Reducing Carbon Input Improved the Diversity of Bacterial Community in Large-Scale Biofloc Shrimp Culture Facilities
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of Shrimp Culturing Facilities
2.2. Experimental Method
2.3. Feed Feeding and Glucose Addition
2.4. Determination of Water Quality Indicators
2.5. Sequencing and Analysis of Microbial Diversity
2.5.1. DNA Extraction and PCR Amplification
2.5.2. Illumina MiSeq Sequencing
2.5.3. Processing of Sequencing Data
3. Results and Discussion
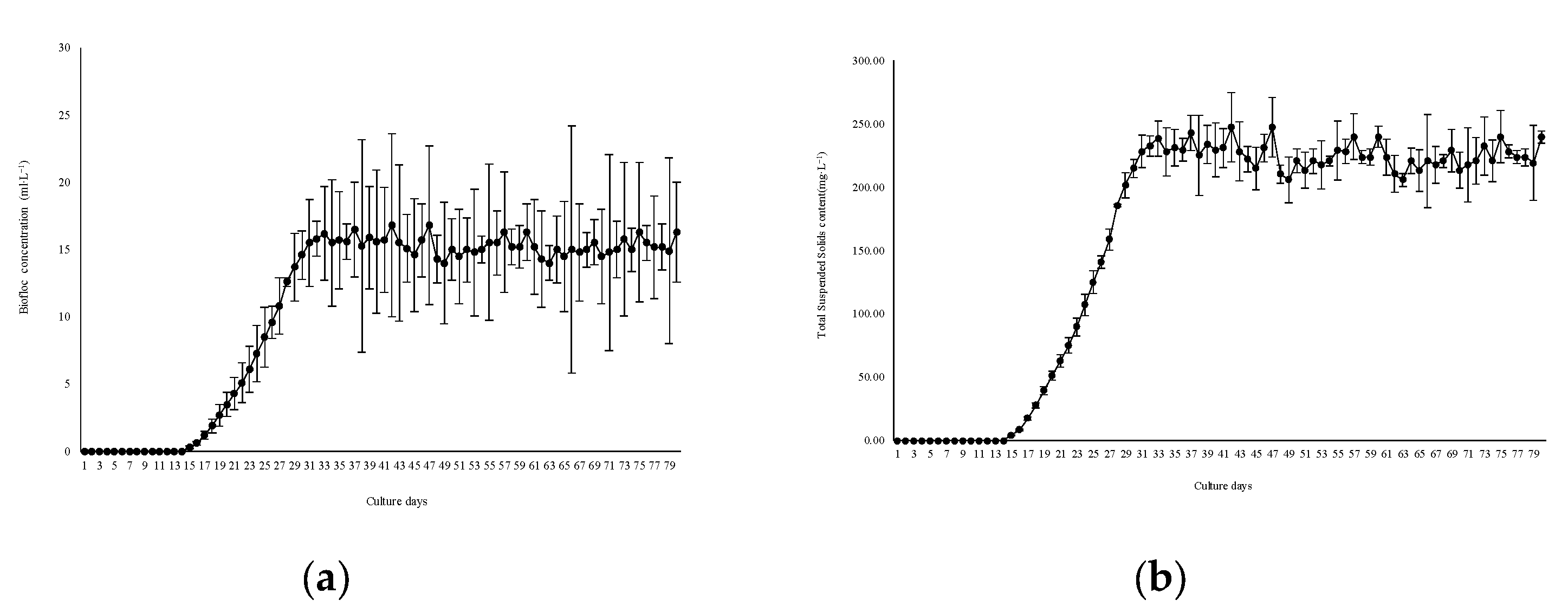
3.1. Biofloc Deposition
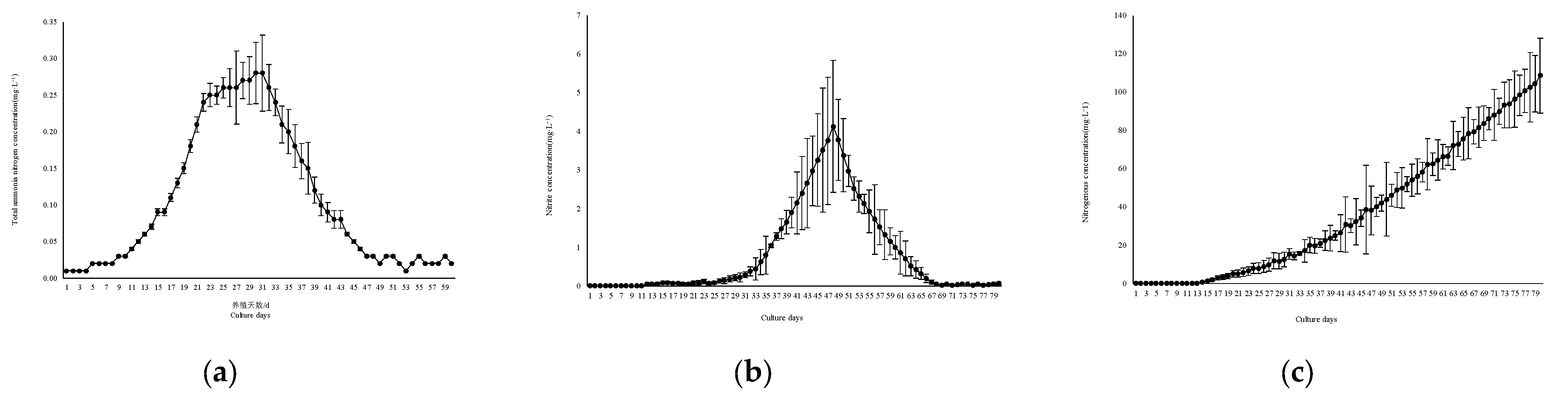
3.2. Concentration Total Ammonia Nitrogen, Nitrite Nitrogen, and Nitrate Nitrogen
3.3. 16 S rDNA Sequencing Analysis
3.3.1. Alpha-Diversity
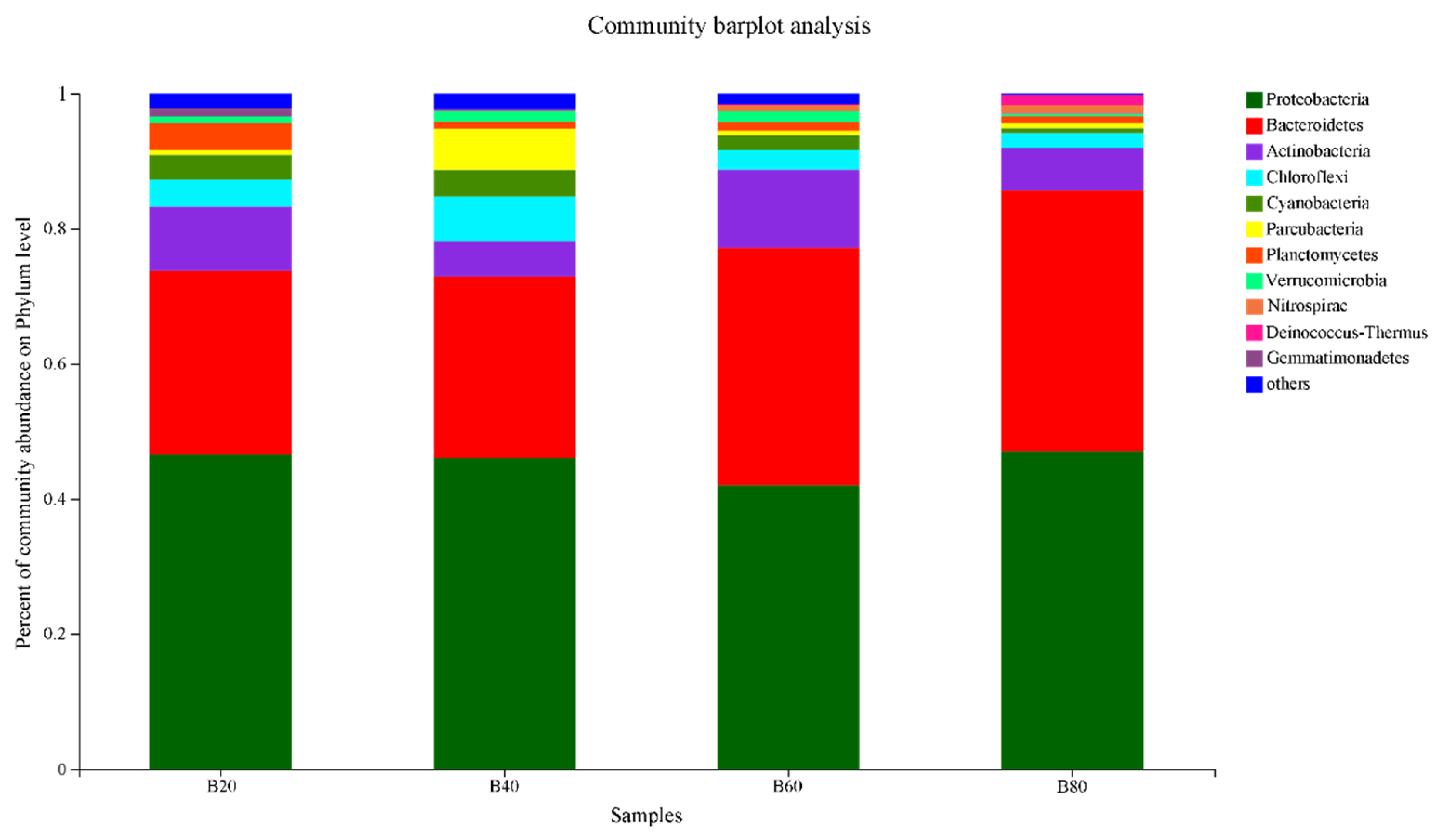
3.3.2. Component Abundances at the Phylum Level
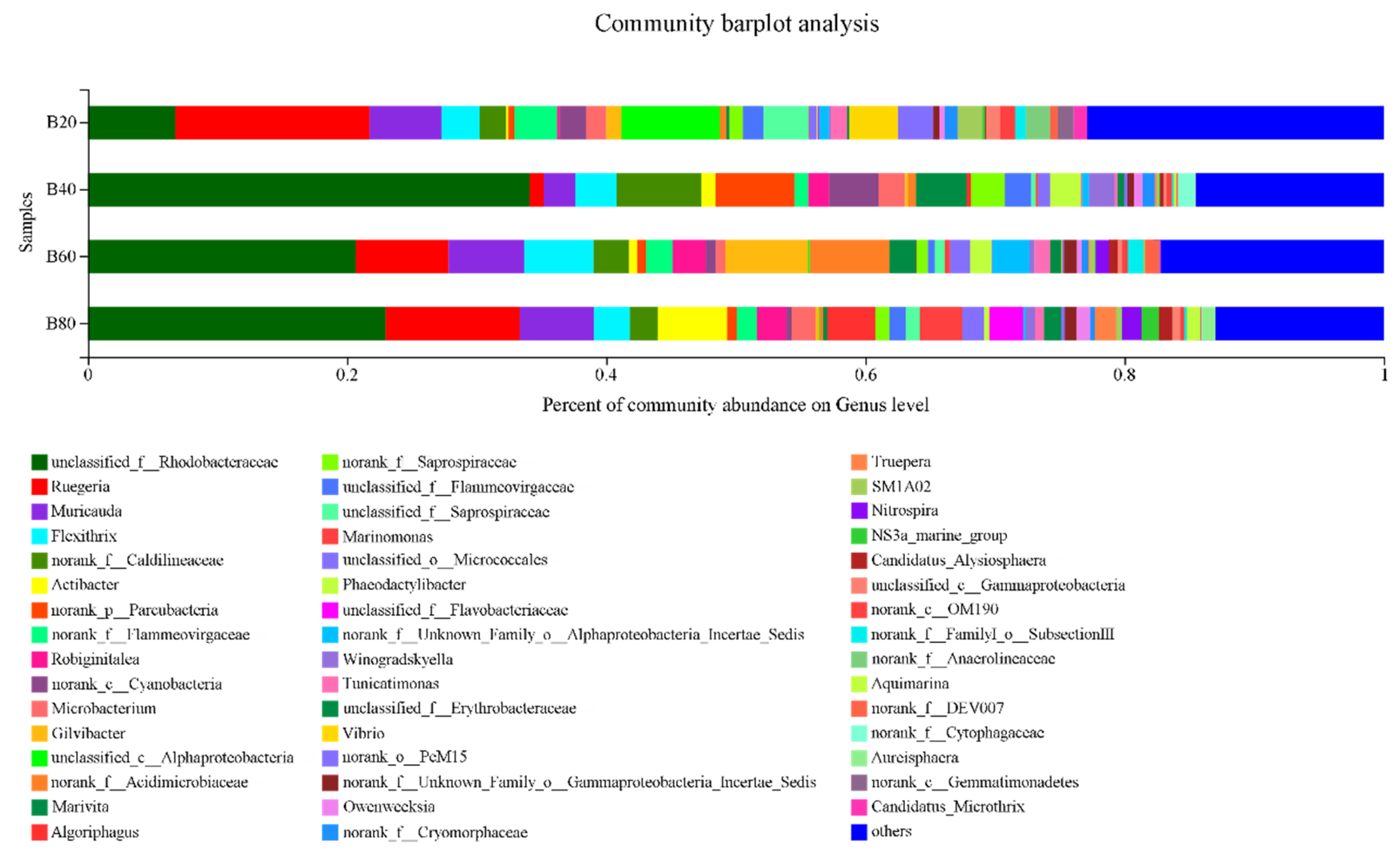
3.3.3. Component Abundances at the Genus Level
3.4. Growth Performance of Shrimp
4. Conclusions
- (1)
- With the Reducing carbon input treatment, the Chao1 index, OTU number, and Shannon index of the bacterial community in large-scale biofloc shrimp culturing facilities increased significantly. The number of OTUs of B80 (572.36 ± 13.26) was significantly higher than that of B60 (489.69 ± 12.97), B40 (423.35 ± 18.46) and B20 (407.67 ± 15.65) (p < 0.05). The Chao1 index of B80 (768.58 ± 36.96) was significantly higher than that of B60 (646.8 ± 52.53), B40 (569.7 ± 46.53) and B20 (516.3 ± 21.35) (p < 0.05). The Shannon index of B80 (5.63 ± 0.16) was higher than that of B60 (4.85 ± 0.13), B40 (4.68 ± 0.21) and B20 (3.65 ± 0.22), with significant difference (p < 0.05). The development of community structure in four periods had different responses to different microecological environments, and there was a strong degree of nitrification process in the B80 and B60 periods. At the end of the experiment, the domestication formed a micro-ecosystem with Proteobacteria as the carrier (46.98% ± 15.82%), Chloroflexi as the skeleton (2.2% ± 0.36%), Nitrospirae (1.35% ± 0.26%) as the main water treatment functional bacteria, and other bacteria as auxiliary nitrogen and phosphorus removal; At the genus level, unclassified_f_Rhodobacteracea (22.97% ± 3.82%), Ruegeria (10.35% ± 1.26%), muricauda (5.73% ± 0.61%), Algoriphagus (3.75% ± 0.85%) and Nitrospira (1.56% ± 0.56%) were the dominant bacteria. Under the synergy of the above bacteria, bioflocs can effectively regulate and control water quality, maintain a low level of ammonia nitrogen and nitrite nitrogen without drainage, and maintain a relatively stable shrimp culturing ecosystem.
- (2)
- Large-scale biofloc culture facilities have strong feasibility in shrimp culture and production. biofloc content (0~16.8 ± 4.3) ml/L, TSS concentration (0~247.46 ± 27.3) mL/L, total ammonia nitrogen concentration (0~0.28 ± 0.052) mg/L, nitrite nitrogen concentration (0~4.13 ± 1.42) mg/L, nitrate nitrogen concentration (108.57 ± 19.6) mg/L were all within the safe concentration range of Litopenaeus vannamei; The survival rate and unit yield of shrimp were (65.32 ± 6.85)% and (4.15 ± 1.58) kg/m3 respectively, which were similar to the reported yields of small-scale Litopenaeus vannamei circulating water and biofloc culture experiments. However, due to the reduction of the aisle and reserved operation space, this facility has the advantage of land saving.
- (3)
- The facilities of the biofloc shrimp culture system need to be further optimized. The uniformity of the water and the power allocation of the system are not necessarily the optimal solutions, which need further research and maturation. We need to develop new equipment to control the total amount of bioflocs and utilize them as resources. During the domestication of biological flocs, the current difficulties are the high frequency of water measurement and the long domestication cycle. No standardized domestication technology has been formed yet. It is necessary to research the accurate control of ammonia nitrogen concentration and the corresponding key technologies of bacterial nutrition; There are still many unknowns about the ecological function and development mechanism of biofloc microorganisms. Denitrifying biofloc is a new type of denitrifying biofloc system in which aerobic denitrifying bacteria are added to autotrophic nitrifying biofloc. It can convert 82% of feed protein into gaseous nitrogen and leave the aquaculture system. It is the most advantageous way to achieve “zero water exchange”. At present, it is in its infancy. How to select special nutrients and suitable carbon sources, and the mechanism of aerobic nitrogen removal need further research.
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- The Ministry of Agriculture Fisheries Bureau. China Fisheries Statistical Yearbook; China Agriculture Press: Beijing, China, 2021. [Google Scholar]
- Defoirdt, T.; Halet, D.; Vervaeren, H.; Boon, N.; Wiele, T.V.D.; Sorgeloos, P. The bacterial storage compound poly-β-hydroxybutyrate protects artemia franciscana from pathogenic vibrio campbellii. Environ. Microbiol. 2007, 9, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Che, X.; Liu, X.G.; Liu, H.; Wang, X.D. Application of Biofloc Technology in Aquaculture. J. Shanxi Agric. Sci. 2019, 47, 1680–1682. [Google Scholar]
- Gutierrez-Wing, M.T.; Malone, R.F. Biological filters in aquaculture: Trends and research directions for freshwater and marine applications. Aquac. Eng. 2006, 34, 163–171. [Google Scholar] [CrossRef]
- Schryver, P.D.; Crab, R.; Defoirdt, T.; Boon, N.; Verstraete, W. The basics of bioflocs technology: The added value for aquaculture. Aquaculture 2008, 277, 125–137. [Google Scholar] [CrossRef]
- Defoirdt, T.; Boon, N.; Bossier, P.; Verstraete, W. Disruption of bacterial quorum sensing: An unexplored strategy to fight infections in aquaculture. Aquaculture 2004, 240, 69–88. [Google Scholar] [CrossRef]
- Avnimelech, Y. Biofloc Technology—A Practical Guide Book, 2nd ed.; The World Aquaculture Society: Baton Rouge, LA, USA, 2012; pp. 182, 191-193, 217-230, 272. [Google Scholar]
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Burford, M.A.; Thompson, P.J.; Melntosh, R.P. Nutrient and microbial dynamics in high-intensity, zero exchange shrimp ponds in Belize. Aquaculture 2003, 219, 393–411. [Google Scholar] [CrossRef]
- Shi, M.M.; Liu, H.; Long, L.N.; Ruan, Y.J.; Guo, X.S.; Zhu, S.M. Effect of carbon source supply tactics on treatment of aquaculture wastewater with biofloc technology. J. Agric. Mach. 2016, 47, 317–323. [Google Scholar]
- Zhang, Z.; Yang, Z.W.; Ge, H.; Chen, H.; Zhuo, X. Effects of different carbon sources on the biofloc formation, nutritional ingredients and bacterial community and water quality in Litopenaeus vannamei culture tank. J. Fish. China 2019, 43, 639–649. [Google Scholar]
- Zhang, Z.; Yang, Z.W.; Ge, H.; Du, X.P.; Zhuo, X.H.; Xu, Z.H. Impacts of Litopenaeus vannamei on microbial diversity of three biofloc and predictive analysis of tax 4 fun gene function during hatchery period in water. Acta Hydrobiol. Sin. 2019, 43, 786–796. [Google Scholar]
- Ray, A.J.; Farno, C.C.; Bailey, B.; Brelang, V.M. Differences in chemical dynamics between chemoautotrophic and three different heterotrophic biofloc-Based shrimp (Litopenaeus vannamei) culture systems. J. Shellfish. Res. 2011, 30, 546. [Google Scholar]
- Ray, A.J.; Lotz, J.M. Comparing a chemoautotrophic-based biofloc system and three heterotrophic-based systems receiving different carbohydrate sources. Aquac. Eng. 2014, 63, 54–61. [Google Scholar] [CrossRef]
- Tan, H.X.; Pang, Y.; Wang, C.H.; Luo, G.Z.; Liu, W.C. Preliminary study on domesticating nitrifying bio-flocs to rear Litopenaeus vannamei. J. Shanghai Ocean. Univ. 2017, 26, 490–500. [Google Scholar]
- Ferreira, G.S.; Santos, D.; Schmachtl, F.; Machado, C.; Vieira, F.N. Heterotrophic, chemoautotrophic and mature approaches in biofloc system for Pacific white shrimp. Aquaculture 2020, 533, 736099. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Che, X.; Liu, X.G.; Cheng, G.F.; Chen, J.; Liu, H.; Chen, X.L. Construction and experiment of simple industrial recirculating water shrimp culture system. Trans. Chin. Soc. Agric. Eng. 2020, 36, 210–216. [Google Scholar]
- Gao, G.; Zhu, K.L.; Zhang, Q.Q.; Wang, Z.J.; Huang, J. Simplified fermentation of a functional probiotics and the application in prawn (Litopenaeus vannamei) Bio-Floc Breeding. Prog. Fish. Sci. 2017, 38, 140–147. [Google Scholar]
- Zhao, P. The Study and Application of Bioflocs Technology in Seawater Aquaculture; Shanghai Ocean University: Shanghai, China, 2011. [Google Scholar]
- Samocha, T.M.; Wilkenfeld, J.S.; Morris, T.C.; Correia, E.S.; Hanson, T. Intensive raceways without water exchange analyzed for white shrimp culture. Glob. Aquac. Advocate 2010, 13, 22–24. [Google Scholar]
- Qin, H.P.; Wang, B.; Hu, S.K.; Zhao, J.C.; He, Z.H.; Yang, S.P.; Sun, C.B. Changes of microbial diversity in biofloc system during nitrogen transformation. Fish. Mod. 2020, 47, 22–28. [Google Scholar]
- Sakami, T.; Fujioka, Y.; Shimoda, T. Comparison of microbial community structures in intensive and extensive shrimp culture ponds and a mangrove area in Thailand. Fish. Sci. 2010, 74, 889–898. [Google Scholar] [CrossRef]
- Wagner, M.R.A.; Amann, R.I.; Lemmer, H. Probing activated sludge with oligonucleotides specific for Proteobacteria: Inadequacy of culture-dependent methods for describing microbial community structure. Appl. Environ. Microbiol. 1993, 59, 1520–1525. [Google Scholar] [CrossRef]
- Yang, Z.W.; Yang, K.; Zhang, Z. Research on the biofloc bacterial community structure during larval rearing of Litopenaeus vannamei using metagenome sequencing. J. Fujian Fish. 2015, 37, 91–97. [Google Scholar]
- Das, S.; Ward, L.R.; Burke, C. Prospects of using marine Actinobacteria as probiotics in aquaculture. Appl. Microbiol. Biotechnol. 2008, 81, 419–429. [Google Scholar] [CrossRef]
- Zwolinski, M.D. DNA sequencing: Strategies for soil microbiology. Soil Sci. Soc. Am. J. 2007, 71, 592–600. [Google Scholar] [CrossRef]
- Asaduzzaman, M.; Wahab, M.A.; Verdegem, M.C.J. C/N ratio control and substrate addition for periphyton development jointly enhance freshwater prawn Macrobrachium rosenbergii production in ponds. Aquaculture 2008, 280, 117–123. [Google Scholar] [CrossRef]
- Ding, C.Y.; Zheng, Y.; Ren, X.M.; Chen, Z. Changes in bacterial community composition during the remediation of Cd contaminated soils of bioenergy crops. Acta Sci. Circumstantiae 2016, 36, 3009–3016. [Google Scholar]
- Wang, W.; Cai, Z.C.; Zhong, W.H. Research advances in aerobic denitrifiers. Chin. J. Appl. Ecol. 2007, 18, 2618–2625. [Google Scholar]
- Wang, Z.; Luo, G.; Li, J.; Chen, S.Y.; Li, Y.; Li, W.T.; Li, A.M. Response of performance and ammonia oxidizing bacteria community to high salinity stress in membrane bioreactor with elevated ammonia loading. Bioresourse Technol. 2016, 216, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Madrid, V.M.; Aller, J.Y.; Aller, R.C. High prokaryote diversity and analysis of community structure in mobile mud deposits off French Guiana identification of two new bacterial candidate divisions. Environ. Microbiol. 2001, 37, 197–209. [Google Scholar] [CrossRef]
- Uchino, Y.; Yokota, A.; Sugiyama, J. Phylogenetic position of the marine subdivision of Agrobacterium species based on 16S rRNA sequence analysis. J. Gen. Appl. Microbiol. 1997, 43, 243–247. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mitova, M.; Tommonaro, G.; Hentschel, U.; Werner, E.G.; Rosa, S.D. Exocellular cyclic dipeptides from a Ruegeria strain associated with cell cultures of suberites domuncula. Mar. Biotechnol. 2004, 6, 95–103. [Google Scholar] [CrossRef]
- Buchan, A.; Mitchell, A.; Cude, W.N.; Campagna, S. Acyl-Homoserine Lactone-Based Quorum Sensing in Members of the Marine Bacterial Roseobacter Clade: Complex Cell-to-Cell Communication Controls Multiple Physiologies//de Bruijn FJ. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, I&II; John Wiley & Sons, Inc.: New York, NY, USA, 2016; pp. 225–233. [Google Scholar]
- McBride, M.J. The Family Flavobacteriaceae. In The Proka-Ryotes, other Major Lineages of Bacteria and the Archaea, 4th ed.; Rosenberg, E., DeLong, E.F., Lory, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 643–676. [Google Scholar]
- Nielsen, P.H.; Mielczarek, A.T.; Kragelund, C. A con-ceptual ecosystem model of microbial communities in en-hanced biological phosphorus removal plants. Water Res. 2010, 44, 5070–5088. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.Q.; Yan, K.Q.; Song, Z.W. Water purification of marine aquaculture system based on different fillers. Chin. J. Environ. Eng. 2018, 12, 2210–2219. [Google Scholar]
- Tal, Y.; Jem, W.; Schreier, S.B.; Sowers, K.R.; Schreier, H.J. Characterization of the microbial community and nitrogen transformation processes associated with moving bed bioreactors in a closed recirculated mariculture system. Aquaculture 2003, 215, 187–202. [Google Scholar] [CrossRef]
- Yue, X.; Liu, Z.H.; Yu, G.P.; Ji, S.M.; Tang, J.L. Fast start-up and performance of the canon process based on a SBAF system and evolution properties of microorganisms. Environ. Sci. 2017, 38, 5192–5200. [Google Scholar]
- Zhang, L.; Chen, Z.; Wang, L.; Chen, S.; Qu, K.M.; Zhang, P.; Zhu, J. Study on application of recirculating aquaculture system for Litopenaeus vannamei. Fish. Mod. 2019, 46, 7–14. [Google Scholar]
- Deng, Y.N.; Zhao, P.; Sun, Y.Z.; Yang, C.H.; Huang, J. Conditions for bio-floc formation and its effects in closed culture system of Litopenaeus vannamei. Prog. Fish. Sci. 2012, 33, 69–75. [Google Scholar]

| Culture Days | Daily Glucose Addition/Daily Feeding Amount | Daily Baking Soda Addition/Daily Feeding Amount |
|---|---|---|
| 1~20 | 150% | 15% |
| 21~40 | 100% | 15% |
| 41~60 | 50% | 15% |
| 61~80 | 0% | 15% |
| Alpha-Diversity Index | Treatment | |||
|---|---|---|---|---|
| B20 | B40 | B60 | B80 | |
| Number of OTUs | 407.67 ± 15.65 c | 423.35 ± 18.46 c | 489.69 ± 12.97 b | 572.36 ± 13.26 a |
| Chao 1 index | 516.3 ± 21.35 d | 569.7 ± 46.53 c | 646.8 ± 52.53 b | 768.58 ± 36.96 a |
| Shannon index | 3.65 ± 0.22 c | 4.68 ± 0.21 b | 4.85 ± 0.13 b | 5.63 ± 0.16 a |
| Coverage/% | 99.9 ± 0.00 | 99.85 ± 0.00 | 99.88 ± 0.00 | 99.92 ± 0.00 |
| 30 Day-Age | 60 Day-Age | 80 Day-Age | Survival Rate/% | Unit Output/kg·m−3 | |||
|---|---|---|---|---|---|---|---|
| Weight/g | Length/cm | Weight/g | Length/cm | Weight/g | Length/cm | ||
| 0.93 ± 0.12 | 4.78 ± 0.43 | 6.18 ± 0.65 | 8.02 ± 0.65 | 12.96 ± 2.85 | 12.26 ± 2.56 | 65.32 ± 6.85 | 4.15 ± 1.58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Che, X.; Liu, X.; Liu, H.; Li, Y.; Wang, J.; Cheng, G.; Chen, J.; Tang, R.; Chen, X.; et al. Reducing Carbon Input Improved the Diversity of Bacterial Community in Large-Scale Biofloc Shrimp Culture Facilities. Diversity 2022, 14, 778. https://doi.org/10.3390/d14100778
Zhu L, Che X, Liu X, Liu H, Li Y, Wang J, Cheng G, Chen J, Tang R, Chen X, et al. Reducing Carbon Input Improved the Diversity of Bacterial Community in Large-Scale Biofloc Shrimp Culture Facilities. Diversity. 2022; 14(10):778. https://doi.org/10.3390/d14100778
Chicago/Turabian StyleZhu, Lin, Xuan Che, Xingguo Liu, Huang Liu, Yiming Li, Jie Wang, Guofeng Cheng, Jun Chen, Rong Tang, Xiaolong Chen, and et al. 2022. "Reducing Carbon Input Improved the Diversity of Bacterial Community in Large-Scale Biofloc Shrimp Culture Facilities" Diversity 14, no. 10: 778. https://doi.org/10.3390/d14100778
APA StyleZhu, L., Che, X., Liu, X., Liu, H., Li, Y., Wang, J., Cheng, G., Chen, J., Tang, R., Chen, X., & Chen, X. (2022). Reducing Carbon Input Improved the Diversity of Bacterial Community in Large-Scale Biofloc Shrimp Culture Facilities. Diversity, 14(10), 778. https://doi.org/10.3390/d14100778








