Abstract
Genomic assessment of local sheep breeds is relevant to the reconstruction of post-domestication migrations and to filling in gaps in the developmental history and contemporary phylogeographic-differentiation patterns in Eurasia. In this study, we aimed to reveal possible genetic relationships between local sheep breeds in Russia and the Persian Highlands (Iran) based on high-density SNP genotypes. All samples (n = 395) from 11 Iranian and 10 Russian sheep breeds were genotyped by using the Ovine Infinium HD BeadChip (Illumina, San Diego, CA, USA). Principal Component Analysis, maximum-likelihood assessment, and Neighbor-Net graph analysis demonstrated a clear differentiation between Russian sheep breeds of European ancestry from the Iranian local cluster and Russian breeds of Asian origin. Admixture analysis revealed a shared ancestral background, which was detected in several Iranian-local and Russian-local fat-tailed coarse-wool sheep breeds. Our findings contribute to a better understanding of the pattern of historic admixture, which is present in the genomes of many Eurasian sheep breeds.
1. Introduction
The introduction of high-throughput SNP arrays, including DNA chips, significantly accelerated genetic research on livestock species [1,2,3], including sheep [4]. The use of DNA chips allowed for the establishment of population structures and genome-wide diversity in various transboundary [5] and local sheep breeds [6], for the estimation of genomic inbreeding and of the distribution of runs of homozygosity [7,8,9], as well as for the detection of selection signals, by using different approaches [10,11,12].
One of the most impressive consequences of DNA-chip implementation is the generation of medium-density and high-density SNP genotypes for numerous transboundary (cosmopolitan) and local livestock breeds and populations, which can be easily pooled and processed by unified genotyping technology. Thus, an assessment of genome-wide diversity in livestock species has shifted from the local to the global level [1,13,14].
The domestic sheep (Ovis aries) is one of the most significant species of livestock, occupying all continents except for Antarctica, and has traditionally been raised in many countries. Establishing the genome-wide diversity and admixture patterns in sheep populations is relevant, not only to providing new insights into their ancestral origin, but also to finding new ways to preserve the variety of valuable and specific genotypes of this livestock species.
To unlock the complex genetic relationships of modern sheep populations, Kijas J. et al. [1] conducted the first comprehensive study based on 50 k genotypes of 74 sheep breeds from diverse locations. Although a clear geographic differentiation was demonstrated, a high level of historic admixture was reported as well [1]. This pattern is the result of long and complex human-mediated migrations of sheep via different routes, from their domestication center in the Zagros Mountains, to Europe and Asia, and then to the New World and Australia. In addition, subsequent mixture events could have affected the gene pools of local breeds. For example, beginning in the 18th century, Spanish Merino and more recently-developed Merino-derived breeds formed and changed the wool industry in many countries, as well as facilitated the economic development of Australia [15]. Furthermore, recent genomic research provided strong evidence of the admixture of Spanish Merino sheep into several central European breeds [16]. Besides the world mutton industry has been significantly promoted by the highly productive British sheep breeds, which have been widely used to improve native breeds [17]. In addition, the replenishment of the 50 k dataset with SNP genotypes of Balkan-native breeds has allowed for the reconstruction of sheep migrations into Europe, and for the fuller analysis of their subsequent spread in the westerly and northwesterly directions [16]. Particularly, research findings provided evidence of the roles of the Balkan region and Italy as post-domestication migration hubs, from which wool sheep reached Spain and north Italy, with further distribution northwards [16].
Although European breeds have had significant impact on worldwide sheep diversity, a great variety of sheep populations have been created and are raised in Asian countries. Beginning with the genotyping of 15 sheep breeds of Asian ancestry [1], a genomic survey included more breeds from China [18], India [19], Iran [4,20,21], Russia [22], Kyrgyzstan [23], and Kazakhstan [24]. However, only a few attempts were made to understand the existing mixture patterns of Asian breeds. Zhao, Y. X. et al. [18] provided evidence that there were two subsequent waves of migrations of fat-tailed sheep into northern China, corresponding to the migrations of ancestors of Hui Muslims eastward, and of Mongols southward, during the 12th–13th centuries, respectively. The genetic links of the breeds from Central Asia, the Middle East, and Northern Asia, respectively, were reported as well [20,23]. In this regard, establishing possible phylogeographic relationships between Asian breeds from different countries will be useful for gaining new knowledge of the origin of these populations, and for reconstructing sheep-migration routes into Eurasia.
In this regard, the aim of our study was to gain an understanding of the genomic characterization of local sheep breeds in Russia and the Persian Plateau (Iran) in order to uncover possible genetic relationships between these populations.
2. Materials and Methods
2.1. Ethic Statement
The present study does not involve any endangered or protected animals. All relevant procedures were conducted according to the ethical guidelines of the L.K. Ernst Federal Science Center for Animal Husbandry and Shahid Bahonar University of Kerman. Protocol No. 1 (4 April 2022), was approved by the Commission on the Ethics of Animal Experiments of the L.K. Ernst Federal Science Center for Animal Husbandry. The samples from the Romanov sheep were collected by trained personnel under strict veterinary rules in accordance with the rules for conducting laboratory research (tests) in the implementation of the veterinary control (supervision) approved by Council Decision Eurasian Economic Commission No. 80 (10 November 2017). SNP genotypes of several local sheep breeds were taken from the Bioresource collection of the L.K. Ernst Federal Research Center for Animal Husbandry [25].
2.2. Samples and Genotyping
Samples for this study included SNP genotypes of 395 sheep from 11 Iranian and 10 Russian local breeds. Two samples of wild sheep (Asiatic mouflon) were added as the outgroup population for phylogenetic analysis. A short description of sample collection is shown in Table 1.

Table 1.
Short description of the Iranian and Russian sheep populations used in this study.
All SNP genotypes for Russian and Iranian sheep populations were generated by using the Ovine Infinium HD BeadChip (Illumina, San Diego, CA, USA) [26].
To consider the accuracy of SNP genotyping, quality control was conducted by setting a cut-off of 0.5 for the GenCall and GenTrain scores [27] using PLINK v1.90 [28]. Samples with a missing genotype call rate of 0.1 were removed from the analysis. SNPs with a call rate below 0.90, a minor allele frequency (MAF) lower than 0.05, or those located on sex chromosomes were deleted from the analysis.
2.3. Data Processing
Principal component analysis (PCA) was performed in PLINK v1.9 and visualized using the R package “ggplot2” [29].
Estimation of pairwise genetic differentiation values (FST) [30] was conducted using the R package StAMPP [31].
A Neighbor-Net graph, which was based on the matrix of pairwise FST values, was constructed using SplitsTree 4.14.5 software [32].
To identify ancestral background, we performed genomic clustering using Admixture v1.3 software [33] and used the R package “BITE” [34] for visualization. The choice of the most probable number of populations (K) was based on the lowest cross˗validation error compared to other K values as implemented in a standard cross˗validation procedure using Admixture v1.3 software [33].
Inference of population splits and mixtures was performed using the TreeMix program [35]. To set position of the root in the maximum likelihood (ML) tree produced by the TreeMix program, we used wild sheep (n = 2) as the outgroup population. Furthermore, we added one or two migration events in the ML tree to obtain the most frequently found variants with significant gene flow, and ran 10 independent interactions for each event.
3. Results and Discussion
Principal Component Analysis (Figure 1) showed that the PC1, accounting for 14.82% of total genetic variability, clearly separated a cluster including all Iranian breeds and Russian coarse-wool fat-tailed breeds from other studied breeds. The PC2 responsible for 9.01% of the genetic variability differentiated the Russian Romanov breed into the lower left quadrant, while the closely connected group of Russian fine-wool breeds (Salsk, Volgograd, and Groznensk) and the Russian Long-Haired semi-fine wool breed were located in the upper left quadrant. The two wild sheep were located in an intermediate position. The Romanov breed was the most differentiated breed within coarse-wool populations (Figure 1A), while, based on the PC3, Kermani-breed separation was found (Figure 1B).
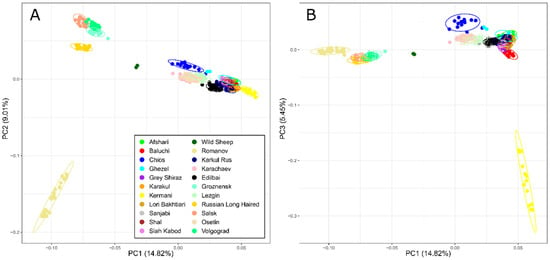
Figure 1.
Principal Component Analysis for Iranian and Russian sheep breeds performed: (A) for the first two principal components (PC1 and PC2); (B) and for the first and third principal components (PC1 and PC3).
A Neighbor-Net graph (Figure 2) showed that all Iranian local sheep breeds, the Russian Karakul breed, and the Edilbai breeds formed a joint cluster. The Chios, Lezgin, Karachaev and Osetin breeds were represented by independent branches, located near the above-mentioned cluster. Wild sheep had the longest independent branch. The Volgograd, Salsk, and Groznensk breeds had their own cluster. A subcluster, which included branches of Russian Long-Haired and Romanov breeds, was attached to the main network.
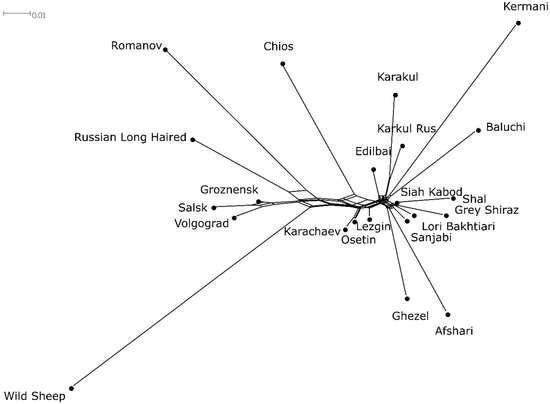
Figure 2.
Neighbor-Net graph for Iranian and Russian local sheep breeds.
Clustering with Admixture software showed that, at K = 2, the Romanov, Volgograd, Salsk, Groznensk, and Russian Long-Haired breeds had a different ancestral background from Iranian local and other Russian breeds (Figure 3).
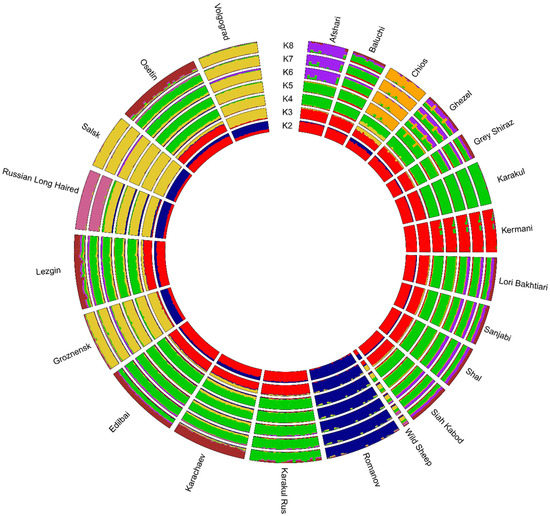
Figure 3.
Clustering of Iranian and Russian local sheep breeds.
Beginning with K = 3, the Romanov breed demonstrated its own specific genomic pattern, which was absent in other studied breeds. In addition, the Volgograd, Salsk, Groznensk, and Russian Long-Haired breeds had a specific ancestral background as well.
At K = 8, with the lowest cross-validation error, the following pattern was observed. The genomic backgrounds found in the Kermani and Chios breeds differed from other Iranian local breeds. The genomic background that was predominant in the Afshari breed (purple color) was found in the Baluchi, Ghezel, Grey Shiraz, Lori Bakhtiari, Sanjabi, Shal, and Siah Kabod breeds, and relevant traces were found in the Edilbai and Lezgin breeds as well. The Iranian Karakul breed showed the most solid background (green color), which was similar to that found in the Russian Karakul breed. The genomic components shared with the Karakul breeds were found in both the Iranian (Baluchi, Ghezel, Grey Shiraz, Lori Bakhtiari, Sanjabi, Shal, and Siah Kabod) and Russian local breeds (Edilbai, Lezgin, Karachaev (traces), and Osetin (traces)). Additionally, the ancestral background predominant in the Karachaev and Osetin breeds was present in the Baluchi, Ghezel, Grey Shiraz, Lori Bakhtiari, Sanjabi, Shal, Siah Kabod, Edilbai, and Lezgin breeds.
A maximum likelihood assessment of population history based on Treemix analysis generally confirmed aspects that had been already detected by Neighbor-Net analysis. However, the Romanov-sheep-breed branch was attached to the branch connecting clusters of Iranian sheep breeds and Russian coarse-wool breeds. The Volgograd, Salsk, and Groznensk breeds had their own cluster, while the Russian Long-Haired breed had its own branch.
With the addition of migrations events, significant replicable effects were revealed when one migration event was allowed (p < 0.05) (Figure 4). A mixture event was revealed in the ancestral population of sheep with the presence of the Merino genomic component (Groznensk, Salsk, and Volgograd) with the Chios breed. We may hypothesize that this pattern might be assumed to be an indirect indication of undocumented admixture events between the Chios breed and breeds with the Merino genetic background.
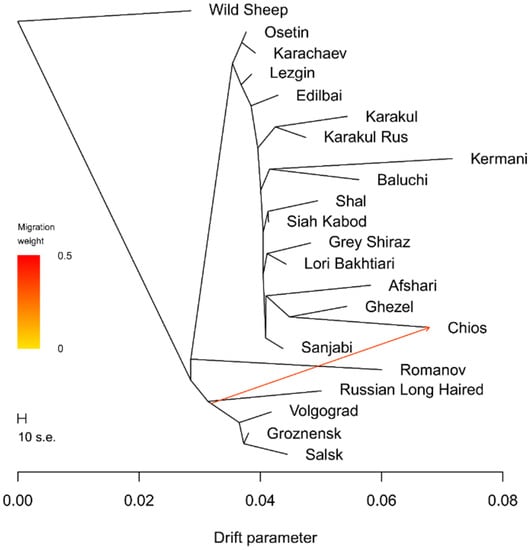
Figure 4.
Phylogenetic network inferred by Treemix plot of the relationships between the Iranian and Russian sheep breeds with one migration event.
Iran and Russia have a rich diversity of native and locally developed sheep breeds, which are a valuable part of the sheep gene pool in Eurasia. Therefore, the genetic relationships of both Iranian and Russian sheep breeds to global sheep populations have been studied previously. Based on 50 k SNP genotypes, the genomic diversity of Russian-local sheep breeds was addressed in the context of 58 worldwide sheep breeds, obtained from publicly available sources [22]. Shared genomic components with Chinese breeds were found in coarse-wool fat-tailed breeds, while fine-wool breeds expectedly jointed the global Merino-derived cluster. A study of the genome-wide admixture between the Iranian and the worldwide coarse-wool sheep breeds revealed the presence of the genetic background of Iranian sheep in Chinese and Kyrgyz populations [20]. However, only three Iranian breeds, including Afshari, Moghani, and Qezel, were presented in the 50 k dataset [1] in the first study, while Russian breeds were absent in the second research work. Thus, here, we performed the first genomic assessment of a joint dataset including high-density SNP genotypes of Iranian and Russian local sheep breeds.
Several major findings were revealed in this study. Firstly, neither PCA, clustering with Admixture software, nor a Neighbor Net graph analysis could establish genetic relationships between the Iranian local and Russian fine-wool breeds (Salsk, Volgograd, and Groznensk) and the semi-fine wool breed (Russian Long-Haired). Further, no admixture events were found between the Russian Romanov coarse-wool breed and Iranian local sheep breeds. The splits between the breeds due to wool type (fine wool and semi-fine wool versus coarse wool), origin (European versus Asian), and tail type (fat tails versus thin tails within the coarse-wool breeds) [22] have been reported previously.
PCA and Admixture results provided a pattern of moderate differentiation of the Iranian Kermani breed from the majority of other investigated sheep populations. The Kermani breed is a dual-purpose fat-tailed sheep that originated in the southeastern parts of Iran, and is well suited to dry and harsh climatic conditions, which are predominant in the Kerman province [36]. The samples of the Kermani breed used in this study were obtained from a population that had been isolated for a long time. The relatively low values of heterozygosity and other diversity indicators, which were calculated earlier [20], implicitly pointed out the isolation of the Kermani sheep group. This population probably has some private alleles that led to the separate clustering shown by the PCA and Admixture analyses, and may also explain the long branch in the Treemix tree and on the Neighbor Net graph. However, Iranian native ancestry is clearly present in the Kermani breed because it joined the Baluchi breed in the Treemix tree and on the Neighbor Net graph, which corresponded to the results obtained by using microsatellites [37].
Furthermore, we found a strong separation of the Russian Romanov breed from other studied breeds from both countries. This genomic pattern might be caused by several major factors, and corresponds to previous results, based on 50 k and 600 k data [10,22]. The Romanov sheep is a highly prolific breed that originated in central Russia (Yaroslavl region, Volga Valley) [38] and gained worldwide fame for its remarkable reproductive traits [39,40]. The presence of specific genetic variants that underlie the reproductive performance of the Romanov breed is being discussed [41]. However, we can hypothesize that accumulation of these specific genomic components might contribute to the observed separation pattern. Nevertheless, a more convincing theory is that the Romanov breed belongs to the group of North European short-tailed sheep, which originally inhabited the area from Russia to Iceland [42]. Based on high-density SNP profiles, Rochus, C. M. et al. (2020) studied the population structure of five Swedish-native sheep breeds and demonstrated the clear differentiation between the North European short-tailed group and other sheep populations, including transboundary and local breeds [6]. These findings are in line with our results because the Romanov breed is the lone representative of the north European short-tailed group in our sample.
Nevertheless, PCA and a Neighbor Net graph suggested genetic links between several Iranian and Russian fat-tailed coarse wool sheep breeds. Furthermore, based on Admixture analysis, components of shared ancestral background (green, brown, and purple colors in Figure 3) were found in these breeds. Although Iran and Russia do not share borders, this genomic pattern might be caused by various factors, some of which can be hypothetically outlined.
Primarily, sheep domestication took place in the Fertile Crescent located between Central Anatolia and the area North of the Zagros Mountains (Iran) [43]. With low probability, co-ancestral elements might be tracked through post-domestication routes from Iranian to Russian sheep breeds. However, recent studies involving ancient DNA samples showed that, based on the sequencing of nuclear and mitochondrial data, Anatolian Neolithic sheep demonstrated higher genomic affinity to modern European than non-European breeds [44]. Considering these latest findings, the exchange of genetic components between Iranian and Russian local sheep breeds had most likely occurred later.
According to Christian, D. [45], the role of the Silk Roads in the preservation of an underlying Eurasian unity has been underestimated. Between about 200 B.C. and the beginning of the Christian era, trading routes passed through modern Xinjiang, through central Asia, then either through Afghanistan to Kashmir and northern India, or to the Mediterranean, which they reached through Iran, through the Caucasus, or even by routes passing north of the Caspian and Black Seas [46]. Livestock were among the valuable goods [47] that the traders carried with them. The genes, which were transported through this great connection, penetrated, and shaped local populations. This process, and subsequent migration events, probably contributed to the formation of the admixture pattern in local Eurasian sheep breeds from non-neighboring countries, such as Iran and China [18,20], Iran and Kyrgyzstan [20], and Russia and Kyrgyzstan [23]. Our findings supported this suggestion.
4. Conclusions
Here, we present the results of a genomic study of Iranian and Russian local sheep breeds based on high-density SNP genotypes. Components of shared ancestral background were identified in several Iranian local and Russian local sheep breeds. Our findings contribute to a better understanding of the pattern of historic admixture, which is present in the genomes of many Eurasian sheep breeds. Additional sampling of sheep breeds from the other countries that were involved in Asia-Europe trading routes is required to reconstruct migrations and subsequent genetic events.
Author Contributions
Conceptualization, T.D., A.E. and N.Z.; methodology, A.D. and O.K.; software, A.D.; validation, T.D., A.E. and N.Z.; formal analysis, A.D.; investigation, O.K.; resources, M.A.F., A.E., N.Z. and T.D.; data curation, M.M.K., M.M., A.E. and A.D.; writing—original draft preparation, T.D. and A.E.; writing—review and editing, T.D. and A.E.; visualization, F.-H.L., A.D.; supervision, T.D., A.E. and N.Z.; project administration, A.E. and T.D.; funding acquisition, A.E. and T.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was jointly financially supported by RFBR (project number 20-516-56002) and Iran National Science Foundation: INSF (Grant number 98028814). Additional SNP-genotypes of Volgograd sheep breed used as comparison group were obtained with support of RSF project No. 21-46-00001.
Institutional Review Board Statement
This study was conducted according to the ethical guidelines of the L.K. Ernst Federal Science Center for Animal Husbandry and Shahid Bahonar University of Kerman. The protocol No. 1 of 4 April 2022, was approved by the Commission on the Ethics of Animal Experiments of the L.K. Ernst Federal Science Center for Animal Husbandry. The samples of Romanov sheep were collected by trained personnel under strict veterinary rules in accordance with the rules for conducting laboratory research (tests) in the implementation of the veterinary control (supervision) approved by Council Decision Eurasian Economic Commission No. 80 (10 November 2017).
Data Availability Statement
Ovine SNP-genotypes presented in this study are available upon request to the authors.
Acknowledgments
The ovine HD BeadChip was developed under the auspices of the International Sheep Genomics Consortium in work underwritten by FarmIQ (http://www.farmiq.co.nz/ (accessed on 1 April 2022), a joint New Zealand government and industry Primary Growth Partnership programme. Feng-Hua Lv was supported by the National Natural Science Foundation of China Project number: 32061133010).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-wide analysis of the world’s sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tosser˗Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.M.; Jamli, S.; et al. Design and characterization of a 52k SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Decker, J.E.; McKay, S.D.; Rolf, M.M.; Kim, J.; Molina Alcalá, A.; Sonstegard, T.S.; Hanotte, O.; Götherström, A.; Seabury, C.M.; Praharani, L.; et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.H.; Xu, S.S.; Shen, M.; Chen, Z.H.; Gao, L.; Lv, F.H.; Xie, X.-L.; Wang, X.-H.; Yang, H.; Liu, C.-B.; et al. Historical Introgression from Wild Relatives Enhanced Climatic Adaptation and Resistance to Pneumonia in Sheep. Mol. Biol. Evol. 2021, 38, 838–855. [Google Scholar] [CrossRef] [PubMed]
- Al-Mamun, H.A.; Clark, S.A.; Kwan, P.; Gondro, C. Genome-wide linkage disequilibrium and genetic diversity in five populations of Australian domestic sheep. Genet. Sel. Evol. GSE 2015, 47, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochus, C.M.; Jonas, E.; Johansson, A.M. Population structure of five native sheep breeds of Sweden estimated with high density SNP genotypes. BMC Genet. 2020, 21, 27. [Google Scholar] [CrossRef] [PubMed]
- Abied, A.; Xu, L.; Sahlu, B.W.; Xing, F.; Ahbara, A.; Pu, Y.; Lin, J.; Berihulay, H.; Islam, R.; He, X.; et al. Genome-Wide Analysis Revealed Homozygosity and Demographic History of Five Chinese Sheep Breeds Adapted to Different Environments. Genes 2020, 11, 1480. [Google Scholar] [CrossRef]
- Dzomba, E.F.; Chimonyo, M.; Pierneef, R.; Muchadeyi, F.C. Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genom. 2021, 22, 7. [Google Scholar] [CrossRef]
- Nosrati, M.; Asadollahpour Nanaei, H.; Javanmard, A.; Esmailizadeh, A. The pattern of runs of homozygosity and genomic inbreeding in world-wide sheep populations. Genomics 2021, 113, 1407–1415. [Google Scholar] [CrossRef]
- Yurchenko, A.A.; Deniskova, T.E.; Yudin, N.S.; Dotsev, A.V.; Khamiruev, T.N.; Selionova, M.I.; Egorov, S.V.; Reyer, H.; Wimmers, K.; Brem, G.; et al. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genom. 2019, 20 (Suppl. 3), 294. [Google Scholar] [CrossRef]
- Tsartsianidou, V.; Sánchez-Molano, E.; Kapsona, V.V.; Basdagianni, Z.; Chatziplis, D.; Arsenos, G.; Triantafyllidis, A.; Banos, G. A comprehensive genome-wide scan detects genomic regions related to local adaptation and climate resilience in Mediterranean domestic sheep. Genet. Sel. Evol. GSE 2021, 53, 90. [Google Scholar] [CrossRef]
- Shihabi, M.; Lukic, B.; Cubric-Curik, V.; Brajkovic, V.; Oršanić, M.; Ugarković, D.; Vostry, L.; Curik, I. Identification of Selection Signals on the X-Chromosome in East Adriatic Sheep: A New Complementary Approach. Front. Genet. 2022, 13, 887582. [Google Scholar] [CrossRef]
- Bovine HapMap Consortium; Gibbs, R.A.; Taylor, J.F.; Van Tassell, C.P.; Barendse, W.; Eversole, K.A.; Gill, C.A.; Green, R.D.; Hamernik, D.L.; Kappes, S.M.; et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science 2009, 324, 528–532. [Google Scholar] [CrossRef] [Green Version]
- Stella, A.; Nicolazzi, E.L.; Van Tassell, C.P.; Rothschild, M.F.; Colli, L.; Rosen, B.D.; Sonstegard, T.S.; Crepaldi, P.; Tosser-Klopp, G.; the AdaptMap Consortium; et al. AdaptMap: Exploring goat diversity and adaptation. Genet. Sel. Evol. GSE 2018, 50, 61. [Google Scholar] [CrossRef] [Green Version]
- Ciani, E.; Lasagna, E.; D’Andrea, M.; Alloggio, I.; Marroni, F.; Ceccobelli, S.; Delgado Bermejo, J.V.; Sarti, F.M.; Kijas, J.; Lenstra, J.A.; et al. International Sheep Genomics Consortium. Merino and Merino-derived sheep breeds: A genome-wide intercontinental study. Genet. Sel. Evol. GSE 2015, 47, 64. [Google Scholar] [CrossRef] [Green Version]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Econogene Consortium; et al. On the origin of European sheep as revealed by the diversity of the Balkan breeds and by optimizing population-genetic analysis tools. Genet. Sel. Evol. GSE 2020, 52, 25. [Google Scholar] [CrossRef]
- Romanov, M.N.; Zinovieva, N.A.; Griffin, D.K. British Sheep Breeds as a Part of World Sheep Gene Pool Landscape: Looking into Genomic Applications. Animals 2021, 11, 994. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Yang, J.; Lv, F.H.; Hu, X.J.; Xie, X.L.; Zhang, M.; Li, W.-R.; Liu, M.-J.; Wang, Y.-T.; Li, J.-Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 2017, 34, 2380–2395. [Google Scholar] [CrossRef]
- Eydivandi, S.; Roudbar, M.A.; Karimi, M.O.; Sahana, G. Genomic scans for selective sweeps through haplotype homozygosity and allelic fixation in 14 indigenous sheep breeds from Middle East and South Asia. Sci. Rep. 2021, 11, 2834. [Google Scholar] [CrossRef]
- Moosanezhad Khabisi, M.; Asadi Foozi, M.; Lv, F.H.; Esmailizadeh, A. Genome-wide DNA arrays profiling unravels the genetic structure of Iranian sheep and pattern of admixture with worldwide coarse-wool sheep breeds. Genomics 2021, 113, 3501–3511. [Google Scholar] [CrossRef]
- Mohamadipoor Saadatabadi, L.; Mohammadabadi, M.; Amiri Ghanatsaman, Z.; Babenko, O.; Stavetska, R.; Kalashnik, O.; Kucher, D.; Kochuk-Yashchenko, O.; Asadollahpour Nanaei, H. Signature selection analysis reveals candidate genes associated with production traits in Iranian sheep breeds. BMC Vet. Res. 2021, 17, 369. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. GSE 2018, 50, 29. [Google Scholar] [CrossRef] [Green Version]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population Structure and Genetic Diversity of Sheep Breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Niu, Z.; Zeng, Z.; Jiang, Y.; Jiang, Y.; Ding, Y.; Tang, S.; Shi, H.; Ding, X. Using High-Density SNP Array to Reveal Selection Signatures Related to Prolificacy in Chinese and Kazakhstan Sheep Breeds. Animals 2020, 10, 1633. [Google Scholar] [CrossRef]
- UNU. Available online: https://www.vij.ru/infrastruktura/unu (accessed on 1 April 2022).
- Kijas, J.W.; Porto-Neto, L.; Dominik, S.; Reverter, A.; Bunch, R.; McCulloch, R.; Hayes, B.J.; Brauning, R.; McEwan, J.; the International Sheep Genomics Consortium. Linkage disequilibrium over short physical distances measured in sheep using a high-density SNP chip. Anim Genet. 2014, 45, 754–757. [Google Scholar] [CrossRef]
- Fan, J.B.; Oliphant, A.; Shen, R.; Kermani, B.G.; Garcia, F.; Gunderson, K.L.; Hansen, M.; Steemers, F.; Butler, S.L.; Deloukas, P.; et al. Highly parallel SNP genotyping. Cold Spring Harb. Symp. Quant. Biol. 2003, 68, 69–78. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 1–16. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Pembleton, L.; Cogan, N.; Forster, J. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Milanesi, M.; Capomaccio, S.; Vajana, E.; Bomba, L.; Garcia, J.F.; Ajmone-Marsan, P.; Colli, L. BITE: An R Package for Biodiversity Analyses. bioRxiv 181610 [Preprint]. 2017. Available online: https://www.biorxiv.org/content/10.1101/181610v1 (accessed on 1 April 2022).
- Pickrell, J.K.; Pritchard, J.K. Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashidi, A.; Mokhtari, M.S.; Safi Jahanshahi, A.; Mohammad Abadi, M.R. Genetic parameter estimates of pre-weaning growth traits in Kermani sheep. Small Rumin. Res. 2008, 74, 165–171. [Google Scholar] [CrossRef]
- Ruiz-Larrañaga, O.; Asadollahpour Nanaei, H.; Montes, I.; Ayatollahi Mehrgardi, A.; Abdolmohammadi, A.; Kharrati-Koopaee, H.; Sohrabi, S.S.; Rendo, F.; Manzano, C.; Estonba, A.; et al. Genetic structure of Iranian indigenous sheep breeds: Insights for conservation. Trop. Anim. Health Prod. 2020, 52, 2283–2290. [Google Scholar] [CrossRef]
- Ernst, L.K.; Dmitriev, N.G.; Paronyan, I.A. Geneticheskie Resursy Sel‘Skokhozyaistvennykh Zhivotnykh v Rossii i Sopredel‘Nykh Stranakh; VNIIGRZH: Saint–Petersburg, Russian, 1994. (In Russian) [Google Scholar]
- Gallivan, C.; Kemp, R.A.; Berger, Y.M.; Young, L.D. Comparison of Finnish Landrace and Romanov as prolific breeds in a terminal-sire crossbreeding system. J. Anim. Sci. 1993, 71, 2910–2918. [Google Scholar] [CrossRef]
- Thomas, D.L. Performance and utilization of Northern European short-tailed breeds of sheep and their crosses in North America: A review. Anim. Int. J. Anim. Biosci. 2010, 4, 1283–1296. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.S.; Gao, L.; Xie, X.L.; Ren, Y.L.; Shen, Z.Q.; Wang, F.; Shen, M.; Eyϸórsdóttir, E.; Hallsson, J.H.; Kiseleva, T.; et al. Genome-Wide Association Analyses Highlight the Potential for Different Genetic Mechanisms for Litter Size Among Sheep Breeds. Front. Genet. 2018, 9, 118. [Google Scholar] [CrossRef]
- Dýrmundsson, O.R.; Niżnikowski, R. North European short-tailed breeds of sheep: A review. Anim. Int. J. Anim. Biosci. 2010, 4, 1275–1282. [Google Scholar] [CrossRef] [Green Version]
- Zeder, M.A. The Origins of Agriculture: New Data, New Ideas. Curr. Anthropol. 2011, 52, S221–S235. [Google Scholar] [CrossRef] [Green Version]
- Yurtman, E.; Özer, O.; Yüncü, E.; Dağtaş, N.D.; Koptekin, D.; Çakan, Y.G.; Özkan, M.; Akbaba, A.; Kaptan, D.; Atağ, G.; et al. Archaeogenetic analysis of Neolithic sheep from Anatolia suggests a complex demographic history since domestication. Commun. Biol. 2021, 4, 1279. [Google Scholar] [CrossRef]
- Christian, D. Silk Roads or Steppe Roads? The Silk Roads in World History. J. World Hist. 2000, 11, 1–26. [Google Scholar] [CrossRef]
- Curtin, P.D. Cross-Cultural Trade in World History; Cambridge University Press: Cambridge, UK, 1985. [Google Scholar]
- Franck, I.; Brownstone, D. The Silk Road, 1st US ed.; Facts on File, Inc.: New York, NY, USA, 1986; p. 39. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).