. Correlation and Path Coefficient Analysis of Fruit Yield Attributes in Forty Genotypes of Okra (Abelmoschus esculentus). Agric. Res. 2021, 11, 15–23. [Google Scholar] [CrossRef]
- Abed, M.Y.; Ibrahim, E.A.; El-Shoura, A.M. Development of Okra (Abelmoschus esculentus L. Moench) Hybrids Derived from Selected Inbreds under Drought Stress. J. Plant Prod. 2020, 11, 61–69. [Google Scholar] [CrossRef]
- Saifullah, M.; Rabbani, M.G. Evaluation, and characterization of okra (Abelmoschus esculentus L. Moench) genotypes. SAARC J. Agric. 2009, 7, 92–99. [Google Scholar]
- Mihretu, Y.; Wayessa, G.; Adugna, D. Multivariate analysis among okra (Abelmoschus esculentus (L.) Moench) collection in South-Western Ethiopia. J. Plant Sci. 2014, 9, 43–50. [Google Scholar]
- Reddy, M.T.; Haribabu, K.; Ganesh, M.; Redyy, K.C.; Begum, H. Genetic divergence analysis of indigenous and exotic collections of okra (Abelmoschus esculentus L. Moench). J. Agric. Technol. 2012, 8, 611–623. [Google Scholar]
- Alake, C.O. Genetic variability and diversity in okra landraces using agromorphological traits and seed elemental minerals. Int. J. Veg. Sci. 2020, 26, 127–149. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Surendran, U.; Gopinath, G.; Chandran, K.M.; Anjali, N.K.; Ct, M.F. Elucidation of stage specific physiological sensitivity of okra to drought stress through leaf gas exchange, spectral indices, growth and yield parameters. Agric. Water Manag. 2019, 222, 92–104. [Google Scholar] [CrossRef]
- Kyriakopoulou, O.G.; Arens, P.; Pelgrom, K.T.; Karapanos, I.; Bebeli, P.; Passam, H.C. Genetic and morphological diversity of okra (Abelmoschus esculentus [L.] Moench.) genotypes and their possible relationships, with particular reference to Greek landraces. Sci. Hortic. 2014, 171, 58–70. [Google Scholar] [CrossRef]
- Kaur, K.; Pathak, M.; Kaur, S.; Pathak, D.; Chawla, N. Assessment of morphological and molecular diversity among okra (Abelmoschus esculentus (L.) Moench.) germplasm. Afr. J. Biotechnol. 2013, 12, 3160–3170. [Google Scholar]
- Massucato, L.R.; Nakamura, K.K.; Ruas, P.M.; Zefa, D.M.; Da Silva, D.J.H.; Gonçalves, L.S.A. Genetic diversity among Brazilian okra landraces detected by morphoagronomic and molecular descriptors. Acta Sci. Agron. 2020, 42, e43426. [Google Scholar] [CrossRef]
- Aladele, S.E.; Ariyo, O.J.; Lapena, R.D.E. Genetic relationships among West African okra (Abelmoschus caillei) and Asian genotypes (Abelmoschus esculentus) using RAPD. Afr. J. Biotechnol. 2008, 7, 1426–1431. [Google Scholar]
- Yuan, C.; Zhang, C.; Wang, P.; Hu, S.; Chang, H.; Xiao, W.; Lu, X.; Jiang, S.; Ye, J.; Guo, X. Genetic diversity analysis of okra (Abelmoschus esculentus L.) by inter-simple sequence repeat (ISSR) markers. Genet. Mol. Res. 2014, 13, 3165–3175. [Google Scholar] [CrossRef] [PubMed]
- Kpodo, F.; Agbenorhevi, J.; Alba, K.; Bingham, R.; Oduro, I.; Morris, G.; Kontogiorgos, V. Pectin isolation and characterization from six okra genotypes. Food Hydrocoll. 2017, 72, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Salameh, N.M. Genetic diversity of okra (Abelmoschus esculentus L.) genotypes from different agroecological regions revealed by amplified fragment length polymorphism analysis. Am. J. Appl. Sci. 2014, 11, 1157–1163. [Google Scholar] [CrossRef]
- Fougat, R.S.; Purohit, A.R.; Kumar, S.; Parekh, M.J.; Kumar, M. SSR based genetic diversity in Abelmoschus species. Indian J. Agric. Sci. 2015, 85, 1223–1228. [Google Scholar]
- Mohammed, W.; Amelework, B.; Shimelis, H. Simple sequence repeat markers revealed genetic divergence and population structure of okra [Abelmoschus esculentus] collections of diverse geographic origin. Aust. J. Crop Sci. 2020, 14, 1032–1041. [Google Scholar] [CrossRef]
- El-Fattah, B.E.S.A.; Haridy, A.G.; Abbas, H.S. Response to planting date, stress tolerance and genetic diversity analysis among okra (Abelmoschus esculentus (L.) Moench.) varieties. Genet. Resour. Crop Evol. 2020, 67, 831–851. [Google Scholar] [CrossRef]
- Ahmed, Z.G.; El-Sayed, M.A. Influence of drought stress on physiological traits of crossed okra varieties. Jordan J. Biol. Sci. 2021, 14, 253–260. [Google Scholar]
- Schafleitner, R.; Kumar, S.; Lin, C.-Y.; Hegde, S.G.; Ebert, A. The okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 2013, 517, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Ouedraogo, M.H.; Sawadogo, N.; Batieno, T.B.J.; Zida, W.M.S.F.; Bougma, A.L.; Barro, A.; Kiebre, Z.; Sawadogo, M. Evaluation of genetic diversity of okra accessions [Abelmoschus esculentus (L. Moench)] cultivated in Burkina Faso using microsatellite markers. Afr. J. Biotechnol. 2018, 17, 126–132. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlex 6.5: Genetic analysis in Excel. Population software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Li, W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef]
- Nagy, S.; Poczai, P.; Cernák, I.; Gorji, A.M.; Hegedűs, G.; Taller, J. PICcalc: An Online Program to Calculate Polymorphic Information Content for Molecular Genetic Studies. Biochem. Genet. 2012, 50, 670–672. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. Dissimilarity Analysis and Representation for Windows. Available online: http://www.darwin.cirad.fr/darwin.html (accessed on 6 September 2022).
- Maechler, M.; Rousseeuw, P.; Struyf, A.; Hubert, M.; Hornik, K.; Studer, M. Package ‘Cluster’. Available online: https://cran.microsoft.com/snapshot/2014-10-10/web/packages/cluster/cluster2013.pdf (accessed on 12 November 2021).
- Gower, J.C. A General Coefficient of Similarity and Some of Its Properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Kumar, S.; Parekh, M.J.; Fougat, R.S.; Patel, S.K.; Patel, C.B.; Kumar, M.; Patel, B.R. Assessment of genetic diversity among okra genotypes using SSR markers. J. Plant Biochem. Biotechnol. 2017, 26, 172–178. [Google Scholar] [CrossRef]
- Petropoulos, S.; Fernandes, Â.; Barros, L.; Ferreira, I.C. Chemical composition, nutritional value and antioxidant properties of Mediterranean okra genotypes in relation to harvest stage. Food Chem. 2018, 242, 466–474. [Google Scholar] [CrossRef]
- Pradip, K.; De Akotkar, D.K.; Pal, A.K. Genetic variability and diversity in okra [Abelmoschus esculentus (L) Moench]. Electron. J. Plant Breed. 2010, 1, 393–398. [Google Scholar]
- Ravishankar, K.V.; Muthaiah, G.; Mottaiyan, P.; Gundale, S.-K. Identification of novel microsatellite markers in okra (Abelmoschus esculentus (L.) Moench) through next-generation sequencing and their utilization in analysis of genetic relatedness studies and cross-species transferability. J. Genet. 2018, 97, 39–47. [Google Scholar] [CrossRef]
- Eshiet, A.J.; Brisibe, E.A. Morphological Characterization and Yield Traits Analysis in Some Selected Varieties of Okra (Abelmoschus esculentus L. Moench). Adv. Crop Sci. Technol. 2015, 3, 197. [Google Scholar] [CrossRef]
- Adejumo, S.A.; Ezeh, O.S.; Mur, L.A. Okra growth and drought tolerance when exposed to water regimes at different growth stages. Int. J. Veg. Sci. 2018, 25, 226–258. [Google Scholar] [CrossRef]
- Mkhabela, S.S.; Shimelis, H.; Gerrano, A.S.; Mashilo, J. Phenotypic and genotypic divergence in Okra [Abelmoschus esculentus (L.) Moench] and implications for drought tolerance breeding: A review. South Afr. J. Bot. 2021, 145, 56–64. [Google Scholar] [CrossRef]
- Rad, S.K.; Madani, H.; Sharifabadi, H.H.; Mahmoudi, M.; Nourmohamadi, G. Effects of different irrigation intervals and sowing time on yield attributing traits of okra (Abelmoschus esculentus L.). Arab. J. Geosci. 2022, 15, 740. [Google Scholar] [CrossRef]


| Accession Code | Accession Number | Geographical Origin |
|---|---|---|
| LS01 | VI033775 | Malaysia |
| LS02 | VI033797 | Malaysia |
| LS03 | VI056457 | Yugoslavia |
| LS04 | VI039651 | Bangladesh |
| LS05 | VI046561 | Thailand |
| LS06 | VI047672 | Bangladesh |
| LS07 | VI050150 | Taiwan |
| LS08 | VI050957 | Zambia |
| LS09 | VI050960 | Zambia |
| LS10 | VI055110 | Malaysia |
| LS11 | VI055119 | Myanmar |
| LS12 | VI055219 | Malaysia |
| LS13 | VI055220 | Malaysia |
| LS14 | VI055421 | Viet Nam |
| LS15 | VI056069 | Cambodia |
| LS16 | VI056079 | Cambodia |
| LS17 | VI056081 | Cambodia |
| LS18 | VI056449 | United States of America |
| LS19 | VI060131 | Mali |
| LS20 | VI060313 | Tanzania |
| LS21 | VI060679 | India |
| LS22 | VI060803 | Turkey |
| LS23 | VI060817 | Brazil |
| LS24 | VI060822 | Nigeria |
| LS25 | VI060823 | Nigeria |
| LS26 | Clemson Spineless | South Africa |
| Marker Name | Forward Primer Sequence | Reverse Primer Sequence | PIC |
|---|---|---|---|
| Okra 111 | GATGGAATTGAGAAACCAGA | TGTGTTCTTCACTCTCGTCA | 0.89 |
| Okra 152 | GCTCTATTGATGGCGAGTAA | AAAGTCATCCAAGGTGACAA | 0.81 |
| Okra 166 | TTCCAGTTGGAGAGGTAAGA | CTTCCATTTCATCGACTTTC | 0.82 |
| AVRDC-Okra17 | ACGAGAGTGAAGTGGAACTG | CTCCTCTTTCCTTTTTCCAT | 0.81 |
| AVRDC-Okra70 | GTAGCTGAACCCTTTGCTTA | CTATCATGGCGGATTCTTTA | 0.98 |
| AVRDC-Okra39 | TGAGGTGATGATGTGAGAGA | TTGTAGATGAGGTTTGAACG | 0.99 |
| AVRDC-Okra64 | AAGGAGGAGAAAGAGAAGGA | ATTTACTTGAGCAGCAGCAG | 0.87 |
| AVRDC-Okra9 | ACCTTGAACACCAGGTACAG | TTGCTCTTATGAAGCAGTGA | 0.85 |
| AVRDC-Okra57 | CGAGGAGACCATGGAAGAAG | ATGAGGAGGACGAGCAAGAA | 0.78 |
| Okra137 | GAGAGAGATTGCTTCGACTG | TAAACTTTAAACTCAGCGGC | 0.80 |
| Marker | Genetic Parameters | ||||||
|---|---|---|---|---|---|---|---|
| Na | Ne | I | Ho | He | FIS | PIC | |
| AVRDC-Okra70 | 3 | 2.47 | 0.97 | 1.00 | 0.60 | −0.68 | 0.60 |
| AVRDC-Okra64 | 2 | 2.00 | 0.69 | 1.00 | 0.50 | −1.00 | 0.50 |
| Okra 152 | 2 | 2.00 | 0.69 | 1.00 | 0.50 | −1.00 | 0.50 |
| Okra 166 | 2 | 2.00 | 0.69 | 1.00 | 0.50 | −1.00 | 0.50 |
| AVRDC-Okra9 | 5 | 2.76 | 1.16 | 1.00 | 0.64 | −0.57 | 0.64 |
| AVRDC-Okra39 | 3 | 2.31 | 0.91 | 1.00 | 0.57 | −0.76 | 0.57 |
| Okra 111 | 2 | 2.00 | 0.69 | 1.00 | 0.50 | −1.00 | 0.50 |
| Okra137 | 3 | 2.58 | 1.01 | 1.00 | 0.61 | −0.63 | 0.61 |
| AVRDC-Okra57 | 2 | 2.00 | 0.69 | 1.00 | 0.50 | −1.00 | 0.50 |
| Average | 2.70 | 2.24 | 0.83 | 1.00 | 0.54 | −0.85 | 0.55 |
| Standard deviation | 1.00 | 0.30 | 0.18 | 0.00 | 0.06 | 0.19 | 0.06 |
| Standard error | 0.34 | 0.15 | 0.10 | 0.10 | 0.06 | 0.06 | 0.02 |
| S.O.V. | df | PH | DTM | FPL | DPL | DPW | NPPP | PYPP | AGB | HI | RW | RSR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Replications | 1 | 23.80 ns | 11.44 ns | 21.61 * | 35.41 ns | 81.39 * | 0.01 ns | 55.20 * | 1423.50 ** | 1153 ns | 1.50 ns | 1.09 ** |
| Incomplete blocks | 1 | 2063.50 ** | 0.08 ns | 9.99 ns | 11.22 ns | 13.89 ns | 3.47 ns | 1.11 ns | 104.56 ns | 790.10 ns | 85.87 * | 0.09 ns |
| Genotype (G) | 26 | 336.40 * | 225.57 * | 15.15 ** | 13.76 ns | 7.54 ns | 7.92 * | 15.97 * | 136.00 * | 664.10 ns | 17.97 * | 0.14 ns |
| Water regime (WC) | 1 | 2231.00 ** | 75.84 ns | 77.13 ** | 13.18 ns | 10.93 ns | 16.56 * | 229.47 ** | 578.52 ns | 4736.10 * | 82.41 * | 0.04 ns |
| G × WC | 25 | 234.60 ns | 89.58 * | 6.82 * | 10.15 ns | 6.99 ns | 4.43 ns | 12.01 * | 55.27 ns | 714.90 ns | 8.91 ns | 0.07 ns |
| Residual | 49 | 139.80 | 48.05 | 3.96 | 11.88 | 8.26 | 3.98 | 6.96 | 75.76 | 429.40 | 10.19 | 0.09 |
| Accession Code | PH (cm) | DTM | FPL (cm) | DPL (cm) | DPW (g Per Plant) | NPPP | PYPP (g Per Plant) | AGB (g Per Plant) | HI (%) | RW (g Per Plant) | RSR | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | DS | NS | |
| LS01 | 71.50 | 64.25 | 80.25 | 95.17 | 3.92 | 7.96 | 11.59 | 7.78 | 6.50 | 2.00 | 3.50 | 8.00 | 3.92 | 7.02 | 13.92 | 13.02 | 17.15 | 54.91 | 5.25 | 7.75 | 0.58 | 0.59 |
| LS02 | 65.37 | 61.62 | 101.00 | 95.67 | 5.19 | 7.13 | 9.13 | 1.75 | 4.00 | 0.00 | 3.50 | 3.50 | 2.58 | 7.83 | 7.08 | 17.83 | 36.67 | 44.47 | 6.00 | 5.25 | 0.83 | 0.30 |
| LS03 | 60.50 | 68.38 | 83.00 | 83.50 | 3.03 | 5.96 | 5.15 | 6.83 | 3.50 | 3.50 | 8.00 | 7.00 | 2.50 | 8.79 | 17.50 | 17.29 | 13.96 | 51.86 | 5.25 | 6.25 | 0.33 | 0.37 |
| LS04 | 49.87 | 52.12 | 86.25 | 86.75 | 6.21 | 7.50 | 8.29 | 8.54 | 1.00 | 2.50 | 6.00 | 5.50 | 4.19 | 6.09 | 9.19 | 12.09 | 47.52 | 56.69 | 0.75 | 3.00 | 0.09 | 0.29 |
| LS05 | 54.50 | 72.50 | 89.00 | 80.25 | 5.73 | 7.25 | 6.50 | 7.08 | 2.00 | 8.00 | 4.00 | 5.50 | 6.17 | 7.33 | 16.17 | 18.33 | 38.30 | 44.78 | 5.25 | 3.75 | 0.32 | 0.27 |
| LS06 | 61.50 | 82.88 | 77.00 | 77.00 | 8.23 | 9.06 | 4.50 | 8.85 | 2.50 | 5.50 | 4.00 | 5.00 | 5.05 | 8.00 | 16.55 | 29.50 | 62.42 | 30.64 | 4.00 | 7.50 | 0.50 | 0.25 |
| LS07 | 58.38 | 70.38 | 86.25 | 89.00 | 8.58 | 4.96 | 7.60 | 9.00 | 3.00 | 6.50 | 4.00 | 3.50 | 7.92 | 2.92 | 16.92 | 14.92 | 65.16 | 29.01 | 5.00 | 4.50 | 0.34 | 0.37 |
| LS08 | 73.50 | 64.00 | 77.00 | 95.00 | 6.00 | 1.50 | 3.50 | 4.67 | 1.50 | 1.00 | 4.50 | 2.50 | 6.58 | 2.63 | 21.58 | 17.12 | 41.57 | 8.97 | 8.75 | 12.25 | 0.49 | 0.96 |
| LS09 | 82.25 | 69.75 | 89.67 | 83.50 | 3.80 | 7.83 | 7.50 | 4.21 | 1.00 | 1.00 | 3.00 | 3.00 | 4.60 | 7.17 | 17.10 | 16.67 | 21.83 | 48.07 | 10.50 | 7.75 | 0.85 | 0.50 |
| LS10 | 68.75 | 88.25 | 95.17 | 90.00 | 10.17 | 11.50 | 0.00 | 8.38 | 0.00 | 1.50 | 3.00 | 3.50 | 14.00 | 13.25 | 32.00 | 37.75 | 43.75 | 35.80 | 4.00 | 7.25 | 0.13 | 0.20 |
| LS11 | 79.00 | 66.50 | 95.50 | 86.00 | 6.51 | 9.75 | 8.33 | 3.63 | 3.00 | 0.00 | 6.00 | 4.00 | 6.76 | 9.23 | 16.76 | 19.23 | 40.34 | 47.33 | 7.75 | 9.00 | 0.46 | 0.47 |
| LS12 | 65.25 | 82.38 | 101.00 | 83.17 | 3.99 | 5.51 | 3.75 | 5.08 | 2.50 | 4.00 | 6.50 | 6.50 | 2.85 | 7.68 | 19.85 | 21.68 | 19.12 | 46.84 | 5.00 | 7.25 | 0.33 | 0.40 |
| LS13 | 74.25 | 49.75 | 80.25 | 98.25 | 2.50 | 7.46 | 5.71 | 6.19 | 2.50 | 0.50 | 1.00 | 5.50 | 2.00 | 6.13 | 17.00 | 9.13 | 10.98 | 79.55 | 6.50 | 4.00 | 0.40 | 0.48 |
| LS14 | 65.50 | 87.25 | 86.42 | 83.17 | 5.93 | 8.09 | 6.44 | 7.38 | 0.50 | 3.00 | 4.50 | 6.00 | 4.48 | 8.56 | 18.97 | 23.06 | 23.56 | 60.68 | 8.00 | 8.25 | 0.42 | 0.49 |
| LS15 | 52.50 | 57.88 | 89.00 | 92.42 | 6.71 | 5.47 | 6.40 | 6.40 | 1.50 | 1.50 | 5.00 | 6.00 | 4.71 | 4.82 | 10.71 | 8.32 | 47.71 | 57.66 | 3.50 | 3.75 | 0.40 | 0.45 |
| LS16 | 53.50 | 78.12 | 95.17 | 92.25 | 3.95 | 7.75 | 7.40 | 6.92 | 3.50 | 3.00 | 2.50 | 4.50 | 2.63 | 11.55 | 13.62 | 24.55 | 18.99 | 47.05 | 4.25 | 8.75 | 0.29 | 0.36 |
| LS17 | 67.00 | 79.00 | 98.00 | 83.50 | 4.23 | 3.88 | 4.75 | 8.08 | 1.00 | 5.00 | 5.50 | 3.50 | 3.69 | 6.00 | 21.19 | 20.00 | 23.54 | 30.08 | 7.00 | 9.25 | 0.49 | 0.46 |
| LS18 | 83.62 | 75.00 | 98.25 | 86.75 | 7.21 | 7.33 | 7.75 | 10.38 | 1.50 | 1.50 | 6.00 | 3.50 | 5.42 | 6.10 | 16.42 | 25.10 | 33.94 | 24.68 | 10.75 | 6.25 | 0.74 | 0.25 |
| LS19 | 54.12 | 72.00 | 95.50 | 83.50 | 1.00 | 0.00 | 4.00 | 5.25 | 0.50 | 0.00 | 1.00 | 1.00 | 0.50 | 0.00 | 6.50 | 12.50 | 16.67 | 0.00 | 5.50 | 9.25 | 1.43 | 0.73 |
| LS20 | 60.25 | 67.12 | 95.16 | 89.67 | 1.81 | 8.46 | 1.88 | 7.63 | 0.00 | 2.50 | 2.50 | 4.00 | 0.75 | 8.08 | 11.25 | 18.08 | 7.89 | 48.95 | 4.50 | 6.25 | 0.45 | 0.42 |
| LS21 | 54.12 | 66.25 | 92.17 | 92.25 | 8.50 | 10.08 | 7.58 | 7.58 | 4.00 | 1.50 | 6.00 | 6.00 | 4.17 | 9.58 | 10.17 | 12.08 | 41.96 | 84.21 | 2.00 | 2.75 | 0.27 | 0.24 |
| LS22 | 63.50 | 119.25 | 89.67 | 92.75 | 5.52 | 8.04 | 5.83 | 7.63 | 0.00 | 6.00 | 3.00 | 6.00 | 1.75 | 11.44 | 10.75 | 37.94 | 12.96 | 30.57 | 6.25 | 11.75 | 0.62 | 0.31 |
| LS23 | 69.75 | 86.75 | 92.17 | 89.50 | 4.94 | 7.29 | 6.13 | 8.79 | 2.50 | 5.00 | 2.50 | 7.00 | 5.88 | 8.00 | 20.87 | 34.50 | 30.75 | 23.72 | 6.50 | 10.00 | 0.35 | 0.28 |
| LS24 | 59.12 | 86.25 | 95.17 | 77.00 | 4.83 | 5.31 | 1.50 | 3.38 | 0.00 | 0.50 | 3.00 | 2.50 | 4.17 | 6.88 | 17.17 | 25.87 | 27.86 | 19.23 | 1.00 | 9.50 | 0.11 | 0.42 |
| LS25 | 59.62 | 83.62 | 101.00 | 90.00 | 0.00 | 6.92 | 0.00 | 3.50 | 0.00 | 0.00 | 1.00 | 3.50 | 0.00 | 7.04 | 12.50 | 26.54 | 0.00 | 35.18 | 1.50 | 10.75 | 0.04 | 0.41 |
| LS26 | 46.12 | 52.62 | 86.75 | 83.50 | 4.67 | 5.63 | 5.45 | 3.05 | 2.00 | 1.50 | 2.00 | 6.00 | 4.00 | 5.24 | 6.00 | 7.24 | 33.33 | 82.28 | 1.75 | 2.50 | 0.15 | 0.22 |
| Mean | 63.59 | 73.23 | 90.61 | 87.67 | 5.12 | 6.83 | 5.64 | 6.46 | 1.92 | 2.58 | 3.90 | 4.71 | 4.28 | 7.21 | 15.30 | 20.01 | 29.92 | 43.20 | 5.25 | 7.10 | 0.44 | 0.40 |
| p-value | ns | ** | * | ns | * | * | ns | ns | * | * | * | ns | * | ns | * | * | ns | * | ns | ns | * | ns |
| SED | 11.73 | 12.84 | 6.29 | 7.49 | 1.73 | 2.13 | 3.19 | 3.77 | 2.41 | 3.25 | 1.85 | 2.17 | 2.29 | 2.95 | 7.63 | 9.81 | 21.16 | 20.58 | 3.10 | 3.29 | 0.37 | 0.17 |
| LSD (5%) | 34.24 | 26.44 | 12.96 | 15.97 | 5.05 | 4.39 | 9.32 | 7.77 | 7.05 | 6.69 | 5.41 | 4.48 | 6.68 | 6.08 | 22.26 | 20.21 | 43.67 | 42.38 | 9.04 | 6.77 | 1.09 | 0.35 |
| CV (%) | 18.48 | 17.53 | 6.94 | 8.54 | 34.77 | 31.22 | 55.83 | 58.42 | 65.12 | 56.10 | 47.68 | 46.12 | 56.79 | 40.93 | 50.20 | 49.02 | 69.23 | 47.63 | 58.87 | 46.35 | 63.74 | 42.66 |
| Traits | PH | DTM | FPL | DPL | DPW | NPPP | PYPP | AGB | HI | RW | RSR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PH | 0.01 ns | 0.06 ns | 0.14 ns | 0.07 ns | 0.10 ns | 0.24 ns | 0.52 ** | −0.19 ns | 0.81 ** | 0.32 ns | |
| DTM | −0.22 ns | −0.20 ns | −0.24 ns | −0.27 ns | −0.05 ns | 0.52 ** | −0.06 ns | −0.29 ns | −0.05 ns | 0.07 ns | |
| FPL | 0.16 ns | 0.12 ns | 0.21 ns | 0.12 ns | 0.43 * | 0.81 ** | 0.36 ns | 0.85 ** | 0.03 ns | −0.23 ns | |
| DPL | 0.24 ns | 0.03 ns | 0.26 ns | 0.71 ** | 0.30 ns | −0.03 ns | −0.35 ns | 0.25 ns | 0.29 ns | 0.28 ns | |
| DPW | 0.41 * | −0.29 ns | 0.06 ns | 0.58 ** | 0.27 ns | 0.04 ns | −0.14 ns | 0.17 ns | 0.03 ns | 0.05 ns | |
| NPPP | −0.03 ns | 0.15 ns | 0.39 ** | 0.24 ns | 0.38 ns | 0.26 ns | 0.19 ns | 0.34 ns | 0.14 ns | −0.15 ns | |
| PYPP | 0.46 ** | 0.08 ns | 0.83 ** | 0.16 ns | 0.15 ns | 0.37 ns | 0.69 ** | 0.67 ** | 0.13 ns | −0.27 ns | |
| AGB | 0.88 ** | −0.14 ns | 0.36 ns | 0.27 ns | 0.30 ns | −0.06 ns | 0.60 ** | 0.13 ns | 0.34 ns | 0.27 ns | |
| HI | −0.47 * | 0.22 ns | 0.50 ** | −0.07 ns | −0.11 ns | 0.61 ** | 0.31 ns | −0.48 * | −0.09 ns | −0.13 ns | |
| RW | 0.70 ** | −0.02 ns | −0.24 ns | −0.12 ns | −0.05 ns | −0.32 ns | −0.08 ns | 0.62 ** | −0.69 ** | 0.58 ** | |
| RSR | −0.18 ns | 0.20 ns | −0.65 ** | −0.29 ns | −0.33 ns | −0.35 ns | −0.57 ** | −0.31 ns | −0.37 ns | 0.49 * |
| Traits | Drought-Stressed | Non-Stressed | |||||
|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC1 | PC2 | PC3 | PC4 | |
| PH | 0.32 | 0.74 | 0.46 | 0.55 | 0.76 | 0.05 | 0.01 |
| DTM | −0.33 | −0.03 | 0.32 | −0.09 | −0.27 | 0.48 | 0.74 |
| FPL | 0.89 | −0.23 | −0.07 | 0.79 | −0.31 | 0.37 | −0.09 |
| DPL | 0.29 | 0.50 | −0.73 | 0.51 | 0.05 | −0.53 | 0.41 |
| DPW | 0.28 | 0.30 | −0.69 | 0.53 | 0.17 | −0.71 | 0.17 |
| NPPP | 0.53 | 0.07 | −0.22 | 0.53 | −0.49 | −0.15 | 0.37 |
| PYPP | 0.87 | −0.20 | 0.30 | 0.85 | −0.01 | 0.46 | −0.05 |
| AGB | 0.58 | 0.01 | 0.70 | 0.64 | 0.71 | 0.19 | −0.02 |
| HI | 0.78 | −0.29 | −0.27 | 0.24 | −0.90 | 0.16 | −0.01 |
| RW | 0.26 | 0.87 | 0.30 | −0.06 | 0.88 | 0.31 | 0.21 |
| RSR | −0.20 | 0.74 | −0.07 | −0.76 | 0.30 | 0.10 | 0.39 |
| Explained variance (eigenvalue) | 3.27 | 2.38 | 2.13 | 3.55 | 3.19 | 1.56 | 1.09 |
| Proportion of total variance (%) | 29.69 | 21.64 | 19.37 | 32.24 | 28.99 | 14.22 | 9.89 |
| Cumulative variance (%) | 29.69 | 51.33 | 70.70 | 32.24 | 61.23 | 75.45 | 85.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mkhabela, S.S.; Shimelis, H.; Gerrano, A.S.; Mashilo, J.; Shayanowako, A. Characterization of Okra (Abelmoschus esculentus L.) Accessions with Variable Drought Tolerance through Simple Sequence Repeat Markers and Phenotypic Traits. Diversity 2022, 14, 747. https://doi.org/10.3390/d14090747
Mkhabela SS, Shimelis H, Gerrano AS, Mashilo J, Shayanowako A. Characterization of Okra (Abelmoschus esculentus L.) Accessions with Variable Drought Tolerance through Simple Sequence Repeat Markers and Phenotypic Traits. Diversity. 2022; 14(9):747. https://doi.org/10.3390/d14090747
Chicago/Turabian StyleMkhabela, Sonto Silindile, Hussein Shimelis, Abe Shegro Gerrano, Jacob Mashilo, and Admire Shayanowako. 2022. "Characterization of Okra (Abelmoschus esculentus L.) Accessions with Variable Drought Tolerance through Simple Sequence Repeat Markers and Phenotypic Traits" Diversity 14, no. 9: 747. https://doi.org/10.3390/d14090747
APA StyleMkhabela, S. S., Shimelis, H., Gerrano, A. S., Mashilo, J., & Shayanowako, A. (2022). Characterization of Okra (Abelmoschus esculentus L.) Accessions with Variable Drought Tolerance through Simple Sequence Repeat Markers and Phenotypic Traits. Diversity, 14(9), 747. https://doi.org/10.3390/d14090747









