Genetic and Population Structure of Croatian Local Donkey Breeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Pedigree Analysis
2.2. Microsatellite Analysis
2.3. Mitochondrial DNA Analysis
3. Results
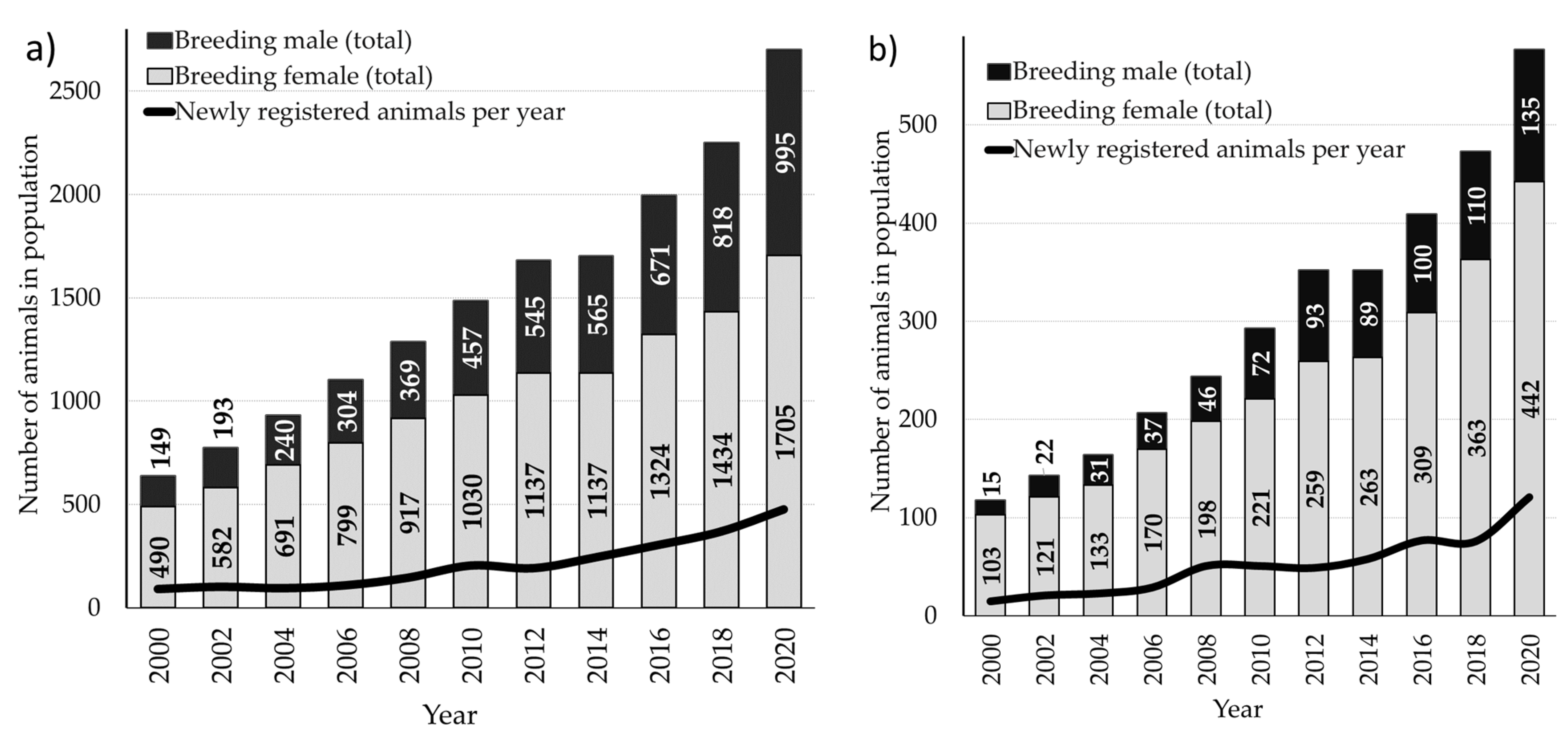
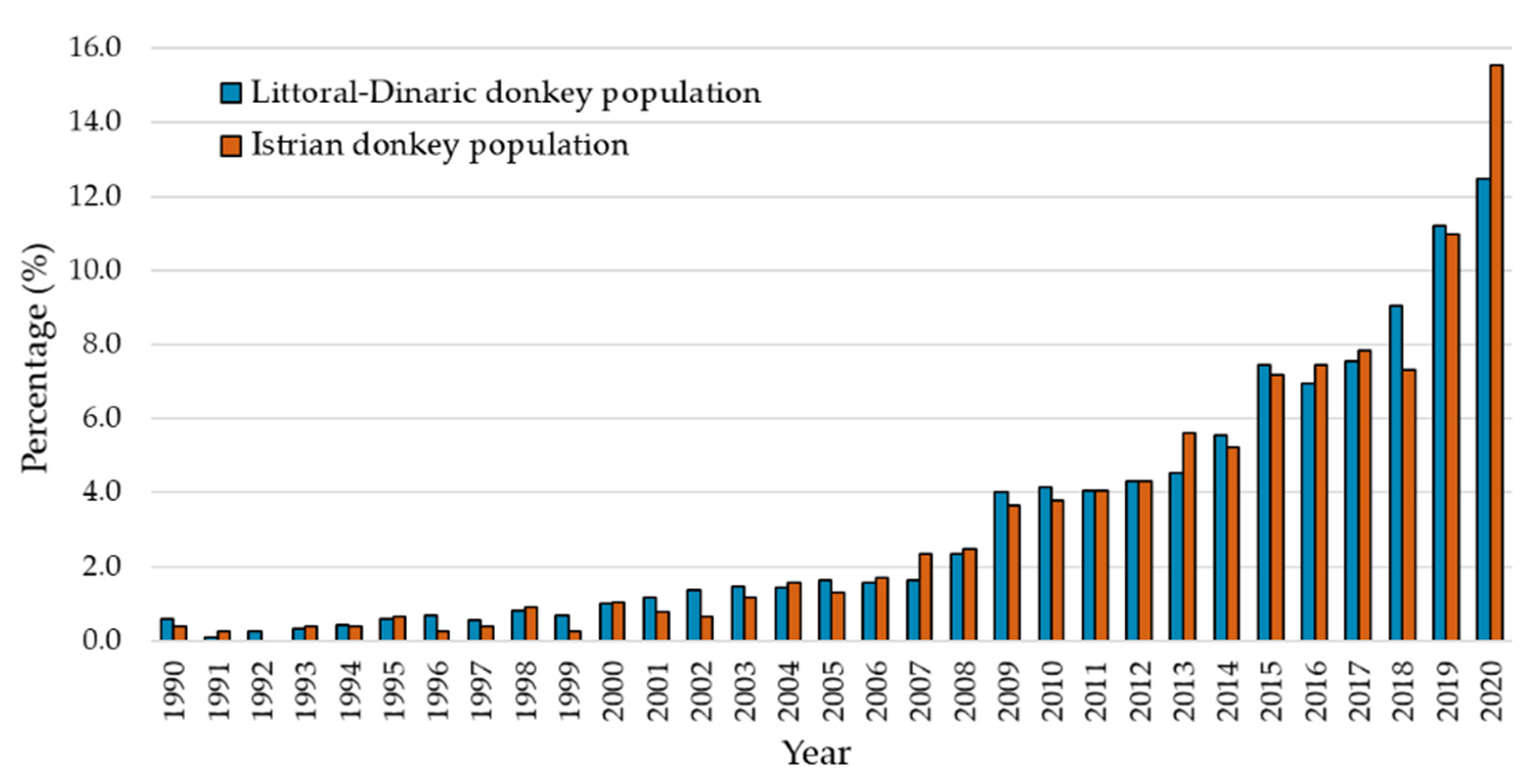
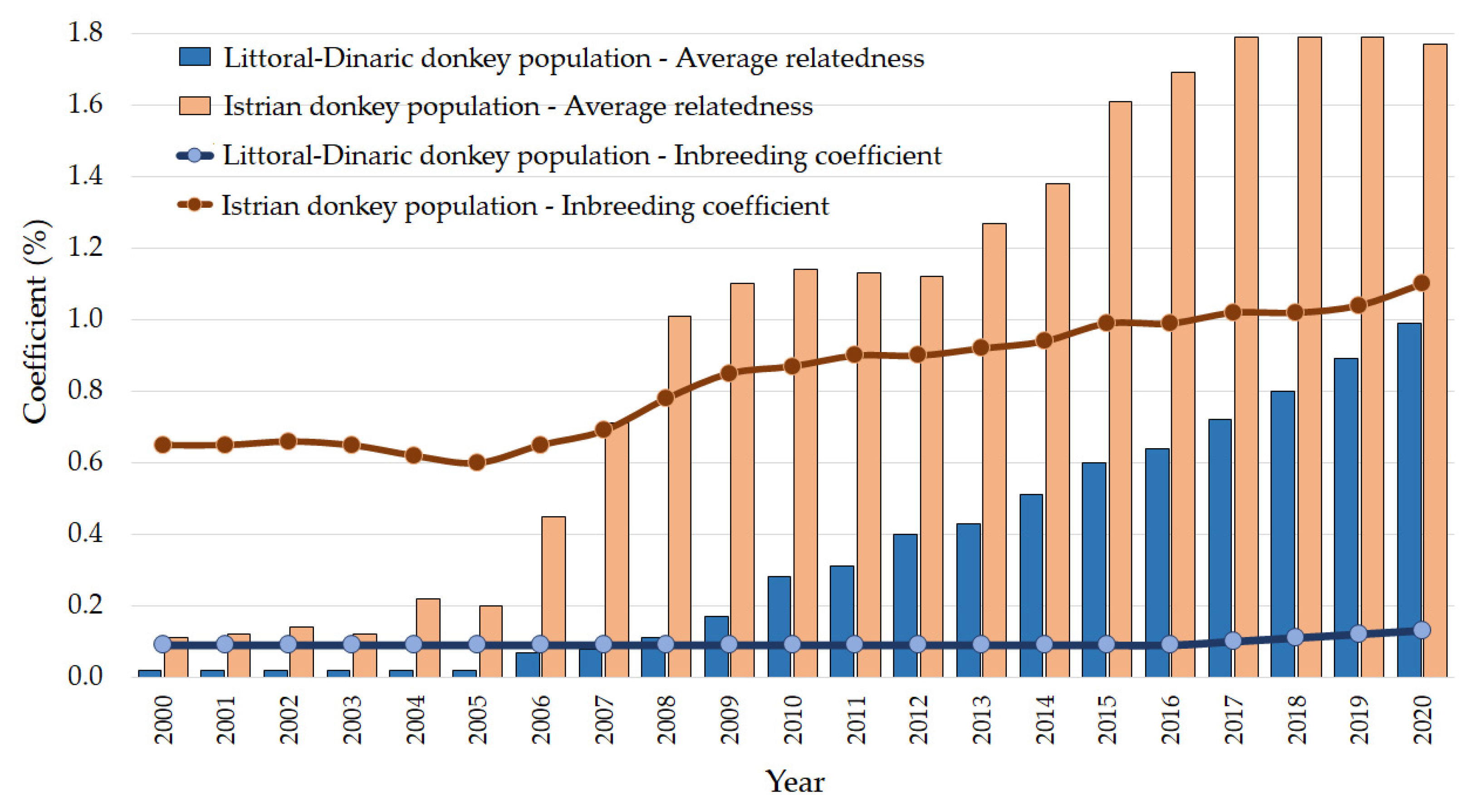
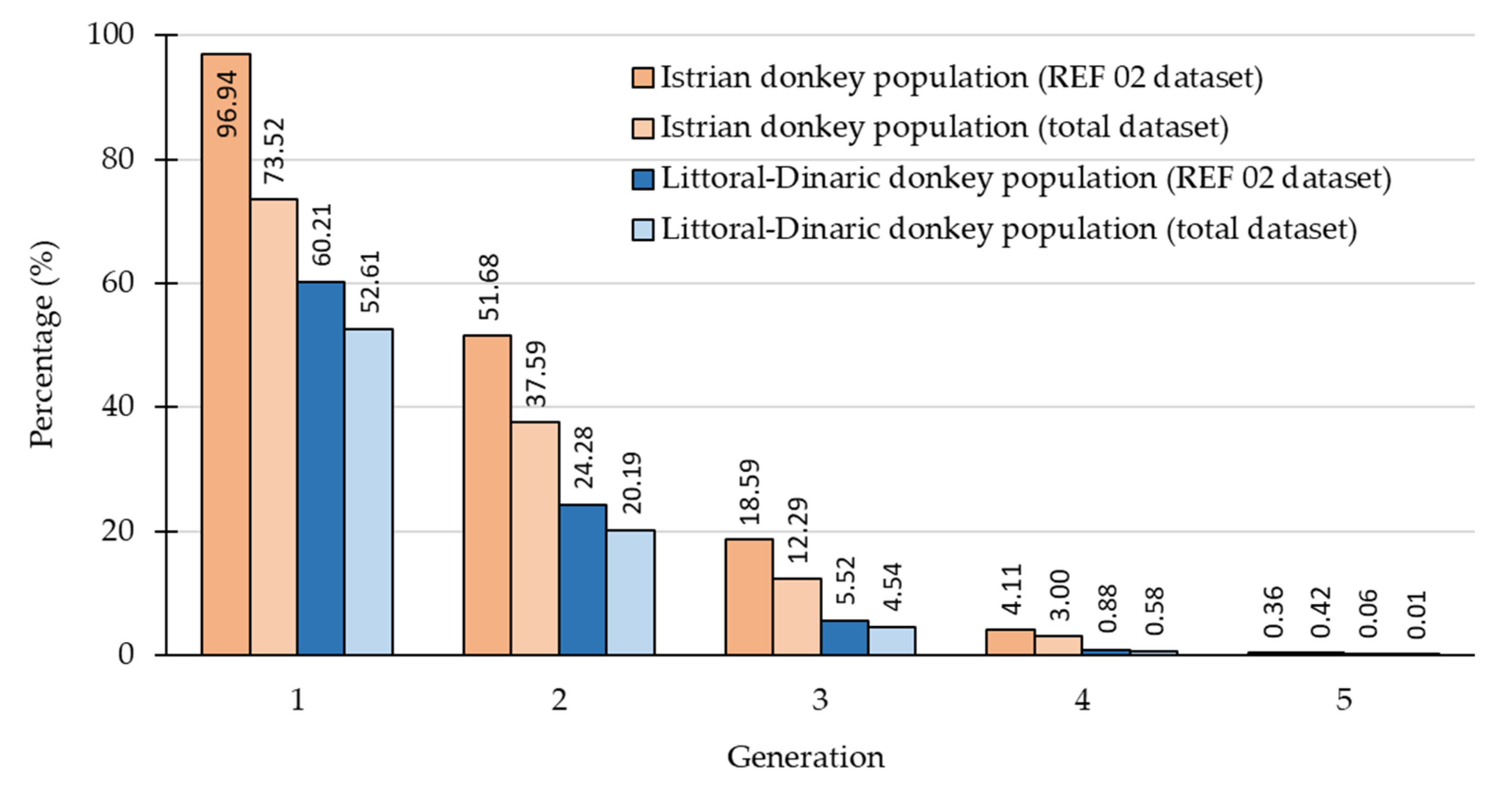
3.1. Pedigree
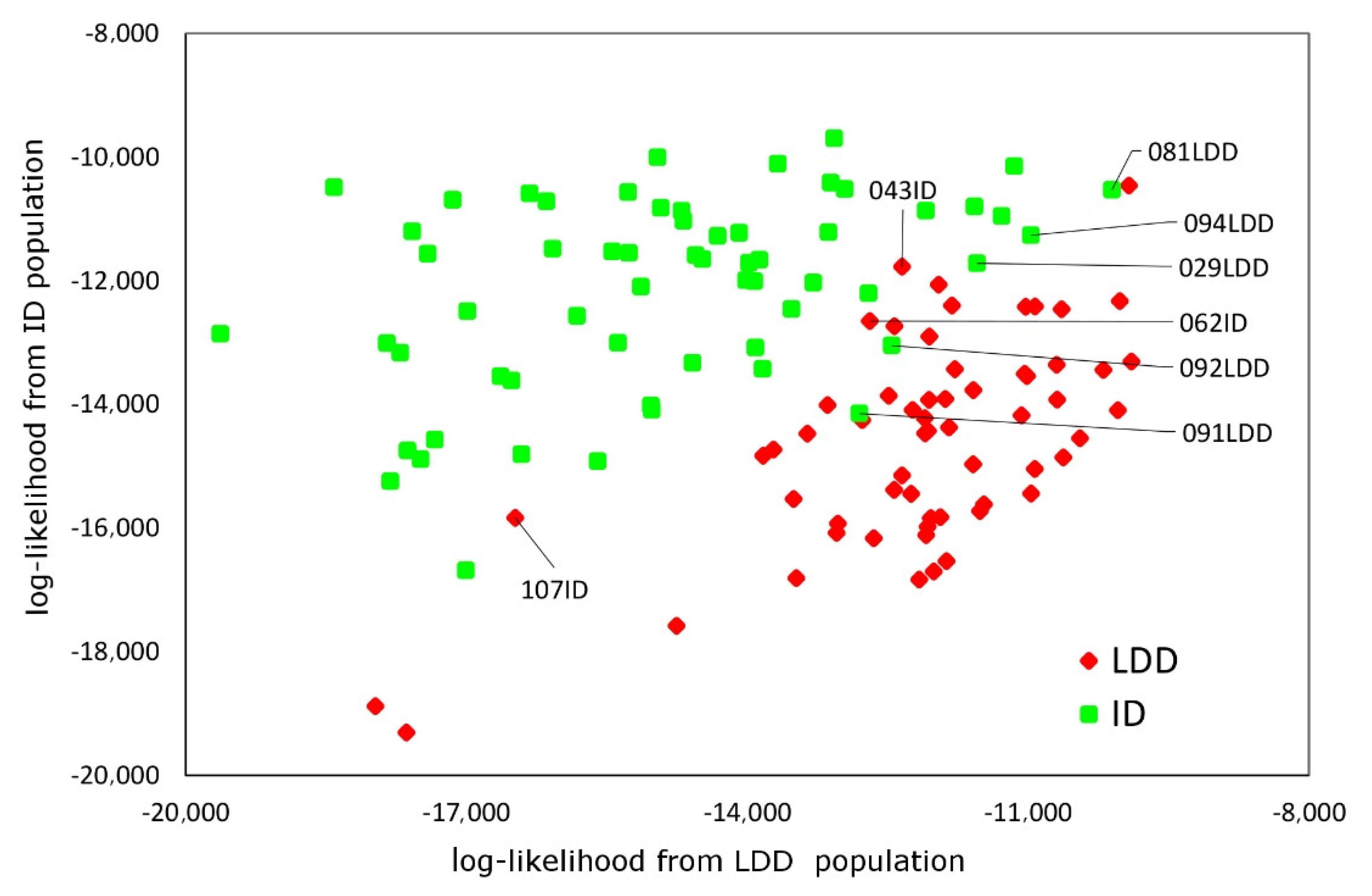
3.2. Microsatellite Variability and Genetic Diversity Indices
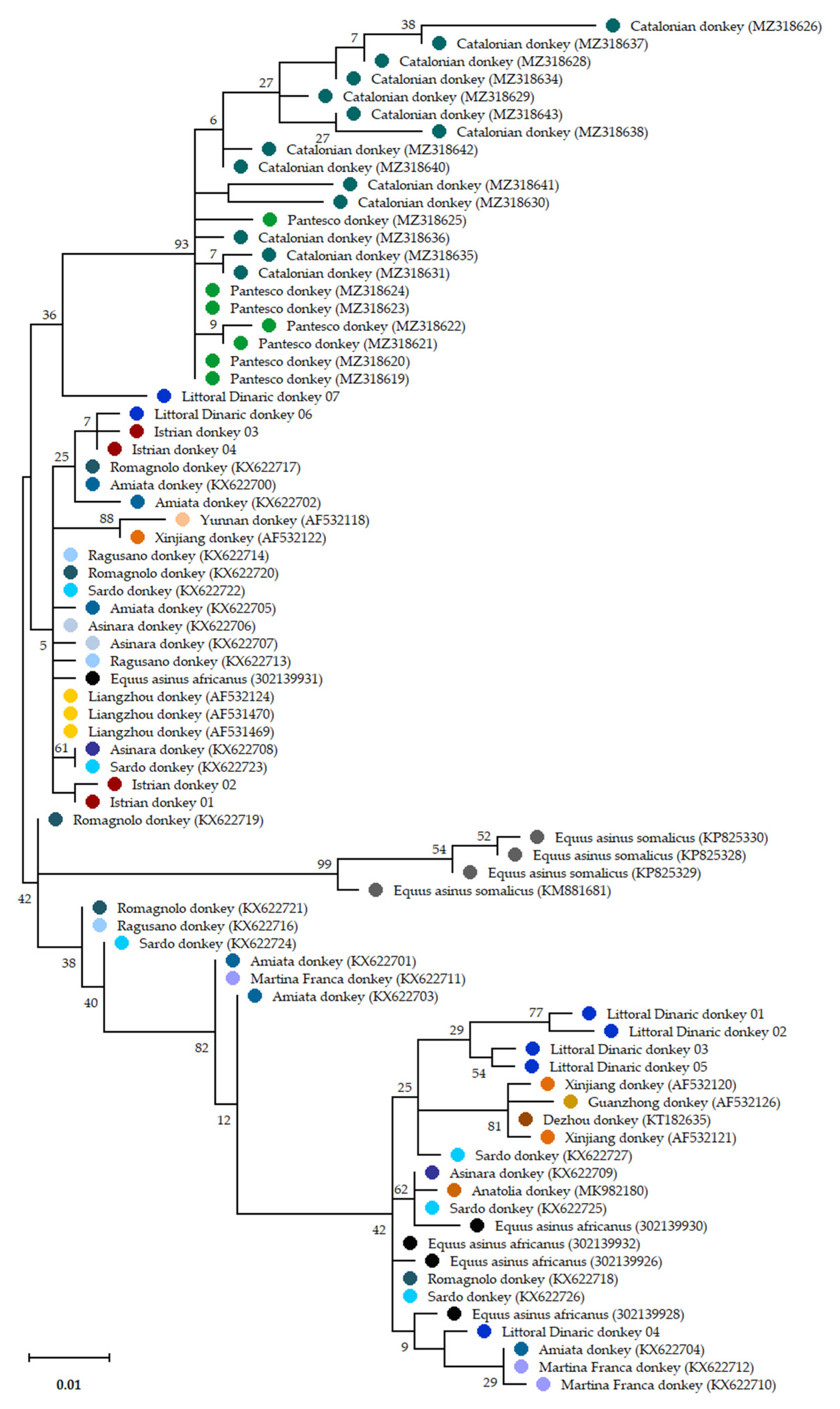
3.3. Mitochondrial Variability and Relationship with Other Donkey Breeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO (Food and Agriculture Organsations). Status and Trends of Animal Genetic Resources; FAO—Commission on Genetic Resources for Food and Agriculture: Rome, Italy, 2019; Available online: http://www.fao.org/3/my867en/my867en.pdf (accessed on 16 January 2022).
- Babić, E. Prilog poznavanju apuljskih magaraca u Dalmaciji. Vet. Arhiv. 1939, 9, 228–252. [Google Scholar]
- Ivanković, A.; Caput, P.; Mioč, B.; Pavić, V. The Phenotype Features of Donkeys in Croatia. Agric. Conspec. Sci. 2000, 65, 99–105. [Google Scholar]
- Vostrý, L.; Capková, Z.; Přibyl, J.; Hofmanová, B.; Vydrová, H.V.; Mach, K. Population structure of Czech cold-blooded breeds of horses. Arch. Anim. Breed. 2011, 54, 1–9. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Marmi, J.; Goyache, F.; Jordana, J. Pedigree information reveals moderate to high levels of inbreeding and a weak population structure in the endangered Catalonian donkey breed. J. Anim. Breed. Genet. 2005, 122, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Cecchi, F.; Ciampolini, R.; Ciani, E.; Matteoli, B.; Mazzanti, E.; Tancredi, M.; Presciuttini, S. Demographic genetics of the endangered Amiata donkey breed. Ital. J. Anim. Sci. 2006, 5, 387–391. [Google Scholar] [CrossRef]
- Rizzi, R.; Tullo, E.; Cito, A.M.; Caroli, A.; Pieragostini, E. Monitoring of genetic diversity in the endangered Martina Franca donkey population. Sci. J. Anim. Sci. 2011, 89, 1304–1311. [Google Scholar] [CrossRef][Green Version]
- Quaresma, M.; Martins, A.M.F.; Rodrigues, J.B.; Colaço, J.; Payan-Carreira, R. Pedigree and herd characterization of a donkey breed vulnerable to extinction. Animal 2014, 8, 354–359. [Google Scholar] [CrossRef][Green Version]
- Santana, M.L.; Bignardi, A.B. Status of the genetic diversity and population structure of the Pega donkey. Trop. Anim. Health Prod. 2015, 47, 1573–1580. [Google Scholar] [CrossRef]
- Navas, F.J.; Jordana, J.; León, J.M.; Barba, C.; Delgado, J.V. A model to infer the demographic structure evolution of endangered donkey populations. Animal 2017, 11, 2129–2138. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Goyache, F. A note on ENDOG: A computer program for analysing pedigree information. J. Anim. Breed. Genet. 2005, 122, 172–176. [Google Scholar] [CrossRef]
- Ivanković, A.; Caput, P. Genetic polymorphism of transferrin and albumin in donkeys in Croatia. Czech J. Anim. Sci. 2000, 45, 49–52. [Google Scholar]
- Ivanković, A.; Kavar, T.; Caput, P.; Mioč, B.; Pavić, V.; Dovč, P. Genetic diversity of three donkey populations in the Croatian coastal region. Anim. Genet. 2002, 33, 169–177. [Google Scholar] [CrossRef]
- Aranguren-Mendez, J.; Jordana, J.; Gomez, M. Genetic diversity in Spanish donkey breeds using microsatellite DNA markers. Genet. Sel. Evol. 2001, 33, 433–442. [Google Scholar] [CrossRef]
- Blasi, M.; Perrotta, G.; Lanza, A.; Iamartino, D.; Pilla, F. Genetic diversity in three Italian donkey populations assessed by microsatellite markers. Ital. J. Anim. Sci. 2005, 4, 127. [Google Scholar]
- Matassino, D.; Cecchi, F.; Ciani, F.; Incoronato, C.; Occidente, M.; Santoro, L.; Ciampolini, R. Genetic diversity and variability in two Italian autochthonous donkey genetic types assessed by microsatellite markers. Ital. J. Anim. Sci. 2014, 13, 53–60. [Google Scholar] [CrossRef]
- Rosenbom, S.; Costa, V.; Al-Araimi, N.; Kefena, E.; Abdel-Moneim, A.S.; Abdalla, M.A.; Bakhiet, A.; Beja-Pereira, A. Genetic diversity of donkey populations from the putative centers of domestication. Anim. Genet. 2015, 46, 30–36. [Google Scholar] [CrossRef]
- Jordana, J.; Ferrando, A.; Miro, J.; Goyache, F.; Loarca, A.; Martínez López, O.R.; Canelón, J.L.; Stemmer, A.; Aguirre, L.; Lara, M.A.C.; et al. Genetic relationships among American donkey populations: Insights into the process of colonization. J. Anim. Breed. Genet. 2016, 133, 155–164. [Google Scholar] [CrossRef]
- Stanisic, L.J.; Aleksic, J.M.; Dimitrijevic, V.; Simeunovic, P.; Glavinic, U.; Stevanovic, J.; Stanimirovic, Z. New insights into the origin and the genetic status of the Balkan donkey from Serbia. Anim. Genet. 2017, 48, 580–590. [Google Scholar] [CrossRef]
- Yatkın, S.; Özdil, F.; Özkan Ünal, E.; Genç, S.; Kaplan, S.; Gürcan, E.K.; Arat, S.; Soysal, M.I. Genetic characterization of native donkey (Equus asinus) populations of turkey using microsatellite markers. Animals 2020, 10, 1093. [Google Scholar] [CrossRef]
- Xu, X.; Gullberg, A.; Arnason, U. The complete mitochondrial DNA (mtDNA) of the donkey and mtDNA comparisons among four closely related mammalian species-pairs. J. Mol. Evol. 1996, 43, 438–446. [Google Scholar] [CrossRef]
- Underhill, P.A.; Shen, P.; Lin, A.A.; Jin, L.; Passarino, G.; Yang, W.H.; Kauffman, E.; Bonné-Tamir, B.; Bertranpetit, J.; Francalacci, P.; et al. Y chromosome sequence variation and the history of human populations. Nat. Genet. 2000, 26, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Rentsch, J.; Hübner, P. PCR-RFLP analysis of mitochondrial DNA: A reliable method for species identification. J. Agric. Food Chem. 1999, 47, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Aranguren-Mendez, J.; Beja-Pereira, A.; Avellanet, R.; Dzama, K.; Jordana, J. Mitochondrial DNA variation and genetic relationships in Spanish donkey breeds (Equus asinus). J. Anim. Breed. Genet. 2004, 121, 319–330. [Google Scholar] [CrossRef]
- Beja-Pereira, A.; England, P.R.; Ferrand, N.; Jordan, S.; Bakhiet, A.O.; Abdalla, M.A.; Mashkour, M.; Jordana, J.; Taberlet, P.; Luikart, G. African origins of the domestic donkey. Science 2004, 304, 1781. [Google Scholar] [CrossRef]
- Lei, C.Z.; Chen, H.; Yang, G.S.; Sun, W.B.; Lei, X.Q.; Ge, Q.L.; Wang, Z.F.; Lu, N.; Gao, X.; Hou, W.T. Study on mitochondrial DNA D-loop polymorphism in Chinese donkeys. Yi Chuan Xue Bao 2005, 35, 481–486. [Google Scholar]
- Chen, S.Y.; Zhou, F.; Xiao, H.; Sha, T.; Wu, S.F.; Zhang, Y.P. Mitochondrial DNA diversity and population structure of four Chinese donkey breeds. Anim. Genet. 2006, 37, 427–429. [Google Scholar] [CrossRef]
- Cozzi, M.C.; Valiati, P.; Cherchi, R.; Gorla, E.; Prinsen, R.T.M.M.; Longeri, M.; Bagnato, A.; Strillacci, M.G. Mitochondrial DNA Genetic Diversity in Six Italian Donkey Breeds (Equus Asinus). Mitochondrial DNA A DNA Mapp. Seq. Anal. 2018, 29, 409–418. [Google Scholar] [CrossRef]
- Pengjia, B.; Guo, X.; Pei, J.; Ding, X.; Wu, X.; Xiong, L.; Liang, C.; Chu, M.; Yan, P. Characterization of the complete mitochondrial genome of the Liangzhou donkey (Equus asinus). Mitochondrial DNA Part B 2019, 4, 1846–1847. [Google Scholar] [CrossRef]
- Ünal, E.Ö.; Özdil, F.; Kaplan, S.; Gürcan, E.K.; Genç, S.; Arat, S.; Soysal, M.I. Phylogenetic Relationships of Turkish Indigenous Donkey Populations Determined by Mitochondrial DNA D-loop Region. Animals 2020, 10, 1970. [Google Scholar] [CrossRef]
- Mazzatenta, A.; Vignoli, M.; Caputo, M.; Vignola, G.; Tamburro, R.; De Sanctis, F.; Roig, J.M.; Bucci, R.; Robbe, D.; Carluccio, A. Maternal Phylogenetic Relationships and Genetic Variation among Rare, Phenotypically Similar Donkey Breeds. Genes 2021, 12, 1109. [Google Scholar] [CrossRef]
- Boichard, D.; Maignel, L.; Verrier, E. The value of using probabilities of gene origin to measure genetic variability in a population. Genet. Sel. Evol. 1997, 29, 5–23. [Google Scholar] [CrossRef]
- Lacy, R.C. Analysis of founder representation in pedigrees: Founder equivalents and founder genome equivalents. Zoo Biol. 1989, 8, 111–123. [Google Scholar] [CrossRef]
- Alderson, L. Alderson, L. A system to maximize the maintenance of genetic variability in small populations. In Genetic Conservation of Domestic Livestock II; Alderson, L., Bodo, I., Eds.; CABI: Wallingford, UK, 1992; pp. 18–29. [Google Scholar]
- Duru, S. Pedigree analysis of the Turkish Arab horse population: Structure, inbreeding and genetic variability. Animal 2017, 11, 1449–1456. [Google Scholar] [CrossRef]
- James, J.W. A note on selection differentials and generation length when generations overlap. Anim. Sci. 1977, 24, 109–112. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Cho, G.J. Standardization and usefulness of ISAG microsatellite markers for individual identification and parentage verification in horse breeds. J. Vet. Clin. 2009, 26, 220–225. [Google Scholar]
- FAO. Molecular Genetic Characterization of Animal Genetic Resources; FAO Animal Production and Health Guidlines. No. 9; FAO: Rome, Italy, 2011; Available online: https://www.fao.org/3/i2413e/i2413e00.pdf (accessed on 16 January 2022).
- Peakall, R.; Smouse, P.E. GENALEX 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 116, 1099–1106. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation (Version 2.9.3.2). 2001. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 24 April 2017).
- Rousset, F. GENEPOP’007: A complete re-implementation of the genepop software for Windows and Linux. Mol. Ecol. 2008, 8, 1103–1106. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX 4.05, Logiciel Sous Windows TM Pour La Génétique des Populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000; Université de Montpellier II: Montpellier, France, 2004. [Google Scholar]
- Cornuet, J.M.; Luikart, G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 1996, 144, 2001–2014. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. Computer note. BOTTLENECK: A computer program for detecting recent reductions in the effective size using allele frequency data. Heredity 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Giontella, A.; Cardinali, I.; Lancioni, H.; Giovannini, S.; Pieramati, C.; Silvestrelli, M.; Sarti, F.M. Mitochondrial DNA Survey Reveals the Lack of Accuracy in Maremmano Horse Studbook Records. Animals 2020, 10, 839. [Google Scholar] [CrossRef]
- Pjontek, J.; Kadlečík, O.; Kasarda, R.; Horný, M. Pedigree analysis in four Slovak endangered horse breeds. Czech J. Anim. Sci. 2012, 57, 54–64. [Google Scholar] [CrossRef]
- Folch, P.; Jordana, J. Demographic characterization, inbreeding and maintenance of genetic diversity in the endangered Catalonian donkey breed. Genet. Sel. Evol. 1998, 30, 195–202. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E. Operation of conservation schemes. In Genebanks and the Conservation of Farm Animal Genetic Resources; Oldenbroek, K.K., Ed.; DLO Institute for Animal Science and Health: Lelystad, The Netherlands, 1999; pp. 9–112. [Google Scholar]
- Caballero, A.; Toro, M.A. Analysis of genetic diversity for the management of conserved subdivided populations. Conserv. Genet. 2002, 3, 289–299. [Google Scholar] [CrossRef]
- Gutiérrez, J.P.; Altarriba, J.; Díaz, C.; Quintanilla, R.; Cañón, J.; Piedrafita, J. Pedigree analysis of eight Spanish beef cattle breeds. Genet. Sel. Evol. 2003, 35, 43–64. [Google Scholar] [CrossRef]
- Stephens, T.D.; Splan, R.K. Population history and genetic variability of the American Shire horse. Anim. Genet. Resour. 2013, 52, 31–38. [Google Scholar] [CrossRef]
- Lutaaya, E.; Misztal, I.; Bertrand, J.K.; Mabry, J.W. Inbreeding in populations with incomplete pedigrees. J. Anim. Breed. Genet. 1999, 116, 475–480. [Google Scholar] [CrossRef]
- Pinheiro, M.; Kjöllerström, H.J.; Oom, M.M. Genetic diversity and demographic structure of the endangered Sorraia horse breed assessed through pedigree analysis. Livest. Sci. 2013, 152, 1–10. [Google Scholar] [CrossRef]
- Di, R.; Liu, Q.; Xie, F.; Hu, W.; Wang, X.-Y.; Cao, X.-H.; Pan, Z.-Y.; Chen, G.; Chu, M. Evaluation of genetic diversity and population structure of five Chinese indigenous donkey breeds using microsatellite markers. Czech J. Anim. Sci. 2017, 62, 219–225. [Google Scholar] [CrossRef]
- Bordonaro, S.; Guastella, A.M.; Criscione, A.; Zuccaro, A.; Marletta, D. Genetic Diversity and Variability in Endangered Pantesco and Two Other Sicilian Donkey Breeds Assessed by Microsatellite Markers. Sci. World J. 2012, 2012, 648427. [Google Scholar] [CrossRef]
- Khanshour, A.M.; Hempsey, E.K.; Juras, R.; Cothran, E.G. Genetic Characterization of Cleveland Bay Horse Breed. Diversity 2019, 11, 174. [Google Scholar] [CrossRef]
- Babić, E. Prilog Poznavanju Domaćih Primorskih Magaraca (A Contribution to the Knowledge of Local Coastal Donkeys). Ph.D. Thesis, Faculty of Agriculture and Forestry in Zagreb, Zagreb, Croatia, 1939. [Google Scholar]
- Maretto, F.; Cassandro, M. Temporal variation in genetic diversity and population structure of Burlina cattle breed. Ital. J. Anim. Sci. 2014, 13, 322–329. [Google Scholar] [CrossRef][Green Version]
| Item | LDD | ID |
|---|---|---|
| Original dataset (TP; total population) | 5030 | 1233 |
| Reference population 1 (REF 01; animal with both parents known) | 2724 | 897 |
| Reference population 2 (REF 02; live animals in 2020) | 3296 | 758 |
| Number of founders contributing to REF 01 population | 1097 | 210 |
| Effective number of founders (fe) | 325 | 70 |
| Effective population size of founders | 914.13 | 121.77 |
| Number of ancestors contributing to REF 01 population | 1023 | 200 |
| Effective number of ancestors (fa) | 316 | 65 |
| Number of ancestors explaining 50% of genetic variability | 141 | 23 |
| Effective number of founders/effective number of ancestors (fe/fa) | 1.03 | 1.08 |
| Number of founder herds in REF 01 population (fh) | 358 | 79 |
| Effective number of founder herds for the REF 01 population | 65.4 | 18.6 |
| Effective population size (Ne) | 74.59 | 81.27 |
| Effective population size REF 02 population (computed via individual increase in inbreeding) | 25.99 | 32.78 |
| Mean inbreeding (F) | 0.99 | 1.77 |
| Mean Average Relatedness coefficient (AR) | 0.13 | 1.10 |
| Genetic Conservation Index (GCI) | 1.76 | 2.42 |
| Mean number of maximum generations | 1.01 | 1.80 |
| Mean number of equivalent generations | 0.56 | 0.88 |
| Mean number of complete generations | 0.74 | 1.24 |
| Pathway | Littoral-Dinaric Donkey | Istrian Donkey | ||
|---|---|---|---|---|
| n | Mean ± SD | n | Mean ± SD | |
| Average age | ||||
| Father—son | 1090 | 7.57 ± 3.902 | 343 | 7.41 ± 4.118 |
| Father—daughter | 1412 | 7.47 ± 3.798 a | 502 | 7.56 ± 4.109 |
| Mother—son | 1218 | 7.50 ± 4.161 | 360 | 7.61 ± 3.913 |
| Mother—daughter | 1571 | 7.79 ± 4.296 b | 534 | 7.42 ± 4.104 |
| Overall | 5291 | 7.59 ± 4.057 | 1739 | 7.50 ± 4.067 |
| Generation interval | ||||
| Father—son | 136 | 7.34 ± 3.652 | 46 | 7.87 ± 4.436 |
| Father—daughter | 402 | 7.30 ± 3.701 | 187 | 7.89 ± 3.912 |
| Mother—son | 166 | 6.94 ± 4.055 | 55 | 7.84 ± 4.032 |
| Mother—daughter | 480 | 7.34 ± 4.162 | 203 | 7.53 ± 3.843 |
| Overall | 1184 | 7.27 ± 3.936 | 491 | 7.73 ± 3.943 |
| Marker | NA | ENA | AR | PIC | HO | HE | FIS | FST | FIT | Nm | F(Null) | HW |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AHT4 | 6 | 3.078 | 5.628 | 0.649 | 0.708 | 0.703 | −0.046 | 0.070 | 0.028 | 5.936 | −0.0052 | ns |
| HMS2 | 5 | 3.020 | 4.970 | 0.632 | 0.675 | 0.690 | −0.005 | 0.052 | 0.048 | 7.839 | 0.0113 | ns |
| HMS7 | 5 | 1.635 | 4.500 | 0.364 | 0.358 | 0.388 | 0.071 | 0.013 | 0.084 | 22.206 | 0.0537 | ns |
| TKY297 | 9 | 4.593 | 8.374 | 0.766 | 0.742 | 0.798 | 0.060 | 0.023 | 0.082 | 15.262 | 0.0374 | ns |
| TKY343 | 4 | 2.505 | 3.993 | 0.563 | 0.542 | 0.626 | 0.106 | 0.063 | 0.163 | 6.443 | 0.0712 | ns |
| ABS23 | 6 | 4.524 | 5.970 | 0.753 | 0.792 | 0.789 | −0.008 | 0.009 | 0.001 | 28.910 | −0.0031 | ns |
| HMS3 | 7 | 3.206 | 5.985 | 0.639 | 0.642 | 0.693 | 0.0476 ** | 0.055 | 0.100 | 7.443 | 0.0363 | ** |
| HTG10 | 9 | 5.667 | 8.721 | 0.810 | 0.850 | 0.834 | −0.026 | 0.013 | −0.013 | 23.014 | −0.014 | ns |
| TKY312 | 5 | 3.116 | 4.751 | 0.636 | 0.692 | 0.691 | −0.015 | 0.025 | 0.011 | 14.530 | 0.002 | ns |
| HMS18 | 9 | 3.370 | 8.425 | 0.668 | 0.750 | 0.711 | −0.069 | 0.024 | −0.043 | 15.144 | −0.0303 | ns |
| HMS6 | 3 | 1.883 | 2.970 | 0.389 | 0.392 | 0.492 | 0.164 | 0.093 | 0.241 | 4.410 | 0.1141 | ns |
| HTG7 | 10 | 4.301 | 9.990 | 0.749 | 0.733 | 0.770 | 0.008 | 0.077 | 0.084 | 5.416 | 0.0273 | ns |
| TKY337 | 4 | 2.230 | 3.939 | 0.501 | 0.550 | 0.559 | −0.028 | 0.083 | 0.057 | 5.049 | 0.0027 | ns |
| Mean | 6.31 | 3.317 | 6.017 | 0.624 | 0.648 | 0.673 | 0.014 | 0.045 | 0.058 | 12.431 |
| Littoral-Dinaric Donkey (LDD, n = 60) | Istrian Donkey (ID, n = 60) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | NA | ENA | HO | HE | FIS | NA | ENA | HO | HE | FIS |
| AHT4 | 6 | 3.379 | 0.783 | 0.704 | −0.104 * | 5 | 2.777 | 0.633 | 0.640 | 0.019 |
| HMS2 | 5 | 3.285 | 0.750 | 0.696 | −0.07 | 5 | 2.755 | 0.600 | 0.637 | 0.067 |
| HMS7 | 4 | 1.798 | 0.367 | 0.444 | 0.182 | 5 | 1.473 | 0.350 | 0.321 | −0.082 |
| TKY297 | 9 | 4.712 | 0.767 | 0.788 | 0.035 | 8 | 4.475 | 0.717 | 0.777 | 0.085 |
| TKY343 | 4 | 2.576 | 0.517 | 0.612 | 0.164 | 4 | 2.433 | 0.567 | 0.589 | 0.046 |
| ABS23 | 5 | 4.514 | 0.783 | 0.778 | 0.002 | 6 | 4.534 | 0.800 | 0.779 | −0.018 |
| HMS3 | 7 | 3.998 | 0.767 | 0.750 | −0.014 * | 5 | 2.414 | 0.517 | 0.586 | 0.126 *** |
| HTG10 | 9 | 6.277 | 0.883 | 0.841 | −0.042 | 7 | 5.056 | 0.817 | 0.802 | −0.01 |
| TKY312 | 4 | 3.416 | 0.683 | 0.707 | 0.042 | 5 | 2.816 | 0.700 | 0.645 | −0.077 |
| HMS18 | 7 | 2.855 | 0.733 | 0.650 | −0.12 | 9 | 3.886 | 0.767 | 0.743 | −0.024 |
| HMS6 | 3 | 1.699 | 0.400 | 0.411 | 0.036 | 3 | 2.067 | 0.383 | 0.516 | 0.265 * |
| HTG7 | 10 | 2.748 | 0.600 | 0.636 | 0.065 | 10 | 5.854 | 0.867 | 0.829 | −0.037 |
| TKY337 | 4 | 1.762 | 0.500 | 0.432 | −0.148 | 4 | 2.698 | 0.600 | 0.629 | 0.055 |
| Mean | 5.92 | 3.31 | 0.656 | 0.650 | −0.002 | 5.85 | 3.33 | 0.640 | 0.653 | 0.029 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanković, A.; Bittante, G.; Šubara, G.; Šuran, E.; Ivkić, Z.; Pećina, M.; Konjačić, M.; Kos, I.; Kelava Ugarković, N.; Ramljak, J. Genetic and Population Structure of Croatian Local Donkey Breeds. Diversity 2022, 14, 322. https://doi.org/10.3390/d14050322
Ivanković A, Bittante G, Šubara G, Šuran E, Ivkić Z, Pećina M, Konjačić M, Kos I, Kelava Ugarković N, Ramljak J. Genetic and Population Structure of Croatian Local Donkey Breeds. Diversity. 2022; 14(5):322. https://doi.org/10.3390/d14050322
Chicago/Turabian StyleIvanković, Ante, Giovanni Bittante, Gordan Šubara, Edmondo Šuran, Zdenko Ivkić, Mateja Pećina, Miljenko Konjačić, Ivica Kos, Nikolina Kelava Ugarković, and Jelena Ramljak. 2022. "Genetic and Population Structure of Croatian Local Donkey Breeds" Diversity 14, no. 5: 322. https://doi.org/10.3390/d14050322
APA StyleIvanković, A., Bittante, G., Šubara, G., Šuran, E., Ivkić, Z., Pećina, M., Konjačić, M., Kos, I., Kelava Ugarković, N., & Ramljak, J. (2022). Genetic and Population Structure of Croatian Local Donkey Breeds. Diversity, 14(5), 322. https://doi.org/10.3390/d14050322













