Abstract
Paphiopedilum armeniacum S. C. Chen et F. Y. Liu is an endangered lady’s slipper orchid species with high horticultural value. As observed for other orchids, mycorrhizal fungi and endophytic bacteria play important roles in the growth and development of P. armeniacum. In the present study, the community structure dynamics across three growth and development stages of cultivated P. armeniacum were investigated. The potential interactions between Tulasnellaceae fungi and core bacterial genera on one hand and the stability of the presumed mycorrhizal fungi communities on the other were analyzed in three growth stages of P. armeniacum to enhance our understanding of endophytic microbial community structure dynamics in the roots at different development stages. Based on sequencing, 3 and 16 phyla and 59 and 269 genera were identified in the fungal and bacterial communities, respectively. The predominant fungi and bacteria were Basidiomycota (62.90%) and Proteobacteria (43.98%), which exhibited changes in abundance and diversity depending on the growth stage of P. armeniacum. Assessment of the entire microbial communities from different growth stages showed that the seedling stage had the highest richness and diversity. The microbial communities recruited by P. armeniacum at the seedling stage were different from those recruited at the vegetative and reproductive growth stages, and the microbial communities recruited in the latter two stages overlapped. Tulasnellaceae were the only dominant fungal symbionts during P. armeniacum growth. Brevibacillus, Mycobacterium, and Sphingomonas, the three core genera, showed significant interactions with the main OTUs of Tulasnellaceae. Putative mycorrhizal fungi in P. armeniacum were relatively stable across different growth environments, and the core mycorrhizal fungi were uncultured Tulasnellaceae (OTU1). This could facilitate the ex situ conservation and commercial development of the endangered orchid.
1. Introduction
Paphiopedilum Pfitzer is the largest genus of the subfamily Cypripedioideae, which comprises about 96–100 species around the globe [1,2]. Paphiopedilum armeniacum S. C. Chen et F. Y. Liu is an endangered orchid distributed in Baoshan, Gongshan, Fugong, Yunlong, Lanping, and Lushui counties in western Yunnan, China, as well as in neighboring Myanmar [3]. It is an attractive and popular horticultural species due to its unique labellum and its bright yellow flowers. Despite their high horticultural value, wild P. armeniacum populations are under a constant threat of extinction due to overcollection and habitat destruction. As a result, P. armeniacum is listed as endangered in the IUCN Red List of Threatened Species, as well as in the Convention on International Trade in Endangered Species of Wild Fauna and Flora Appendix I, and its international trade is prohibited [4].
Similar to other orchid species, mycorrhizae play a vital role in the life cycle and evolutionary history of Paphiopedilum [5,6]. Research in Paphiopedilum is relatively basic compared to that in other orchids [7,8]. Many studies on Paphiopedilum endophytes have mainly focused on the isolation and identification of mycorrhizal fungi. The endophytic fungi isolated from Paphiopedilum are mainly Basidiomycetes, including Tulasnellaceae and Ceratobasidiaceae, as well as the genera Fusarium and Chaetomium in the phylum Ascomycota [8,9]. Among them, Tulasnellaceae are the main symbiotic fungi in Paphiopedilum and promote the growth and development of their host plants [10,11]. In a previous study, the endophytic fungi of P. armeniacum were investigated using cultivable methods, and the authors reported that they were mainly members of uncultured Tulasnellaceae and of the genera Fusarium and Chaetomium, with uncultured Tulasnellaceae fungi accounting for the largest proportion, at 50%, and representing the dominant taxa [12]. In another study, Tulasnella calospora, isolated from the root of P. armeniacum, promoted the growth of host tissue culture seedlings [13]. Furthermore, following the investigation of mycorrhizal fungi of P. armeniacum using cultivation-independent methods, the fungi were characteriezd by six internal transcribed spacer (ITS) sequence types and were clustered into three major clades following the phylogenetic analysis of 5.8 S sequences of ribosomal DNA [14].
Fungi colonization can occur at various stages of plant growth [15]. At least for the germination process, orchids rely entirely on orchid mycorrhizal fungi (OMF) for the resources required for growth and development [16]. OMF specificity patterns in orchid seedlings and adults vary among orchid species [17]. Some saprophytic orchids species, such as Chamaegastrodia inverta, are associated with a restricted group of OMF throughout their lifecycle [18,19,20]. However, in Cephalanthera and other autotrophic terrestrial orchids, the fungi detected in the seedlings represent only a subset of the broader range observed in germinating seeds and mature plants; furthermore, mycorrhiza can shift from relationships with a single fungus to relationships with various fungi [18,21,22,23]. However, it remains unclear whether fungal taxa shift across the life history stages in Paphiopedilum.
Similar to plant–fungus symbiosis, bacterial associations are vital for orchid growth and development [24,25]. Numerous orchid endophytic bacteria enhance plant growth, fitness, and several other ecological or evolutionary processes in orchids. Previous studies have demonstrated that bacteria isolated from orchids can enhance plant growth or resistance to pathogens [26]. In addition, in a previous study, interactions between endophytic fungi and endophytic bacteria had significant growth promotion effects on tissue culture seedlings of Dendrobium candidum [27]. Furthermore, Mycorrhiza Helper Bacteria (MHB) closely associated with mycorrhizal fungi have a stimulating effect on mycelial growth and mycorrhiza formation [28]. Suárez identified several bacterial strains associated with Serendipita spp., the orchid mycorrhizal fungus [29].
The objectives of the present study were (1) to investigate the root endophytic microbiome communities associated with cultivated P. armeniacum at different developmental stages, based on Illumina sequencing approaches; (2) analyze the relationships between core bacterial groups and mycorrhizal fungi of cultivated P. armeniacum; (3) compare the differences of presumed mycorrhizal fungi of P. armeniacum observed in this study with those reported in previous studies by phylogenetic analysis.
2. Materials and Methods
2.1. Sample Collection and Surface Sterilization
To assess endophytic microbial community structures across different development stages of cultivated P. armeniacum, 15–20 plants in each stage were collected randomly. We collected 1–2 healthy roots from each plant. All plants were obtained from seed germination by tissue culture in vitro. The seedling-stage plants were tissue culture plants transplanted into the greenhouse for 1 month, with a height of about 7–9 cm, and are referred to as phase S. The vegetative growth-stage plants had been planted in the greenhouse for two years and are referred to as phase M. Finally, the reproductive growth-stage plants had experienced their first bloom and are referred to as phase L (Figure 1).
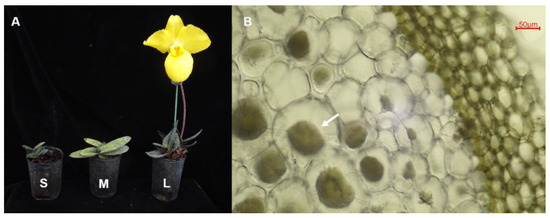
Figure 1.
Different developmental stages of Paphiopedilum armeniacum (A) and freehand section of about 30 μm showing intracellular fungal pelotons in the roots (B). S: seedling stage; M: vegetative growth stage; L: reproductive growth stage. The white arrow indicates the intracellular fungal pelotons in the root.
All the samples were collected on 3 April 2019 from a glass greenhouse with a water curtain and blowers for cooling and ventilation in Xingyi, Guizhou, southwest China. The plants were potted in a bark substrate for orchids, under no more than 800 μmol·m−2·s−1 of natural light maintained by a sunshade net. The average temperature and relative humidity were 10–32 °C and 70–90%, respectively. In the process of cultivation, stability and proportion of the substrate were maintained as far as possible. Meanwhile, part of the original substrate was retained during the repotting process.
After careful excavation from the culture substrate, the roots were transported to the laboratory at 4 °C. All the vegetative- and reproductive-stage roots were checked microscopically for mycorrhizal colonization (Figure 1), and the root segments with mycorrhizal colonization were selected for molecular analyses. The distinct intracellular fungal pelotons were not obvious in the seedling stage, hence fifteen seedling-stage roots were randomly selected. After microscopy, they were surface-sterilized immediately (30 s submergence in 75% ethanol, followed by a 30 s rinse step in sterile distilled water, 5 min submergence in 1% sodium hypochlorite, and five 30 s rinse steps in sterile distilled water). The sterilized roots from each of the growth stages were then cut separately into 2–3 mm small pieces, evenly mixed in equal proportions, and divided into three replicates for molecular analysis. To confirm the sterilization process was successful, the final water rinse was also collected for molecular analysis as a negative control.
2.2. DNA Extraction and PCR Amplification
DNA was extracted from samples and negative control using the QIAamp DNA Stool Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. DNA quality was checked by 1% agarose gel electrophoresis, and DNA concentrations and purity were determined using a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, NC, USA). Primers 799F (5′- AACMGGATTAGATACCCKG -3′) and 1193R (5′- ACGTCATCCCCACCTTCC-3′) targeting the V5–V7 regions of 16S rRNA genes and Primers ITS3F (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4-OF (5′- GTTACTAGGGGAATCCTTGTT-3′) targeting the fungal ITS-2 regions were used for PCR [30,31]. The library was constructed by two-step PCR amplification. The first step involved amplifying the target fragment with specific primers. The initial PCR reactions were performed in 50 μL reaction volumes with 1–2 μL of DNA template, 1 μL of dNTPs at 10 mM, 5× reaction buffer, and 1U of Phusion DNA Polymerase (New England Biolabs, Ipswich, MA, USA). PCR conditions consisted of initial denaturation at 94 °C for 2 min, followed by 29 (16S rRNA) or 35 (ITS) cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s, with a final extension at 72 °C for 5 min. The PCR products were recovered using an AxyPrep DNA gel recovery kit (AXYGEN Inc., Union city, CA, USA) and quantified using an FTC-3000TM Real-Time PCR instrument; the samples were mixed according to a mole ratio similar to that for secondary PCR amplification. The role of the secondary PCR amplification was to add the connector and the sequence primer and barcode required by the Illumina platform to both ends of the target fragment. The PCR reactions were carried out in 40 μL reaction volumes with 5 μL of DNA template, 1 μL of dNTPs at 10 mM, 8 μL of 5× reaction buffer, and 1U of Phusion DNA polymerase. The cycling conditions consisted of one cycle at 94 °C for 2 min, followed by eight cycles at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s, followed by a final extension cycle at 72 °C for 5 min. The subsequent sequencing analysis was not performed on the negative control, as the target band was not detected in the first PCR amplification.
2.3. Statistical Analyses
Raw fastq files were demultiplexed based on the corresponding barcode. Paired-end reads for all samples were run through Trimmomatic v0.35 to remove low-quality base pairs using the following parameters: SLIDINGWINDOW: 50:20 MINLEN: 50 [32]. FLASH v1.2.11 was then used to merge trimmed reads with default parameters. Low-quality contigs were removed using the screen.seqs command based on the following filtering parameters: maxambig = 0, minlength = 200, maxlength = 485, maxhomop = 8. The fragments were analyzed using a combination of Mothur v1.33.3 [33], UPARSE (usearch v8.1.1756, [http://drive5.com/uparse/ (accessed on 16 November 2021)], and R v3.2.3). The demultiplexed reads were clustered into operational taxonomic units (OTUs) at 97% sequence identity using the UPARSE pipeline (http://drive5.com/usearch/manual/uparsecmds.html (accessed on 16 November 2021)). The representative sequences in each OTU were obtained and mapped to Silva128 and UNITE (2019_version8) reference databases using the Ribosomal Database Project classifier (https://sourceforge.net/projects/rdp-classifier/ (accessed on 16 November 2021)) for the bacterial and fungal communities, respectively. OTUs classified as mitochondria or chloroplasts, or less than 1 sequences, were filtered from the datasets. Downstream analysis and visualization based on OTU tables was implemented in R v3.6.0 (https://cran.R-project.org/ (accessed on 16 November 2021)), including alpha- and beta-diversity analysis using QIIME 2 and vegan 2.5-6 package in R [34]. Differences were compared mainly based on Tukey’s test. Visualization was carried out using the ggplot2 3.2.1 package in R [35] and imageGP (http://www.ehbio.com/ImageGP (accessed on 16/11/2021)). All the raw sequences from the samples were deposited in the NCBI SRA database under BioProject PRJNA793335 for bacteria and PRJNA793341 for fungi.
2.4. Covariation Network Analysis of Core Bacterial Genera and Mycorrhizal Fungi
To explore the interactions between root core bacterial communities and mycorrhizal fungi of P. armeniacum, a covariation network diagram was constructed based on the correlation coefficients between core bacterial genera and mycorrhizal fungi.
The OTUs that appeared in all samples were defined as “core OTUs” [36,37]. Only core bacterial genera accounting for more than 1% of the total reads were used in the calculations. The Spearman rank correlation coefficient between two different populations was calculated using R v3.6.0, and the co-variation networks of different populations were constructed using igraph v1.2.4.2 [38]. The correlation P value less than 0.05 was plotted.
2.5. Comparative Analysis of Tulasnella in Paphiopedilum armeniacum
To analyze the stability of the Tulasnellaceae fungal communities in P. armeniacum samples, we generated a presumed Tulasnellaceae dataset of P. armeniacum using data gathered from a previous study [14], in addition to data obtained from the present study. Phylogenetic trees were generated using ITS sequences of each of the sequences obtained from BLAST searches (Fasta S1). All sequences were aligned in MAFFT v7.311 [39] and manually adjusted in MEGA X [40]. MEGA X was used to construct a maximum likelihood (ML) tree under the K2+G+I model, with 1000 bootstrap replicates. Phylogenetic trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 16 November 2021)). Branches that received bootstrap support for ML greater than 50% or equal 0.70 were considered significantly supported.
3. Results
A total of 423,337 and 379,593 high-quality reads were obtained from the ITS2 and 16S rRNA gene sequencing data, respectively. After discarding non-bacterial, non-fungi, mitochondrial, chloroplast, and low-abundance OTUs, we obtained 199 and 828 fungal and bacterial OTUs, respectively, at 97% similarity. After subsampling, 813 bacterial OTUs and 195 fungi OTUs were observed (Table S1). The observed OTUs and the numbers of unique or shared OTUs identified in different development stages are illustrated in Figure 2. The seedling stage had the highest number of observed OTUs (Figure 2A,C). Overall, 432 and 157 (bacterial and fungal) OTUs were unique to different stages (118/94, 244/26, and 70/37, in the seedling, vegetative, and reproductive stage, respectively), whereas 176 and 17 OTUs were shared among all three stages (Figure 2B,D).
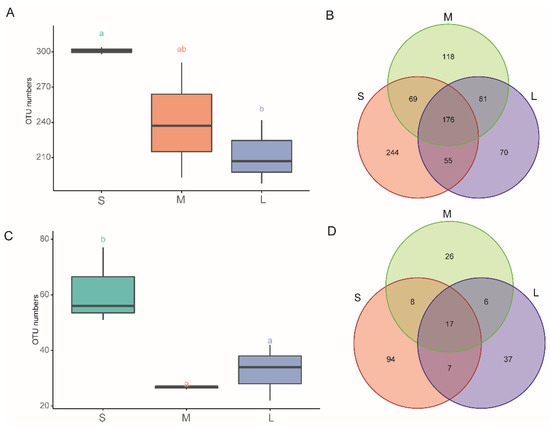
Figure 2.
OTU numbers and Venn analysis of exclusive and shared Operational Taxonomic Units (OTUs) in different samples. (A) Number of OTUs in the bacterial community; (B)Venn diagram of the bacterial community; (C) number of OTUs in the fungal community; (D) Venn diagram of the fungal community. S, M, and L represent the seedling stage, vegetative growth stage, and reproductive growth stage of Paphiopedilum armeniacum, respectively. Lowercases a and b in the graphs indicate significant differences between the different samples, determined using Tukey’s test (p < 0.05).
3.1. Diversity Analysis in Different Samples
Microbiota diversity and richness in different samples were investigated using Chao1 and Shannon indexes based on ITS and V3–V4 sequence data. According to the Chao1 index results, the seedling stage samples had higher species richness than the samples from the other stages in both the fungal and the bacterial dataset (Figure 3A,C). In addition, significant differences were observed in fungal richness among the samples (Figure 3C, p < 0.05). Similar results were observed based on the Shannon index values, with higher diversity in fungal and bacterial communities in seedling-stage samples than in samples from the other two stages, with significant differences in bacterial diversity among the samples (Figure 3B, p < 0.05).
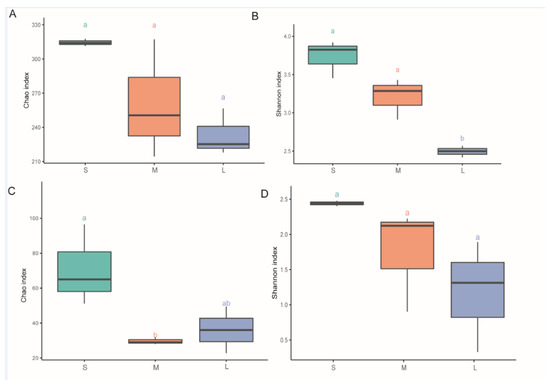
Figure 3.
Diversity indices in different developmental phases of Paphiopedilum armeniacum. (A,B) Chao and Shannon indices of the bacterial community, respectively; (C,D) Chao and Shannon indices of the fungal community, respectively. S, M, and L represent the seedling stage, vegetative growth stage, and reproductive growth stage of Paphiopedilum armeniacum, respectively. Lowercase letters a and b in the graphs indicate significant differences between the different samples, determined based on Tukey’s test (p < 0.05).
In the bacterial microbiomes, measures of within-sample diversity (α-diversity indices) showed decreases in microbial richness and diversity with the development of P. armeniacum based on the Chao1 index and Shannon index, respectively, showing that in both bacterial and fungal communities, higher richness and diversity were observed in the seedling stage than in the other development stages.
The relationships among the microbial communities in the different samples were investigated using Principal Coordinate Analysis (PCoA) based on Bray–Curtis distance matrix and unweighted UniFrac distances. The PCoA results indicated that the microbiota in the seedling stage samples was distinct from that in the other stages.
The two axes explain 76.2% of the total variation in V3–V4 sequences (Figure 4A), as well as 81.9% of the total variation in ITS sequences (Figure 4C). In the heatmap, different distances were observed between the microbial communities of individual samples (Figure 4). In the case of the fungal community, the seedling-stage samples tended to cluster closer, while samples in the other stages formed another cluster. It was not obvious in hierarchical clustering analysis of the bacterial community, but the PCoA also highlighted differences between the seedling stage and the other two stages.
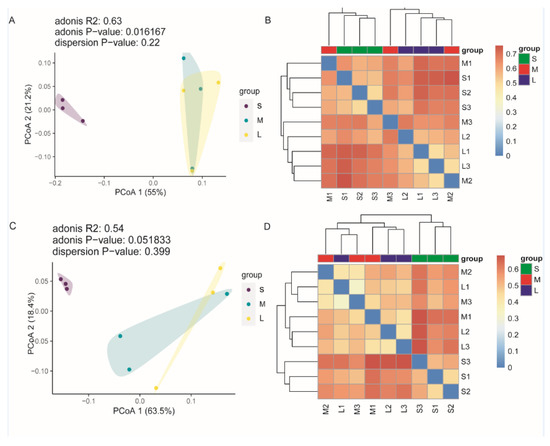
Figure 4.
Principal coordinate analysis (PCoA) of microbial community structures based on the Bray–Curtis distance matrix and a heatmap based on unweighted UniFrac distances. (A,B) PCoA analysis and unweighted UniFrac (UUF) heatmap analysis of the bacterial community, respectively; (C,D) PCoA analysis and UUF heatmap analysis of the fungal community, respectively.
3.2. Taxonomic Assignment of the Microbial Community Composition
In total, 3 and 16 phyla and 9 and 27 classes were identified in the fungal and bacterial communities, respectively. In the bacterial communities (Figure 5A, Table S1), the dominant phyla (>1%) were Proteobacteria, Firmicutes, and Actinobacteria, with average relative abundances of 43.98%, 38.51%, and 15.81%, respectively. In the fungal communities (Figure 5B, Table S1), the dominant phyla across all samples were Ascomycota and Basidiomycota, with average relative abundances of 62.94% and 18.60%, respectively. The average relative abundance of Proteobacteria in the seedling stage was higher than that in the other stages, while the average relative abundance of Actinobacteria displayed an opposite trend (Figure S1). Fungal phyla’s relative abundances changed considerably in the course of P. armeniacum development. The average relative abundance of Basidiomycota increased during P. armeniacum development, whereas the average relative abundance of Ascomycota decreased significantly in the course of development (Figure S1).
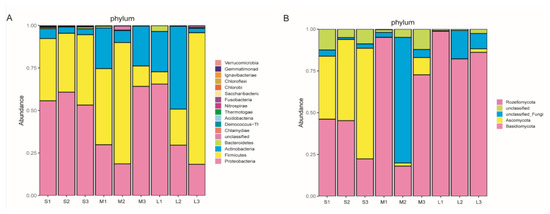
Figure 5.
Microbial community structure at different developmental stages of Paphiopedilum armeniacum at the phylum level. (A) Relative abundance of bacterial taxa of each group. (B) Relative abundance of fungal taxa of each group.
A detailed analysis of the relative distributions of the sequences from the phylum to genus levels revealed significant variations in the course of Paphiopedilum armeniacum development (Table S2). Among the bacterial genera, the sequences assigned to Caldibacillus, Paenibacillus, Uruburuella, and Vibrio were significantly enriched in the seeding stage. Conversely, among the fungal genera, unclassified Tulasnellaceae showed a significantly increase during P. armeniacum development (Table S2).
3.3. Covariation Network Analysis of Core Bacterial Genera and Mycorrhizal Fungi
Co-occurrence networks can display interrelationships among various microorganisms and reveal effects of microorganisms overlooked in difference comparisons based on abundance [41]. The core microbiota potentially performs critical functions within its habitats. In the present study, there were obvious interactions between bacteria and fungi based on the covariation networks of core bacterial genera and mycorrhizal OTUs (Figure 6). Mycobacterium was significantly correlated with seven genera, whereas Sphingomonas and Brevibacillus were significantly correlated with six genera. Excluding Cupriavidus, there were significant correlations among other core bacterial genera. OTU1, OTU3, OTU11, and OTU33 were significantly correlated with bacterial genera, with OTU33 being significantly correlated with six bacterial genera. Mycobacterium, Sphingomonas, Brevibacillus, and OTU33, the four core genera, interacted with each other and influenced seven bacterial genera and three OTUs of Tulasnella species. The covariation network analysis results showed significant interactions between core bacterial genera and OTUs of Tulasnella species.
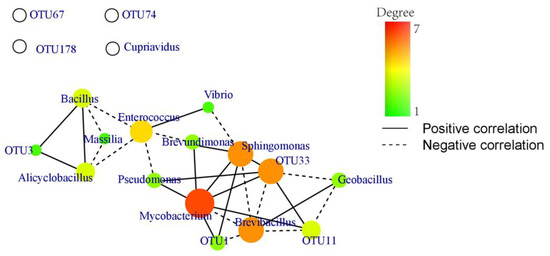
Figure 6.
Covariation network of core bacterial genera and outs of Tulasnella species in Paphiopedillum armeniacum. Degree: the number of microbial genus associated with other genus. The value of degree increases gradually from green to red.
3.4. Comparison of the Mycorrhizal Fungi of P. armeniacum in Xingyi Greenhouse and in Other Habitats
Seven Tulasnellaceae OTUs, designated OTU1, OTU3, OTU11, OTU33, OTU67, OTU74, and OTU178, which accounted for 39.54% of the total reads, were observed in the present study. The sequences were BLAST searched against the GenBank database (National Center for Biotechnology Information) for identification and the sequences deposited in the GenBank database (Accession No: OL891761-67) (Table 1). Phylogenetic analysis showed that all the sequences were roughly divided into four groups (Figure 7). OTU1 (type1) and OTU3 (type7) appeared closely related to mycorrhizal fungi from different habitats of P. armeniacum observed in previous studies [14]. OTU11, OUT33, and FJ786646(type20) formed one branch; OTU178 resulted similar to OTU1, whereas OTU67 formed an independent branch. The seven OTUs were distributed distinctly among the different growth phases of P. armeniacum (Table 1). OTU1 was observed in all developmental stages of P. armeniacum and was the only mycorrhizal fungus in the seedling stage. OTU11 and OTU178 were only observed in the reproductive growth stage, whereas OTU74 was only observed in the vegetative growth stage.

Table 1.
List of mycorrhizal fungi operational taxonomic units (OTUs) in Paphiopedillum armeniacum, and relative abundance of each OTU at each development stage.
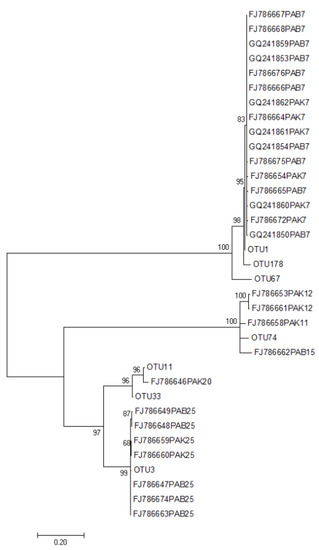
Figure 7.
Maximum likelihood (ML) tree based on internal transcribed spacers (ITS) nuclear ribosomal DNA sequences of Tulasnellaceae fungi obtained from all samples and the GenBank database. Bootstrap (BS) values from 1000 replications in the ML analysis are indicated at the branches of the nodes. Scale bar is shown to infer evolutionary distances. PAB represent Paphiopedillum armeniacum from Baoshan (wild); PAK represents P. armeniacum from Kunming (greenhouse). The sequences with NCBI serial numbers are mycorrhizal fungi sequences obtained from P. armeniacum by Huang [14] and typing of the sequences.
4. Discussion
In the present study, the authors investigated microbial community structure dynamics across three growth and development stages of cultivated P. armeniacum. In addition, Tulasnellaceae fungi of P. armeniacum at different growth stages were analyzed to gain insights into their mycorrhizal features. Most studies on the root biomes of orchids have mainly focused on specific stages of growth, with little attention directed at trends during the development of cultivated orchids [5,6,42,43,44,45].
The composition of endophyte communities is governed by host genotype and developmental stage, as well as by the environment. However, the significate of the effects may differ between studies [42]. When compared to environmental factors, greater influence of the host plant was observed in root endophytes in some research. Xiong et al. demonstrated that microbiome assembly along the soil–plant continuum is shaped predominantly by compartment niche and host species rather than by site or fertilization practice [43]. Strong cultivar-dependent variations in the fungal and bacterial microbiome were found for Cannabis sativa [44,45]. In order to minimize the impact of the substrate on endophytes, the stability and proportion of the substrate were maintained as far as possible. Meanwhile, part of the original substrate was retained during the potting process in this study. Host effects have been reported in many studies. The common OTUs of different growth stages and the genus of the core bacteria indicated the influence of host selection by P. armeniacum to a certain extent.
Higher richness and diversity were observed in the seedling stage than in the other development stages for both bacterial and fungal datasets, indicating that P. armeniacum may rely on interactions with more fungi or bacteria during its early growth than in the later growth stages, which is similar to what observed for Gymnadenia conopsea, in which microbial diversity in root samples was lower in the reproductive growth stage than in the vegetative growth stage [46]. The PCoA analysis results revealed that microbial community taxa were clustered at the seedling stage; no obvious clustering was observed at the other two stages. The results suggest that the microbial communities recruited by P. armeniacum at the initial growth stages were distinct from those in the following two stages, and there was a certain degree of overlap in microbial communities during the vegetative growth stage and the reproductive growth stage; some of these microorganisms could participate in essential processes.
Many endophytic bacteria isolated from orchid tissues play important functions for their hosts, including plant growth promotion [25,47,48,49,50]. However, little is known about orchid-associated microbial community structure in the course of growth and development of P. armeniacum. Across the three development stages of P. armeniacum defined in the present study, Proteobacteria, Firmicutes, and Actinobacteria were the dominant groups, which is consistent with findings reported for other orchids [51,52]. The bacterial genera that exhibited the greatest differences among the three development stages were Caldibacillus, Paenibacillus, Uruburuella, and Vibrio, which also represent the genera most commonly found as bacterial endophytes [53,54]. The genus Caldibacillus includes thermophilic and aerobic or facultative anaerobic bacilli, and some species in the genus exhibit cellulase activity [55]. Bacteria in the genus Paenibacillus, which can produce indole-3-acetic acid, are commonly reported to promote plant growth and increase root development [47,56]. In addition, some Vibrio strains have been reported to participate in nitrogen fixation or salicylic acid synthesis [57,58]. Changes in relative abundance at the genus level indicated that P. armeniacum may selectively modulate microbial communities to meet its requirement during growth.
Interactions between fungi and bacteria are beneficial for the growth of orchids [27,49,59]. The obvious interactions between OTUs from Tulasnellaceae and core genera indicated that fungi are also key factors influencing bacterial community composition, especially in the orchid microbiota. Whereas orchids fully or partially rely on mycorrhizal fungi, some bacteria, such as MHB, commonly occur in ectomycorrhiza and in arbuscular mycorrhizal associations, which facilitate mycorrhiza formation [60,61]. In the co-occurrence network analysis in the present study, Brevibacillus, Mycobacterium, and Sphingomonas were core bacteria. In previous studies, Brevibacillus spp. were considered an MHB, whereas Mycobacterium spp. and Sphingomonas spp. were reported to have plant growth promotion effects [62,63,64].
As show in the results, a lot of non-mycorrhizal microorganisms were detected in the roots of P. armeniacum (Tables S1 and S2). The composition of non-mycorrhizal fungi, which were highly abundant in the seedling stage, began to converge throughout the vegetative stage and stabilized during the reproductive phase. This indicated that P. armeniacum screened for non-mycorrhizal fungi depending on the development of growth. Some non-mycorrhizal microorganisms are helpful for seed germination or seedling growth of orchids [9,21]. Fusarium, which was found in the root of P. armeniacum, is reported to form hyphae inside the cells and presented germinative potential, favoring seed germination or seedling growth of orchids [65,66]. Whether these strains are strictly mycorrhizal fungi requires further verification by methods such as stable isotope analyses.
Tulasnellaceae is considered a dominant group among the mycorrhizal fungi of orchids [67,68]. Researchers identified Tulasnellaceae through uncultured techniques as potentially mycorrhizal in orchids. In the present study, Tulasnellaceae were the only dominant fungal symbionts in P. armeniacum and were associated with the three growth and development stages. The compositions and levels of specificity of fungal partners can vary [69,70,71]. In the present study, different growth stages of P. armeniacum harbored different types and amounts of Tulasnellaceae OTUs. The seedling stage was only associated with one Tulasnellaceae OTU (OTU1), while five and six Tulasnellaceae OTUs were observed in the vegetative growth and reproductive growth stages, respectively. Among the seven Tulasnellaceae OTUs detected in the present, only OTU1 (Type7) was observed in all three development stages. Considering the findings of previous studies on mycorrhizal fungi of P. armeniacum [12,13,14], OTU1 may be the core group in P. armeniacum mycorrhizal fungi in different growth environments, geographical locations, and developmental stages and is also the first group to be recruited from the environment. OTU3 (Type25) is the main mycorrhizal group that was present in other habitats. The closest matches in GenBank for OTU11 and OTU33 were isolated from Paphiopedilum micranthum in Xingyi greenhouse, which indicated that OTU11 and OTU33 in P. armeniacum might be derived from horizontal transfer in the environment. OTU11, OTU74, and OTU178 were only associated with one stage of development. OTU67 and OTU178 may be novel taxa recruited from the environment, or they may not have been detected before due to the limited techniques used in previous research. Mycorrhizal fungi were similar in wild and cultivated P. armeniacum, indicating that the cultivated P. armeniacum may survive when transplant in the wild. Meanwhile, the similarity in mycorrhizal fungi makes ex situ conservation or even propagation by means of mycorrhization of axenically grown seedlings possible. Considering the important roles of bacteria and fungi in orchid growth and development [25,49], a more comprehensive study should be carried out in the wild P. armeniacum.
Due to overharvest, habitat loss, and degradation, wild populations of P. armeniacum have declined drastically. Conservation and restoration of the populations of this endangered orchid could facilitate the maintenance of its biodiversity as well as ecosystem stability. It is essential to comprehensively consider the structures of the resident microbial communities in this endangered orchid [72,73,74]. Meanwhile, although Illumina-based sequencing has some certain benefits for understanding the non-culturable endophytes of P. armeniacum, culture-depended methods are also necessary. In the future, more culturomics research is needed to provide strain resources in ecology and conservation research and practice.
5. Conclusions
In the present study, the diversity of microbial communities at different cultivated P. armeniacum development stages was examined based on Illumina sequencing approaches. Microbial community diversity and composition varied across the development stages. The seedling stage presented the highest microbial community richness and diversity, which indicated that P. armeniacum may need the interaction with more fungi or bacteria during the seedling stage. In addition, the core bacterial genera and the main OTUs of Tulasnellaceae showed significant interactions, according to the covariation network analysis results. Tulasnellaceae were the only dominant fungal symbionts during P. armeniacum growth. The results of the present study could facilitate the ex situ conservation and commercial development of endangered P. armeniacum populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14050321/s1, Fasta S1: Sequences of Tulasnellaceae displayed in phylogenetic trees; Table S1: All OTU sequences, profiling, and annotations. Table S2: Significant variations in genera in the course of Paphiopedilum armeniacum development; Figure S1: Shifts in microbial (bacterial and fungal) taxonomic composition at the main phylum level in the course of P. armeniacum development.
Author Contributions
X.C. performed the experiments and wrote the manuscript; T.W. and Y.C. analyzed the data; X.W. prepared the plant materials; N.Y. reviewed and supervised the research. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Fundamental Research Funds of CAF (CAFYBB2019SY001) and National Natural Science Foundation of China (No. 31800523).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data used in the study is available upon request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cribb, P. The Genus Paphiopedilum, 2nd ed.; Natural History Publications: Kota Kinabalu, Malaysia, 1998; pp. 254–260. [Google Scholar]
- Wu, Z.Y.; Raven, P.H.; Hong, D.Y. Flora of China, Volume 25 (Orchidaceae); Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2009; pp. 33–44. [Google Scholar]
- Chen, S.C.; Liu, F.Y. Notes on some species of Paphiopedilum from Yunnan. Acta Bot. Yunnanica 1982, 4, 163–167. [Google Scholar]
- Cites, Appendices I, II and III. Available online: http://www.cites.org/eng/app/appendices.php (accessed on 22 June 2021).
- Han, J.Y.; Xiao, H.F.; Gao, J.Y. Seasonal dynamics of mycorrhizal fungi in Paphiopedilum spicerianum (Rchb. f) Pfitzer—A critically endangered orchid from China. Glob. Ecol. Conserv. 2016, 6, 327–338. [Google Scholar] [CrossRef]
- Parthibhan, S.; Ramasubbu, R. Mycorrhizal and endophytic fungal association in Paphiopedilum druryi (Bedd.) Stein—A strict endemic and critically endangered orchid of the Western Ghats. Ecol. Genet. Genom. 2020, 16, 100059. [Google Scholar] [CrossRef]
- Liu, Z.J.; Chen, X.Q.; Chen, L.J.; Lei, S.P. The Genus Paphiopedilum in China; Science Press: Beijing, China, 2009. [Google Scholar]
- Wang, X.G.; Yan, H.X.; Li, X.L.; He, J.Z.; Zhou, Z.G. Identification and Growth Promoting Analysis of Mycorrhizal Fungi from Paphiopedilum hirsutissimun (Orchidaceae). Southwest China J. Agric. Sci. 2021, 34, 119–125. [Google Scholar] [CrossRef]
- Khamchatra, N.; Dixon, K.W.; Tantiwiwat, S.; Piapukiew, J. Symbiotic seed germination of an endangered epiphytic slipper orchid, Paphiopedilum villosum (Lindl.) Stein. from Thailand. S. Afr. J. Bot. 2016, 104, 76–81. [Google Scholar] [CrossRef]
- Sutthinon, P.; Rungwattana, K.; Suwanphakdee, C.; Himaman, W.; Lueangjaroenkit, P. Endophytic Fungi from Root of Three Lady’s Slipper Orchids (Paphiopedilum spp.) in Southern Thailand. Chiang Mai J. Sci. 2021, 48, 853–866. [Google Scholar]
- Yang, W.-K.; Li, T.-Q.; Wu, S.-M.; Finnegan, P.M.; Gao, J.-Y. Ex situ seed baiting to isolate germination-enhancing fungi for assisted colonization in Paphiopedilum spicerianum, a critically endangered orchid in China. Glob. Ecol. Conserv. 2020, 23, e01147. [Google Scholar] [CrossRef]
- Yi, S. Mycorrhizal Fungi and Symbiotic Seed Germination of Four Paphiopedilum Species; Beijing Forestry University: Beijing, China, 2017; pp. 19–29. [Google Scholar]
- Zhu, X.-M.; Hu, H.; Li, S.-Y.; Yan, N. Interaction between Endophytic Fungi and Seedlings of Two Species of Paphiopedilum during Symbiotic Culture. Plant Divers. Resour. 2012, 34, 171–178. [Google Scholar] [CrossRef]
- Yuan, L.; Yang, Z.L.; Li, S.-Y.; Hu, H.; Huang, J.-L. Mycorrhizal specificity, preference, and plasticity of six slipper orchids from South Western China. Mycorrhiza 2010, 20, 559–568. [Google Scholar] [CrossRef]
- Meng, Y.-Y.; Zhang, W.-L.; Selosse, M.-A.; Gao, J.-Y. Are fungi from adult orchid roots the best symbionts at germination? A case study. Mycorrhiza 2019, 29, 541–547. [Google Scholar] [CrossRef]
- Rasmussen, H.N. Recent developments in the study of orchid mycorrhiza. Plant Soil 2002, 244, 149–163. [Google Scholar] [CrossRef]
- Rasmussen, H.N.; Dixon, K.; Jersakova, J.; Těšitelová, T. Germination and seedling establishment in orchids: A complex of requirements. Ann. Bot. 2015, 116, 391–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidartondo, M.I.; Read, D.J. Fungal specificity bottlenecks during orchid germination and development. Mol. Ecol. 2008, 17, 3707–3716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, R.D.; Barrett, M.D.; Dixon, K.W.; Hopper, S.D. Do mycorrhizal symbioses cause rarity in orchids? J. Ecol. 2011, 99, 858–869. [Google Scholar] [CrossRef]
- Pecoraro, L.; Wang, X.; Venturella, G.; Gao, W.; Wen, T.; Gafforov, Y.; Gupta, V.K. Molecular evidence supports simultaneous association of the achlorophyllous orchid Chamaegastrodia inverta with ectomycorrhizal Ceratobasidiaceae and Russulaceae. BMC Microbiol. 2020, 20, 236. [Google Scholar] [CrossRef]
- Meng, Y.-Y.; Shao, S.-C.; Liu, S.-J.; Gao, J.-Y. Do the fungi associated with roots of adult plants support seed germination? A case study on Dendrobium exile (Orchidaceae). Glob. Ecol. Conserv. 2019, 17, e00582. [Google Scholar] [CrossRef]
- McCormick, M.K.; Whigham, D.F.; O’Neill, J. Mycorrhizal diversity in photosynthetic terrestrial orchids. New Phytol. 2004, 163, 425–438. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Brys, R.; Cammue, B.P.A.; Honnay, O.; Lievens, B. Mycorrhizal associations and reproductive isolation in three closely related Orchis species. Ann. Bot. 2011, 107, 347–356. [Google Scholar] [CrossRef] [Green Version]
- Tsavkelova, E. Bacteria Associated with Orchid Roots. In Bacteria in Agrobiology: Plant Growth Responses; Springer Science & Business Media: Berlin, Germany, 2011; pp. 221–258. [Google Scholar]
- Kaur, J.; Sharma, J. Orchid Root Associated Bacteria: Linchpins or Accessories? Front. Plant Sci. 2021, 12, 661966. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Egorova, M.A.; Leontieva, M.R.; Malakho, S.G.; Kolomeitseva, G.L.; Netrusov, A.I. Dendrobium nobile Lindl. seed germination in co-cultures with diverse associated bacteria. Plant Growth Regul. 2016, 80, 79–91. [Google Scholar] [CrossRef]
- Zhang, P.; Zhong, Y.-F.; Song, X.-Q.; Wang, X.-M.; Wang, J. Interaction Effect of Symbiotic Microorganisms on Seedling Growth of Dendrobium catenatum Lindley (Orchidaceae). Plant Sci. J. 2013, 31, 73–79. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Garbaye, J. Mycorrhiza helper bacteria: A promising model for the genomic analysis of fungal-bacterial interactions. New Phytol. 2005, 168, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Novotná, A.; Suárez, J.P. Molecular detection of bacteria associated with Serendipita sp., a mycorrhizal fungus from the orchid Stanhopea connata Klotzsch in southern Ecuador. Bot. Lett. 2018, 165, 307–313. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and Function of the Bacterial Root Microbiota in Wild and Domesticated Barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waud, M.; Busschaert, P.; Ruyters, S.; Jacquemyn, H.; Lievens, B. Impact of primer choice on characterization of orchid mycorrhizal communities using 454 pyrosequencing. Mol. Ecol. Resour. 2014, 14, 679–699. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The Vegan Package. Commun. Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. Int. J. Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kim, P.-J.; Price, N.D. Genetic Co-Occurrence Network across Sequenced Microbes. PLOS Comput. Biol. 2011, 7, e1002340. [Google Scholar] [CrossRef] [PubMed]
- Rasche, F.; Velvis, H.; Zachow, C.; Berg, G.; Van Elsas, J.D.; Sessitsch, A. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J. Appl. Ecol. 2006, 43, 555–566. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.; Li, P.; Wang, G.; Wu, C.; Ge, A.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Comeau, D.; Novinscak, A.; Joly, D.L.; Filion, M. Spatio-Temporal and Cultivar-Dependent Variations in the Cannabis Microbiome. Front. Microbiol. 2020, 24, 491. [Google Scholar] [CrossRef] [Green Version]
- Wei, G.; Ning, K.; Zhang, G.; Yu, H.; Yang, S.; Dai, F.; Dong, L.; Chen, S. Compartment Niche Shapes the Assembly and Network of Cannabis sativa-Associated Microbiome. Front. Microbiol. 2021, 12, 714993. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.-C.; Qin, L.-Y.; He, H.-Y.; Yu, X.-L.; Yang, M.-Z.; Zhang, H.-B. Dynamics of fungal communities during Gastrodia elata growth. BMC Microbiol. 2019, 19, 158. [Google Scholar] [CrossRef]
- Tsavkelova, E.A.; Cherdyntseva, T.A.; Botina, S.G.; Netrusov, A.I. Bacteria associated with orchid roots and microbial production of auxin. Microbiol. Res. 2007, 162, 69–76. [Google Scholar] [CrossRef]
- Faria, D.C.; Dias, A.C.F.; Melo, I.S.; Costa, F.E.D.C. Endophytic bacteria isolated from orchid and their potential to promote plant growth. World J. Microbiol. Biotechnol. 2013, 29, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.A.T.; Tsavkelova, E.A.; Zeng, S.; Ng, T.B.; Parthibhan, S.; Dobránszki, J.; Cardoso, J.C.; Rao, M.V. Symbiotic in vitro seed propagation of Dendrobium: Fungal and bacterial partners and their influence on plant growth and development. Planta 2015, 242, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Altinkaynak, H.; Ozkoc, I. Isolation and molecular characterization of plant growth promoting bacteria from the rhizosphere of orchids in Turkey. Rhizosphere 2020, 16, 100280. [Google Scholar] [CrossRef]
- Pei, C.; Mi, C.; Sun, L.; Liu, W.; Li, O.; Hu, X. Diversity of endophytic bacteria of Dendrobium officinale based on culture-dependent and culture-independent methods. Biotechnol. Biotechnol. Equip. 2017, 31, 112–119. [Google Scholar] [CrossRef] [Green Version]
- Alibrandi, P.; Schnell, S.; Perotto, S.; Cardinale, M. Diversity and Structure of the Endophytic Bacterial Communities Associated With Three Terrestrial Orchid Species as Revealed by 16S rRNA Gene Metabarcoding. Front. Microbiol. 2020, 11, 604964. [Google Scholar] [CrossRef]
- Lin, S.-Y.; Hameed, A.; Liu, Y.-C.; Wen, C.-Z.; Lai, W.-A.; Hsu, Y.-H.; Young, C.-C. Bacillus lycopersici sp. nov., isolated from a tomato plant (Solanum lycopersicum L.). Int. J. Syst. Evol. Microbiol. 2015, 65, 2085–2090. [Google Scholar] [CrossRef]
- Ali, A.; Shahzad, R.; Khan, A.L.; Halo, B.A.; Al-Yahyai, R.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.-J. Endophytic bacterial diversity of Avicennia marina helps to confer resistance against salinity stress in Solanum lycopersicum. J. Plant Interact. 2017, 12, 312–322. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.P.; Monk, C. Purification and characterization of a cellulase (CMCase) from a newly isolated thermophilic aerobic bacterium Caldibacillus cellulovorans gen. nov., sp. nov. World J. Microbiol. Biotechnol. 2004, 20, 85–92. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and Plant-Microbe Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [Green Version]
- Urdaci, M.C.; Stal, L.J.; Marchand, M. Occurrence of nitrogen fixation among Vibrio spp. Arch. Microbiol. 1988, 150, 224–229. [Google Scholar] [CrossRef]
- Bakker, P.A.H.M.; Ran, L.X. Mercado-Blanco, J. Rhizobacterial salicylate production provokes headaches! Plant Soil 2014, 382, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yam, T.W.; Meng, Q.; Zhu, J.; Zhang, P.; Wu, H.; Wang, J.; Zhao, Y.; Song, X. The dual inoculation of endophytic fungi and bacteria promotes seedlings growth in Dendrobium catenatum (Orchidaceae) under in vitro culture conditions. Plant Cell, Tissue Organ Cult. (PCTOC) 2016, 126, 523–531. [Google Scholar] [CrossRef]
- Aspray, T.J.; Jones, E.E.; Davies, M.W.; Shipman, M.; Bending, G.D. Increased hyphal branching and growth of ectomycorrhizal fungus Lactarius rufus by the helper bacterium Paenibacillus sp. Mycorrhiza 2013, 23, 403–410. [Google Scholar] [CrossRef] [PubMed]
- LabbE, J.L.; Weston, D.; Edunkirk, N.; Pelletier, D.A.; Tuskan, G.A. Newly identified helper bacteria stimulate ectomycorrhizal formation in Populus. Front. Plant Sci. 2014, 5, 579. [Google Scholar] [CrossRef] [PubMed]
- Frey-Klett, P.; Garbaye, J.; Tarkka, M. The mycorrhiza helper bacteria revisited. New Phytol. 2007, 176, 22–36. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Cao, Z.; Zhao, K.; Wang, S.; Chen, M.; Hu, X. Growth-promoting Sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 2014, 7, 611–620. [Google Scholar] [CrossRef]
- Pan, L.; Chen, J.; Ren, S.; Shen, H.; Rong, B.; Liu, W.; Yang, Z. Complete genome sequence of Mycobacterium Mya-zh01, an endophytic bacterium, promotes plant growth and seed germination isolated from flower stalk of Doritaenopsis. Arch. Microbiol. 2020, 202, 1965–1976. [Google Scholar] [CrossRef]
- Jiang, J.; Zhang, K.; Cheng, S.; Nie, Q.; Zhou, S.-X.; Chen, Q.; Zhou, J.; Zhen, X.; Li, X.T.; Zhen, T.W.; et al. Fusarium oxysporum KB-3 from Bletilla striata: An orchid mycorrhizal fungus. Mycorrhiza 2019, 29, 531–540. [Google Scholar] [CrossRef]
- Sisti, L.S.; Flores-Borges, D.N.A.; De Andrade, S.A.L.; Koehler, S.; Bonatelli, M.L.; Mayer, J.L.S. The Role of Non-Mycorrhizal Fungi in Germination of the Mycoheterotrophic Orchid Pogoniopsis schenckii Cogn. Front. Plant Sci. 2019, 10, 1589. [Google Scholar] [CrossRef]
- Jacquemyn, H.; Brys, R.; Waud, M.; Busschaert, P.; Lievens, B. Mycorrhizal networks and coexistence in species-rich orchid communities. New Phytol. 2015, 206, 1127–1134. [Google Scholar] [CrossRef]
- Rafter, M.; Yokoya, K.; Schofield, E.J.; Zettler, L.W.; Sarasan, V. Non-specific symbiotic germination of Cynorkis purpurea (Thouars) Kraezl., a habitat-specific terrestrial orchid from the Central Highlands of Madagascar. Mycorrhiza 2016, 26, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Martos, F.; Munoz, F.; Pailler, T.; Kottke, I.; Gonneau, C.; Selosse, M.-A. The role of epiphytism in architecture and evolutionary constraint within mycorrhizal networks of tropical orchids. Mol. Ecol. 2012, 21, 5098–5109. [Google Scholar] [CrossRef] [PubMed]
- Yagame, T.; Ogura-Tsujita, Y.; Kinoshita, A.; Iwase, K.; Yukawa, T. Fungal partner shifts during the evolution of mycoheterotrophy in Neottia. Am. J. Bot. 2016, 103, 1630–1641. [Google Scholar] [CrossRef]
- Huang, H.; Zi, X.-M.; Lin, H.; Gao, J.-Y. Host-specificity of symbiotic mycorrhizal fungi for enhancing seed germination, protocorm formation and seedling development of over-collected medicinal orchid, Dendrobium devonianum. J. Microbiol. 2018, 56, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Xing, X.; Ma, X.; Men, J.; Chen, Y.; Guo, S. Phylogenetic constrains on mycorrhizal specificity in eight Dendrobium (Orchidaceae) species. Sci. China Life Sci. 2017, 60, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: San Diego, CA, USA, 1997. [Google Scholar]
- Gebauer, G.; Preiss, K.; Gebauer, A.C. Partial mycoheterotrophy is more widespread among orchids than previously assumed. New Phytol. 2016, 211, 11–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).