Abstract
Eranthis Salisb. (Ranunculaceae) is a herbaceous plant genus, including few species disjunctively distributed throughout the temperate zone from Southeastern Europe to Eastern Asia. Until recently, only Eranthis sibirica DC. was known in South Siberia, being considered endemic and tertiary relict. Not long ago, Eranthis tanhoensis Erst was also described in Siberia. We report here a reconstruction of the phylogenetic relationships between the Siberian Eranthis species based on nuclear (ITS) and plastid (trnL + trnL-trnF + trnH-psbA) DNA. The phylogeographic structure of Siberian Eranthis is distinguished by the presence of the two “eastern” and “western” supergroups, which most likely formed as a result of disjunction caused by active mountain uplifts during the late Neogene–early Quaternary and subsequent progressive Pleistocene cooling. The eastern supergroup combines lineage I, containing populations from the eastern Khamar-Daban Ridge, the Eastern Sayan Mountains, and the Tannu-Ola Ridge, and lineage II containing western Khamar-Daban populations. The western supergroup includes only lineage III, containing Western Sayan populations. Our data clearly show that E. tanhoensis is nested in the E. sibirica clade, thereby indicating that its description as a separate species is unjustified, as it compromises the monophyletic status of E. sibirica. Therefore, we suggest here to consider E. tanhoensis as a synonym of E. sibirica.
1. Introduction
Eranthis Salisb., Trans. Linn. Soc. London 8: 303 (1807) (Ranunculaceae) is a genus of herbaceous plants that includes few species disjunctively distributed throughout the temperate zone from Southeastern Europe to Eastern Asia [1,2,3]. Eranthis hyemalis (L.) Salisb. having its natural range in Europe is a type species of the genus, and some aspects of phylogeny and history of the Eranthis genus have already been described in several studies [1,2,3,4,5]. Until recently, only Eranthis sibirica DC. was known in South Siberia, being considered endemic to this region and a tertiary relict plant [6]. This species belongs to the Shibateranthis (Nakai) Tamura section uniting species with white sepals [1]. According to the few known molecular phylogenetic reconstructions, the East Asian Eranthis byunsanensis B.Y.Sun, Eranthis pinnatifida Maxim., Eranthis pungdoensis B.U.Oh, and Eranthis stellata Maxim. appear to be most closely related to E. sibirica [3,4]. E. sibirica was described in the Khamar-Daban Ridge located close to the southern and southeastern coast of Lake Baikal. The species range there was known to extend for about 180 km from the Kultuchnaya River (Kultuk settlement) in the West to the Mysovka River (town of Babushkin) in the East [6]. The other known sites of its occurrence are associated with the foothills and mountain areas of the Eastern Sayan, the Western Sayan [7,8], and the Tannu-Ola Ridge [9]. Our previous studies have revealed genetic heterogeneity in the Khamar-Daban populations [10], which has allowed us to suggest for the first time that the differences between the populations from the western and eastern sides of the ridge are significant, and may even be as high as to correspond to interspecific difference levels [11]. According to A.S. Erst and colleagues [5], our findings inspired them to look for a new Siberian Eranthis species, with Eranthis tanhoensis Erst having been recently described as a narrow endemic of the Khamar-Daban, mostly based on morphological and, in part, genetic differences from E. sibirica. The data on localities and the distribution map presented by the authors clearly show the distribution patterns of the two closely related Siberian species. In particular, the specimens obtained from the Khamar-Daban populations to the East from the Solzan River (city of Baikalsk), previously identified as E. sibirica, were re-described as a new E. tanhoensis species. Therefore, E. sibirica distribution in the Khamar-Daban was limited to a range fragment stretching for around 40 km from west to east in the westernmost part of the ridge. Specimens from the distant range fragments, i.e., the Eastern Sayan and the Western Sayan Mountains and the Tannu-Ola Ridge remained belonging to E. sibirica. Our previous preliminary genetic studies showed, however, that E. sibirica specimens from the Eastern Sayan populations appeared to be more closely related to the eastern Khamar-Daban populations (E. tanhoensis now) than the western ones (still E. sibiraca) [12]. In our opinion, the main limitations of the original study suggesting a new species in the Eranthis genus [5] were the narrow coverage of E. sibirica populations for molecular genetic studies, and strong reliance on the morphological criteria, i.e., morphological differences between the two clades (E. sibirica and E. tanhoensis) when identifying specimens from different isolates as belonging to the two above-mentioned taxa without performing any genetic analysis. In fact, the study was carried out using E. sibirica samples from only three locations in the Khamar-Daban Ridge (the Burovshina, the Sludyanka, and the Utulik rivers). Plant specimens obtained from the distant range fragments (the Eastern and Western Sayan Mountains and the Tannu-Ola Ridge) were assigned to E. sibirica based primarily on morphology, as they were not included in the molecular phylogenetic study. Another limitation of the study was the absence of indications as to the public availability of the E. tanhoensis and E. sibirica nucleotide sequences used to reconstruct the phylogeny, which limited the possibilities of correlating our data and the data obtained in the discussed study. It is also worth noting that, in their subsequent study [4], the authors partially addressed the indicated limitations, which, however, did not help to make the picture clearer. The authors made an attempted to introduce one more Siberian Eranthis species, apparently, for the part of the E. sibirica range relating to the Western Sayan and the Tannu-Ola Ridge (according to Table S2 of [4]). The species was designated as undescribed “Eranthis sp.” on the phylogenetic tree, and the description was not validly published. This study was based on two voucher specimen sequences from the Western Sayan (denoted as Eranthis sp.), and only by one specimen for both E. tanhoensis and E. sibirica from the Khamar-Daban. There were again no genetic data for the populations from the Eastern Sayan and the Tannu-Ola. In addition, the plastid DNA marker sequences deposited in GenBank [4] had a number of skipped data portions (see below), which made them difficult to analyze.
In such a way, we may summarize that the phylogenetic relationships between the Siberian Eranthis species are currently not quite obvious, and the phylogeographic structure shows certain gaps. Considering the relict status of Siberian Eranthis species, we believe that understanding phylogeographical structure and relationships between populations from different fragments may shed light both on their evolutionary history, and the vegetation dynamics in the region during the Pleistocene–Holocene on the whole. Moreover, Siberian Eranthis species are rare, and E. sibirica, as many other nemoral relics, is listed in the regional Red Data Books of Russia [10]. Erst A. S. with colleagues [5] suggested considering E. tanhoensis an endangered species (EN), according to IUCN’s Extent of Occurrence criteria (IUCN 2019). The Convention on Biological Diversity (CBD, 1992) accepted that the biological diversity lying at the genetic level can be a basis for the conservation of endangered species [13]. Therefore, we believe that by providing an understanding of genetic diversity and diversifications, our phylogeographical study can provide essential information for the evidence-based conservation priorities of the Siberian Eranthis taxa.
In our study, we performed a phylogenetic reconstruction based on nuclear and plastid DNA to address the indicated limitations of the previous studies. We actually aimed to: (i) describe phylogeographic patterns and phylogenetic relationships between the populations of Siberian Eranthis using a broadened sample, and (ii) explain the phylogenetic relationships between the Siberian Eranthis taxa and clarify the taxonomic status of E. tanhoensis. Looking ahead, and in a certain way predating the discussion of our findings, further in the text we will use the original name E. sibirica for all populations, specifying individually where specimens correspond to E. tanhoensis, according to A.S. Erst and colleagues [5].
2. Materials and Methods
2.1. Plant Material Collection
Eranthis plants for DNA sequencing were collected in 16 localities distributed between the four presumptive isolated fragments of the South Siberia range: the eastern (EK) and western (WK) sides of the northern macroslope of the Khamar-Daban Ridge, the Eastern Sayan Mountains (ES), the Western Sayan Mountains (WS), and the Tannu-Ola Ridge (TO) (Figure 1, Table 1).
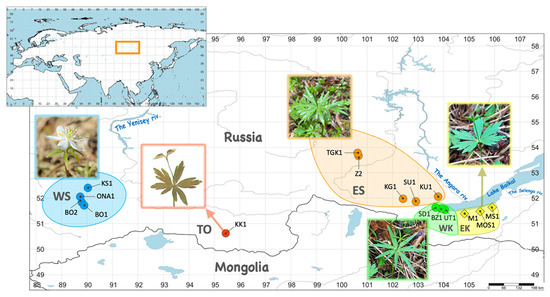
Figure 1.
Eranthis sample collection map for DNA analysis. Collection sites (localities) are indicated with colored circles (correspond to E. sibirica according to Erst et al., 2020 [5]) or diamonds (correspond to E. tanhoensis according to Erst et al., 2020 [5]). Abbreviated locality names and numbers correspond to those listed in Table 1. The areas encircled with lines of different colors show the provisional boundaries of the presumable Eranthis range fragments in South Siberia. The contour color and two-letter abbreviations indicate the fragments to which collection sites belong, as follows: EK (yellow)—eastern part of the Khamar-Daban Ridge, ES (orange)—the Eastern Sayan Mountains, TO (red)—the Tannu-Ola Ridge, WS (blue)—the Western Sayan Mountains, WK (green)—western part of the Khamar-Daban Ridge. Map was drawn using SimpleMappr [14], map projection: geographic; datum WGS84.

Table 1.
The list of Eranthis sampling for DNA analysis.
All mountain ranges mentioned above, belong to a single South Siberian mountain system and have a common geological and biogeographical history. The collection regions are characterized by a semihumid or even humid climate and are considered to play the role of refugia for the survival of mesophytic remnants of broad-leaved forests, such as E. sibirica [15,16]. Samples from the TO fragment (KK1 locality) were obtained from the NSK herbarium, all other samples were collected from living plants in the field and related vouchers were deposited in IRKU herbarium. The images of IRKU herbarium vouchers are available on-line in Irkutsk State University digital herbarium at the National Depository Bank of Live Systems (https://plant.depo.msu.ru/open/public/en/search?collection=IRKU, accessed on 1 August 2022). Plants collected in EK fragment correspond to E. tanhoensis according to their distribution and morphology as described by A.S. Erst and colleagues [5]. Plant samples collected in other localities correspond to E. sibirica according to the distribution map presented in the mentioned study. The material for DNA analysis was sampled from at least six individuals from each natural population, and from a single herbarium voucher specimen in the case of the KK1 locality. Each sample was put in an individual filter paper bag (23 g·m−2), dried and stored in silica gel until DNA isolation.
2.2. DNA Isolation
Total DNA was isolated from silica-dried leaf tissue following the cetyltrimethylammonium bromide (CTAB) method [17], with some modifications. Up to 100 mg of dried tissue from each sample was ground using the automatic homogenizer MiniLys (Bertin Instruments, Montigny-le-Bretonneux, France) in leakage-preventing O-ring screw cap tubes containing two steel beads (3 and 5 mm diameter). Obtained plant powder was collected on the tube bottom by brief centrifugation and diluted in 600 µL of 2% (m/v) CTAB extraction buffer, with 100 mM Tris-HCl (pH 8.0), 20 mM ethylenediaminetetraacetic acid (EDTA, pH 8.0), 1.4 M NaCl, 3% (w/v) polyvinylpyrrolidone (PVP-40), and 1% (v/v) β-mercaptoethanol, mixed well and incubated in a temperature-controlled shaker at 60 °C for 1 h with stirring at 400 rpm. Then, the tubes were briefly spun down, 500 μL of chloroform–isoamyl alcohol mixture at the ratio of 24:1 (v/v) was added to the homogenate, and the samples were mixed well and centrifuged at 14,000× g for 15 min at 4 °C. The upper aqueous phase was transferred to a new tube, an equal volume of chloroform-isoamyl alcohol mixture was added, and the mixture was centrifuged as described above. The upper phase was treated with 40 µg of RNase A (Thermo Fisher Scientific, Vilnius, Lithuania) for 30 min at 37 °C, and DNA was then precipitated by the addition of 0.8 volume of isopropanol to 1 volume of the water phase, followed by incubation for at least 1 h at −20 °C. A DNA pellet was obtained by centrifugation at 14,000× g for 15 min at 4 °C, washed twice with 70% ethanol, air-dried at room temperature in the laminar flow hood, resuspended in 100 µL of nuclease-free water preheated at 60 °C, and stored at −20 °C.
2.3. PCR, Cloning, and Sequencing
For phylogenetic reconstruction, the ITS1-ITS2 region of nuclear DNA (ncDNA) and the trnL (UAA) intron region, trnL-trnF, and trnH-psbA intergenic spacers of plastid DNA (ptDNA) were used as molecular markers. A complete ITS region was amplified using the forward ITS1-P2 [18] and the reverse ITS4 [19] primers, complementary to rDNA 18S and 26S flanking regions. PCR was performed in 25 µL of a reaction mixture containing 1× Q5 Reaction Buffer, and 0.5 units of Q5 High-Fidelity DNA Polymerase (New England BioLabs, Ipswich, MA, USA) with final concentrations of 200 µM of each dNTP and 500 nM of each primer. Amplification conditions for the ITS region were 98 °C for 30 s, 30 cycles for 98 °C for 10 s, 52 °C for 20 s, and 72 °C for 15 s each, and final elongation for 2 min at 72 °C. In the case of the NSK herbarium sample (KK1 locality) insufficient amplification of the ITS region using Q5 polymerase we observed because of DNA degradation probably caused by the long-term storage and treating. GoTaq Flexi DNA Polymerase (Promega, Madison, WI, USA) was used to amplify the ITS region from the KK1 sample and ptDNA regions from all the samples in the study. Using this polymerase, the ITS region could be successfully amplified using the primers indicated above. Combinations of the c + d and e + f primers [20] were used to amplify the trnL and trnL-trnF regions, respectively. The trnH-psbA spacer region was amplified using a trnH2 [21] and psbAF [22] primer pair. The reaction mixture contained 1× Green GoTaq Flexi Buffer, 0.5 units of GoTaq polymerase, 2.5 mM of MgCl2, 250 µM of each dNTP, and 250 nM of each primer in the final volume of 20 µL. The amplification conditions for both DNA regions and all primer pairs were 95 °C for 2 min, 35 cycles of 95 °C for 20 s, 52 °C (ITS), 53 °C (trnL-trnF, trnH-psbA), and 58 °C (trnL) for 30 s, and 72 °C for 1 min each, followed by final elongation for 5 min at 72 °C. Amplicons were either directly purified from the PCR mixture (ITS) using the GeneJET Purification Kit (Thermo Fisher Scientific, Vilnius, Lithuania) or first visualized in 1% agarose gel stained with ethidium bromide after electrophoresis and then gel-purified (ptDNA) using a GeneJET Gel Extraction Kit (Thermo Fisher Scientific, Lithuania). Purified amplicons were either directly sequenced (ptDNA) or first cloned in Escherichia coli cells (ITS). In order to reduce PCR-mediated recombination between ITS clones and to improve PCR accuracy, a proofreading polymerase was used together with lower initial template concentrations (not more than 5 ng per reaction) and PCR cycle number (not more than 30 cycles) as recommended by Lahr and Katz [23]. For molecular cloning, amplicons obtained for at least two samples per each locality were ligated into plasmid vectors pMiniT 2.0 (New England Biolabs, USA) in the case of blunt-end products or pTZ57R/T (Thermo Fisher Scientific, Lithuania) in the case of products with single 3′-A overhangs. Ligation was carried out according to the manufacturer’s recommendations using the insert-to-vector molar ratio of 3:1 in 5 µL of reaction mixture containing 12.5 ng of pMiniT 2.0 or 27.5 ng of pTZ57R/T. Further, 50 µL of One Shot TOP10 E. coli chemically competent cells (Invitrogen, Waltham, MA, USA) was heat shock-transformed at 42 °C for 35 s using 2.5 µL of the obtained ligation mixture. After transformation, the cell culture was incubated in an LB/SOC liquid medium at 37 °C for 1.5 h with horizontal stirring at 250 rpm and plated onto LB agar containing 100 mg·L−1 ampicillin. In the case of the pTZ57R/T vector, 40 µL (20 mg·L−1) of X-Gal solution was surface-spread over agar plates to enable blue-white screening for identification of the colonies carrying the insert. In the case of the pMiniT 2.0 vector carrying a toxic minigene in the cloning site, all grown colonies were considered to contain the insert. Eight colonies from each plate were picked with a sterile pipette tip and inoculated into 5 mL of liquid LB medium containing 100 mg·L−1 of ampicillin. Cells were grown overnight at 37 °C with stirring at 250 rpm. In the case of a low transformation efficiency, all colonies were used for further analysis. Plasmids were isolated from overnight culture using a GeneJet Plasmid Miniprep Kit (Thermo Fisher Scientific, Lithuania). Isolated plasmids and original amplicons were Sanger-sequenced using a BigDye Terminator Cycle Sequencing Kit v. 3.1 (Applied Biosystems, Austin, TX, USA) in a 3500 Genetic Analyzer (Applied Biosystems and Hitachi, Tokyo, Japan). All amplicons and plasmids were sequenced in both forward and reverse directions using the same region-specific primers that were used for PCR.
2.4. Sequence Alignment and Phylogenetic Analysis
Raw sequencing data were edited using SnapGene Viewer software v. 2.6.2 (GSL Biotech, San Diego, CA, USA) and deposited in GenBank of National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov, accessed on 7 September 2022). Sequences belonging to the closely related East Asian Eranthis taxa were used as a reference. E. sibirica and E. tanhoensis sequences available in GenBank (http://www.ncbi.nlm.nih.gov/genbank, accessed on 15 July 2022) were also included in the analysis. Since the original study suggesting E. tanhoensis as a new species [5] did not provide any information on the public availability of the E. tanhoensis and E. sibirica sequences used to reconstruct phylogeny, with no information having been found in GeneBank as well, the sequences used in the latter study carried out by the same authors [4] were used in the present work. The data on the taxa and sequences used in the analysis are summarized in Table 2.

Table 2.
The taxa and DNA sequences used for the phylogenetic reconstructions.
Multiple alignments of nucleotide sequences using the MUSCLE application with a gap opening penalty of 500 and an extension penalty of 4.0 were performed in MEGA v. 7.0.16 [24] followed by manual editing. The left and right ends of alignment were trimmed to correspond to the minimal length of the reference sequences obtained from GenBank. ITS and ptDNA sequences were analyzed both separately and together. A phylogenetic analysis based on the alignment of the multiple ITS region variants revealed by molecular cloning and sequencing was carried out in two variants: (a) a network constructed using all the identified ITS variants and (b) a phylogram based on only the main ITS variants. For joint (ITS + ptDNA) analysis, the most abundant ITS variants (shown in bold in the figure) for each locality were used.
The phylogenetic analysis using ptDNA sequences was based on the combined alignment of the sequences of trnL, trnL-trnF, and trnH-psbA fragments obtained for the plant specimens from each locality. Every inversion/insertion/deletion in the ptDNA alignment was considered as a single evolutionary event. They were binary encoded (with “1” indicating presence and “0” indicating absence of a gap or inversion) and included as separate binary data at the end of the matrix. Only one 21-letter inversion event was observed in the trnH-psbA region, which was coded and then removed from the final alignment. Additionally, 48 indels (positions no. 79, 80, 81, 82, 83, 233–255, 279, 507–513, 526–687, 617–622, 678–682, 707, 805–812, 818–822, 824–826, 882, 883–884, 885–889, 890, 891, 892, 902–903, 908–911, 932–933, 934, 938–946, 947, 948–955, 956–958, 959–962, 963–968, 969, 1059–1064, 1078, 1096–1097, 1098, 1099, 1100–1105, 1106–1111, 1112, 1113, 1114, 1115, 1116, 1117–1120, 1122–1123, 1148, 1201) in the final alignment numbering 1277 positions were coded.
Because of the absence of an amplification product corresponding to the trnH-psbA region in the KK1 specimen, this sample could not be included in the main phylogenetic analysis based on ptDNA and joint (ITS + ptDNA) datasets. However, an additional phylogenetic analysis was carried out based on trnL + trnL-trnF sequences only (i.e., the trnH-psbA sequence was removed from the ptDNA dataset) into which the mentioned sample was included. Additionally, the E. sibirica and E. tanhoensis ptDNA sequences from GenBank (http://www.ncbi.nlm.nih.gov/genbank, accessed on 15 July 2022) had long fragments of missing data in the regions containing the multiple indels that we encoded as a binary set. Therefore, these sequences could not be used in the main analysis because they could not be matched to the binary matrix used for the sequences identified in the present study. For this reason, an alternative phylogenetic analysis was performed based on the alignment, including the full set of nucleotide sequences from which the indicated multiple indel regions were completely deleted and without using a binary matrix. All alternative branches inferred from these additional analyses are shown as superstructures on the main trees.
Phylogenetic reconstructions were obtained independently by the Bayesian inference method (BI) based on the matrixes combining the nucleotide alignments and binary (gaps) datasets in MrBayes v. 3.2.5 [25], and the maximum likelihood method (ML) based on multiple nucleotide sequence alignments in MEGA independently. The best-fit model of nucleotide substitutions based on the lowest Bayesian Information Criterion (BIC) calculated using the “find best DNA/protein models” tool in MEGA (Neighbor-Joining tree to use and ML as a statistical method were applied as the settings) was selected and then used to perform the analysis. Nucleotide frequencies calculated using the “find best DNA/protein models” tool were also included to optimize the models implemented in MrBayes in the case of the Bayesian inference analysis.
A BI analysis of nucleotide datasets was performed using the models implemented in MrBayes with optimized parameters to better correspond with the models used in the ML analysis (see below). In particular, for the ITS dataset, an HKY-like model [26] with fixed equal stationary state frequencies was used to match the Kimura two-parameter model (K80) [27]. The analysis of the ptDNA dataset was performed by specifying separately the model and parameters for each partition of the DNA dataset using the “applyto” option. In particular, for the analysis of ptDNA markers, we used the HKY-like model with base frequencies, the optimized Tamura three-parameter model (T92) [28] with gamma-distribution of substitution rate variation among sites (+G) for trnL or no rate variation for trnL-trnF and trnH-psbA datasets. The base frequencies (A, C, G, and T) were fixed as 0.34, 0.16, 0.16, and 0.34 for trnL, 0.33, 0.17, 0.17, and 0.33 for trnL-trnF, and 0.34, 0.16, 0.16, and 0.34 for trnH-psbA. Binary data (indels + inversion) were analyzed using the F81-like model [29] implemented in MrBayes with the equal stationary state frequencies to match the JC69 model [30]. Analysis of the joint ITS + ptDNA dataset was performed using the above-mentioned models and parameters individually set for each DNA region using the “applyto” option. For each dataset, two simultaneous and completely independent Markov chain Monte Carlo (MCMC) analyses were run with four parallel chains up to 10,000,000 generations, with sampling every 100 generations and diagnostic calculation every 1000 generations. The first 25% of the samples from the cold chain were discarded (relburnin = yes and burninfrac = 0.25). The standard deviation of split frequencies below 0.01 was regarded as a sufficient convergence level, and reaching this value was considered as a criterion of reaching the chain stationary state. The fluctuations of the cold chain likelihood in the stable range were also taken into account for the estimate of reaching stationarity. The sampled trees from both analyses were pooled, and 50% majority-rule consensus trees were constructed from 150,002 trees for ITS, 71,106 trees for ptDNA, and 13,829 for the ITS + ptDNA joint dataset to estimate clade posterior probability values (PP). The final phylogenetic trees were edited in FigTree v. 1.4.3 [31].
For the ML analysis, the K80 model for the ITS dataset and T92 model for the ptDNA and ITS + ptDNA joint datasets were used. In all analyses, the initial tree for the heuristic search was inferred using the Neighbor-Joining method based on a pairwise distance matrix estimated using the maximum composite likelihood (MCL) method. All aligned positions, including the indels, were used in the analysis. A bootstrap test for phylogeny including 1000 replicates was used.
The final phylogenetic trees are presented in the figures as BI phylograms, with the additional indication of Bootstrap values (BS) for the clades on corresponding ML trees. E. stellata was used to root the trees, taking into account the reconstruction of phylogenetic relationships between Eranthis species carried out by K.-L. Xiang and colleagues [4]. The data matrices and trees obtained in the study are available in TreeBASE by the following link: http://purl.org/phylo/treebase/phylows/study/TB2:S29659 (accessed on 1 August 2022).
Minimum spanning networks (MSN) based on the full-size sampling of ITS variants were constructed using the epsilon parameter equal to “0”, with the aid of PopART software v. 1.7 [32].
3. Results
3.1. Phylogenetic Analysis Based on Nuclear DNA
The evolution history of the nuclear genome was estimated by analyzing the polymorphism of the ITS1-ITS2 region of ribosomal DNA. At the start, we encountered the problem of having double peaks at a number of positions in the ITS sequences, which we obtained when directly sequencing PCR products, and this made it impossible to unambiguously decipher each individual ITS nucleotide sequence. We suppose that these double signals may have appeared due to the high level of intragenomic polymorphism known for this region [33]. To be able to split the double peaks, the molecular cloning of the ITS region was carried out. As a result, multiple ITS variants were identified, which were used to construct a minimum spanning network (Figure 2a) and phylogram (Figure 2b). BI and ML phylogenetic analyses showed that all Siberian ITS ribotypes (R) formed a well-supported clade (node A). The intragroup structure seemed to be rather complex, with several well-defined ribotype groups being presented. The first one united the ribotypes from the Eastern Sayan (ES) and the eastern Khamar-Daban (EK) fragments (node B, PP, 0.96). The second embraced exclusively the ribotypes obtained from the western Khamar-Daban (WK) (node C, PP, 1.00; BS, 95). The third comprised ribotypes from the fragments of Khamar-Daban (EK and WK) and the Eastern Sayan (node D, PP, 0.99). The fourth (node E, PP, 0.98) group mainly contained the ribotypes from ES and the Tannu-Ola (TO) fragments of the Eranthis range, with a ribotype belonging to WS also found in this group. The ribotypes from the Khamar-Daban (EK and WK) were not found here.
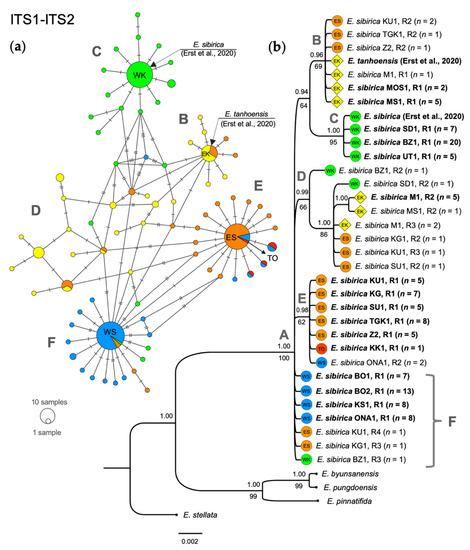
Figure 2.
Diversity and phylogenetic relationships between ITS ribotype variants (R) in E. sibirica from different localities. The color patterns and names correspond to those in Figure 1. Capital letters indicate the groups discussed in the text. (a) Minimum spanning network based on multiple ITS variants identified by molecular cloning. Different ribotypes are presented as colored circles connected by lines, where hatch marks correspond to the numbers of the evolutionary events (substitutions). Circle size corresponds to the number of identical ribotypes sampled (see the circular scale); (b) BI phylogram of main ITS ribotypes. Posterior probabilities are indicated above the branches, Bootstrap values of respective clades on the ML tree below the branches. The number of ribotype copies found by molecular cloning is indicated in brackets next to the branch names. ITS variants used for the further joint ITS + ptDNA analysis (Figure 4) are indicated in bold. The scale bar represents the number of expected changes (substitutions) per site corresponding to a unit of branch length.
Unresolved branches leading to Western Sayan (WS) ribotypes, and to a number of ribotypes from other localities, represented the unstructured group F. These ribotypes were identical forming a dominant circle in the area F of the obtained network (Figure 2a).
The estimated genetic distances, as can be seen in the tree (Figure 2b), show that these ribotypes forming a non-structured group F were in fact the closest to the Siberian common ancestor (node A) among all other ribotype groups.
According to our data, the ribotypes belonging to E. tanhoensis according to A.S. Erst and colleagues [5] were nested in the clade defined by node B, together with the Eastern Sayan ribotypes of E. sibirica. The E. sibirica ribotypes identified by A.S. Erst and colleagues [4,5] combined with E. sibirica ribotypes from the western side of the Khamar-Daban Ridge (WK, node C). Some low-copy ITS variants belonging to different fragments of the Siberian range formed an additional structure within the network (Figure 2a), and might be transitional variants between the main homologous variants (Figure 2b).
3.2. Phylogenetic Analysis Based on Plastid DNA
The phylogenetic reconstruction based on ptDNA was overall similar to the ITS tree, but showed a better defined structure (Figure 3). In particular, similarly to the ITS tree, the clade defined by node A (PP, 1.00; BS, 86) included all the identified Siberian Eranthis plastotypes (P).
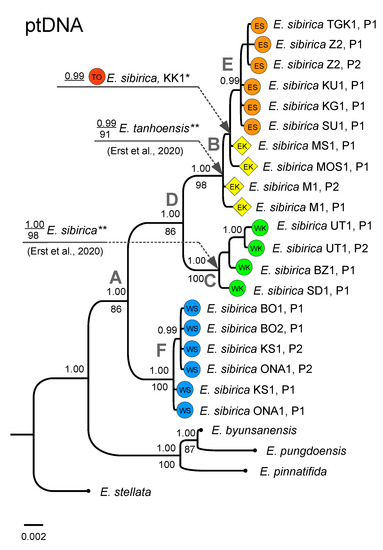
Figure 3.
BI phylogram based on ptDNA. The color patterns and names correspond to those in Figure 1. Posterior probabilities ≥ 0.95 are indicated above branches, Bootstrap values for the corresponding clades on the ML tree (if relevant) below the branches. Capital letters at the nodes correspond to the groups discussed in the text. The scale bar indicates the number of expected changes (substitutions or/and indels and inversions) per site corresponding to a unit of branch length. *—branch position was inferred using the trnL + trnL-trnF dataset only; **—branch positions were inferred from the independent analysis based on the alignment with completely deleted multiple indel regions and not including the binary matrix (see Section 2.4).
The internal structure was also similar to that of the ITS tree, and included group B (PP, 1.00; BS, 98), which embraced the plastotypes from the EK fragment of the Khamar-Daban corresponding to E. tanhoensis [5] and a sub-group formed by the ES E. sibirica plastotypes (PP, 0.99). The plastotype belonging to the TO fragment of the E. sibirica range (the KK1 locality) nested in a sub-clade (PP, 0.99) within the above-mentioned group. The clade defined by node C (PP, 1.00; BS, 100) exclusively contained the plastotypes belonging to the WK fragment, similarly to the ITS tree. Our data show that the plastotypes belonging to E. sibirica, according to A.S. Erst and colleagues [4,5], are found in this group (PP, 1.00; BS, 98). In the case of the ptDNA-based phylogeny, the sequences belonging to the WS populations were united into a well-supported clade F (PP, 1.00; BS, 100) clearly separated from the clade encompassing the plastotypes from the more “eastern” localities (node D, PP, 1.00; BS, 86). We have also found that at least two plastotypes may be simultaneously present (denoted as P1 and P2 in the tree) in the same population. In particular, the intrapopulation ptDNA polymorphism of Eranthis was observed for the M1, KS1, ONA1, UT1 and Z2 localities.
3.3. Combined Phylogenetic Analysis
The topology of the phylogenetic tree built based on the joint dataset (ITS + ptDNA) was well-defined (Figure 4a) and similar to that of the ptDNA tree (Figure 3). Based on our results, at least three main haplotype (H) groups of Siberian Eranthis could be distinguished. Haplogroup I (PP, 1.00; BS, 91) embraced the haplotypes belonging to the populations from EK, ES, and TO fragments. Haplogroup II (PP, 1.00; BS, 100) combined the haplotypes found in the WK populations. Haplogroup III (PP, 1.00; BS, 100) was formed from the haplotypes found in the WS fragment of the Eranthis range. Our results also show that E. tanhoensis [4,5] was nested in haplogroup I, and the haplotype belonging to E. sibirica [4,5] was nested in haplogroup II. Based on the distribution of the mentioned haplogroups (Figure 4b), the “eastern” (PP, 1.00; BS, 87) and “western” (PP, 1.00; BS, 100) supergroups might be distinguished. In particular, the eastern supergroup comprised the lineages I and II, and the western one included only the lineage III. Haplogroup I showed some internal structure. It included the subgroup embracing the ES and TO haplotypes (PP, 0.98) with the nested clade containing the haplotypes from the northernmost Eranthis localities (Z2 + TGK1, PP, 0.99), and two subgroups embracing the western (M1, PP, 1.00; BS, 98) and the eastern (MOS1 + MS1, PP, 1.00; BS, 87) haplotypes within the EK fragment were distinguished.
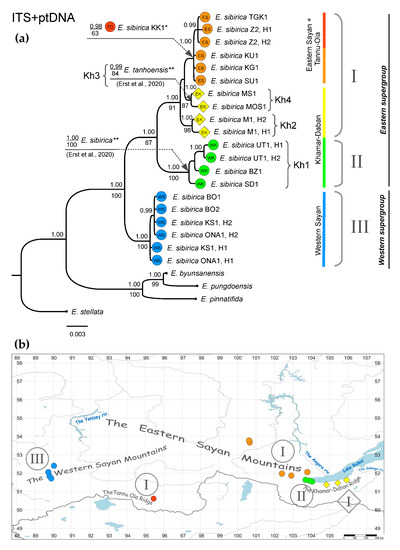
Figure 4.
Phylogeny and phylogeographical patterns of Eranthis in South Siberia based on the combined ITS + ptDNA dataset. The color patterns and names correspond to those in Figure 1. Roman numerals indicate the main identified haplogroups/lineages. (a) BI phylogram. Posterior probabilities ≥ 0.95 are indicated above the branches, Bootstrap values for the corresponding clades on the ML tree (if relevant), below branches. The scale bar indicates the number of expected changes (substitutions or/and indels and inversions) per site corresponding to a unit of branch length. *—branch position was inferred using the combined dataset of ITS and only trnL + trnL-trnF for ptDNA; **—branch positions were inferred from the independent analysis using ptDNA alignment with completely deleted multiple indel regions and not including the binary matrix (see Section 2.4). Kh1–Kh4—the numbers of the Pleistocene microrefugia on the Khamar-Daban Ridge discussed in Section 4.1. (b) Geographical distribution of the haplogroups (lineage) identified in this study. Gray circles indicate the geographical patterns of E. sibirica distribution and the diamond symbol indicates the pattern of E. tanhoensis according to A.S. Erst and colleagues [5].
4. Discussion
4.1. Phylogeographic Patterns of Eranthis in South Siberia
According to our findings, the present phylogeographic structure of Siberian Eranthis includes two, western and eastern, supergroups. The phylogeny reconstruction inferred from molecular genetic data suggested a Northeast Asian origin for the Siberian Eranthis [4], and our data show that the western supergroup appears to be closer to the common Siberian ancestor than the eastern one. A high abundance of ribotypes closely related to the ancestral ones in the western populations, as well as a short distance to the common ancestor inferred from studying the plastotype, may also serve as an evidence. The western lineage includes the populations located on the left side of the main Yenisei riverbed, which are separated from the populations in the eastern supergroup by hundreds of kilometers. The Western Sayan may have played the role of the main gate in the Eranthis expansion from Northeast Asia to South Siberia across the Mongolian Plateau. The nearest known location where the populations included in the eastern supergroup can be found is the eastern Tannu-Ola, the northern slopes of which are the left watershed of the Yenisey Basin, and at the same time face the Eastern Sayan. The other sites of Eranthis findings are located to the east of the Yenisei basin in the Baikal Siberia mountains, i.e., the Eastern Sayan and the Khamar-Daban, and are geographically separated from each other. These mountain ranges housing Eranthis belong to a single South Siberian mountain system, and therefore have a common geological and biogeographical history.
Molecular clock estimates suggest that Siberian Eranthis segregated from its Northeast Asia sister group around 8–29 Ma as a result of the uplift of the Mongolian Platenau region as a consequence of the India–Eurasia and Arabia–Eurasia collisions [4]. Being a nemoral tertiary relict, E. sibirica was confined, at that time, to deciduous broad-leaved forests widespread on the territory of South Siberia in the Early Cenozoic [15]. The former continuity of the Siberian Eranthis range is evidenced by the preservation of the ribotypes closely related to the ancestral ones (Figure 2, group F) in the genomes of the populations from distant fragments of the range, despite the concerted evolution decreasing the intra-individual variability known for this multicopy DNA region [34]. By the end of the Neogene, due to the progressive worsening of climatic conditions, broad-leaved forests were replaced by coniferous forests, which included nemoral elements [15]. The analysis of the bottom sediments of Lake Baikal provided evidence of the dominance of a nemoral dark coniferous–broad-leaved complex in the Cis-Baikal region until the middle of the Late Pliocene, demonstrating increasingly more boreal features at the end of the Pliocene–Eopleistocene [35]. Playing the role of refugia, the mountains of southern Siberia were of great importance for the survival of mesophytic remnants of broad-leaved forests, such as E. sibirica at that time [16].
We believe that the current phylogeographic structure implies that the Eranthis range could have been split at least twice in South Siberia during the Late Cenozoic. We assume that the first fragmentation event might have started at the end of the Pliocene—the beginning of the Quaternary. This presumable time is not far from the time of segregation within the Siberian Eranthis, estimated to have occurred within the range of 2–13 Ma based on molecular clock [4]. The authors of the study suggested that the divergence within Siberian Eranthis was provoked by the secondary uplift and expansion of the Mongolian Plateau region, around 5–10 Ma. However, from our point of view, the events that took place a little later and directly in the South Siberian mountains played a decisive role. We believe that vast cooling late in the Neogene might have caused a reduction in E. sibirica paleorange, while mountain building in South Siberia and the formation of the Yenisei valley, which were remarkably intense at that time [36], created conditions for Western Sayan populations’ separation from the Eastern ones. Progressive cooling and glaciations in the Pleistocene led to a complete geographical isolation of these two groups. However, we cannot completely exclude the existence of gene flows between the two groups, or at least between the Western Sayan populations and eastern populations inhabiting the Eastern Sayan and the Tannu-Ola, since our study revealed the presence of the common ITS paralogous variants (Figure 2, node E). Since ITS is a marker of biparental inheritance, while ptDNA marks only the maternal lineage, the absence of common plastotypes may indicate that presumable contacts between populations more probably occur through pollen transfer, rather than some physical overlapping of their ranges.
The fragmentation of the eastern part of the range most probably happened later than the primary segregation of the western and eastern parts, with the populations still being in contact for some time during the Pleistocene. The evidence for this may be given by the haplotypes identified in the populations from the Tannu-Ola, the Eastern Sayan, and the Khamar-Daban. These haplotypes cluster into the same eastern supergroup, suggesting that the population they belong to did not have enough evolutionary time to accumulate a sufficient number of mismatches to diverge. However, the current phylogeographic structure indicates that the genetic separation between the Eastern Sayan populations and the Khamar-Daban ones might have already started. The presence of the Eastern Sayan subgroup in the ptDNA and combined ITS + ptDNA trees, as well as the high abundance of the ITS paralog that is absent in the Khamar-Daban populations (Figure 2, node E), may be supporting evidence. Individuals from the Tannu-Ola are apparently closer to the East Sayan than to the Khamar-Daban ones. This is indicated by the Tannu-Ola ribotypes (biparentally inherited) clustering together with the Eastern Sayan variants (Figure 2, node E). Since we were not able to obtain an amplification product for the trnH-psbA region in the specimen from the Tannu-Ola, the data on its phylogenetic relationship with the samples from other populations have a low resolution, being based on a reduced dataset. For this reason, we can only assume that the Tannu-Ola plastotype (maternally inherited) is similar to those belonging to the Eastern Sayan populations and those from the eastern Khamar-Daban (Figure 3). As in previous studies [4,5], we confirm a very high genetic divergence between the eastern and western populations within a relatively small Eranthis range on the Khamar-Daban Ridge. Moreover, we found that the haplotypes of the populations occurring on the eastern side of the ridge combined with those characteristic of the Eastern Sayan and the Tannu-Ola populations into a single haplogroup, designated as lineage I (Figure 4). The haplotypes of the populations from the western side of the Khamar-Daban Ridge form lineage II. We assume that this genetic divergence might be a consequence of the geographical isolation that occurred during the Pleistocene cooling. The question of the extent of the Pleistocene glaciation in South Siberia is still unresolved, although it is believed that the Pleistocene cooling did not lead to the formation of a solid ice sheet here, with glaciations progressing by mountains and valley type allowing species to survive in refugia [37]. The analysis of the patterns of the present day distribution of nemoral relict species that we performed earlier has allowed us to suggest the existence of at least four Pleistocene microrefugia on the northern macroslope of the Khamar-Daban Ridge, confined to the floodplains of large rivers [37]. Lineage II relates to the populations geographically belonged to the first microrefugium (Kh1) confined to the Babha and the Utulic rivers in the western side of the Khamar-Daban (Figure 4). Haplotype subgroups in lineage I may belong to the second microrefugium (Kh2) confined to the Snezhnaya River (Figure 4, M1 locality), to the third microrefugium (Kh3) confined to the Pereemnaya River (haplotype of E. tanhoensis obtained by Erst et al., 2020 [4,5]), and to the fourth, the easternmost, microrefugium (Kh4) confined to the Mishikha River (MOS1 and MS1 localities) [37]. Apparently, the westernmost Eranthis populations belonging to lineage II are most strongly isolated from all the other populations. It may be also be suggested that the isolation process is still ongoing, which may together account for the large number of accumulated genetic mismatches. Similar phylogeographic patterns, that is, division into eastern and western phylogenetic groups within the Khamar-Daban Ridge, were also described for another nemoral relict species, Anemone baicalensis Turcz. [38]. The genetic diversity patterns of the species in their natural populations are an essential requisite to designing a successful sampling strategy to create species conservation proposals, e.g., in seed banks [13]. Phylogeographic criteria for priorities-creation for species conservation are considered especially useful in the case of plants with disjunct distribution [13], such as E. sibirica. Although the development of concrete conservation approaches is not covered by our aims, we believe that the haplotypes from all three identified phylogeographic groups (EK + ES, WK, WS) may play an important role in the conservation of Siberian Eranthis.
4.2. Phylogeny of Siberian Eranthis and Taxonomic Status of E. tanhoensis
The results obtained using ptDNA and ITS + ptDNA datasets suggest that there are at least three phylogenetic lineages of Eranthis in South Siberia. As was discussed above, the phylogenetic structure of Siberian Eranthis correlates well with the geographical distribution of the described lineages, which unite into the western and eastern supergroups. At the same time, biparentally inherited ITS ribotypes do not show clear segregation between the indicated groups. Nevertheless, geographically structured ribotype frequency differences can be observed between the groups, as well the presence of ITS paralogs, which are found only in populations belonging to the eastern lineages. The low-copy ITS variants described in specimens belonging to different lineages may be transitional variants between the distant fragments of the Eranthis range.
Our data clearly show that E. tanhoensis, which was described as a new species from the populations previously considered as E. sibirica, is nested in haplogroup I. This lineage includes the Khamar-Daban specimens, which are considered being E. tanhoensis (marked with yellow diamonds in all trees) together with the samples from the Eastern Sayan populations, which were assigned to E. sibirica by A.S. Erst and colleagues based on their morphology [5]. Our findings suggest that the morphometric analysis, carried out by the authors [5] and aimed at separating E. sibirica and E. tanhoensis into two distinct species initially based on the assumption that the Eastern Sayan (e.g., from the KU1 locality) and Tannu-Ola populations belong to the common group with E. sibirica and not with E. tanhoensis, cannot be considered fully valid and has to be revised. The same applies to the published description of E. tanhoensis, and the key to “Eranthis species from Asiatic Russia” [5]. Moreover, the incorporation of E. tanhoensis compromises the monophyly of E. sibirica, which partially presents now in clade I, and entirely constitutes clades II and III. It is worth noting that in their next study, the authors of the discussed work referred to the populations from Western Sayan as an undescribed species “Eranthis sp.” [4]. The introduction of a new species could have partially, but not completely, resolved the E. sibirica monophyly issue. Still, the description of the new species has not been validly published. The monophyly issue has not been completely resolved for the following reasons: (i) the authors, as previously, did not solve the problem of combining the Eastern Sayan and Tannu-Ola populations of E. sibirica with E. tanhoensis according to genetic data, and (ii) according to the geographic coordinates providing in Supplementary Materials presented in Table S2 of the discussed study [4] the authors assigned the specimens from the Tannu-Ola Ridge to the undescribed Eranthis sp., together with the samples from the Western Sayan, instead of combining these specimens with populations from the Eastern Sayan and eastern Khamar-Daban. Thus, until these issues are resolved, E. tanhoensis Erst has to be considered as a synonym of E. sibirica DC.
Regarding the taxonomic issue of Siberian Eranthis, we may express our position in the following way. The debates between “lumpers” and “splitters” are as old as taxonomy itself, and, apparently, it should be admitted that the taxonomic level of a particular group can be arbitrary, and may prove to be less important than the actual demonstration of its monophyly. Nevertheless, the phylogenetic structure demonstrated in this study makes it possible to define three related monophyletic groups of Siberian Eranthis, which may, in principle, be designated as separate species. The rapid integration of molecular biology approaches into classic botany has allowed clearly defined phylogeographic structures to be established for a number of species. At the same time, the mere presence of a phylogeographic structure does not provide sufficient evidence for describing new species in every case. The high variability of morphometric characters, which makes it challenging to divide Siberian Eranthis into the groups corresponding to phylogenetic lineages, as well as the observed incomplete lineage sorting and the presence of ancestral and transitional ITS variants in different localities, may serve as strong counterarguments against subdividing Siberian Eranthis into different species. It should be noted that the study of A.S. Erst and colleagues [5] was not the first attempt to identify new taxa within the E. sibirica range. For instance, previous descriptions of E. uncinata Turcz. growing at higher altitudes are considered unjustified because of the lack of consistency of morphological differences with E. sibirica [39], while the two varieties, E. sibirica DC. var. nuda Schipcz. and E. sibirica DC. var. glandulosa Schipcz., have not been validly published [5,40]. At the moment, we believe that only E. sibirica in the classical sense, i.e., inhabiting the entire South Siberia range of Eranthis, can be recognized as a monophyletic taxon. Despite the controversial issues of the species concept, it is commonly assumed in conservation that we should protect exact species [13]. In such a way, we believe that understanding the taxonomic composition of the Siberian group of Eranthis will be important in conservation development.
5. Conclusions
The phylogeographic structure of Siberian Eranthis shows the presence of the two “eastern” and “western” supergroups, with the latter group being closer to the common Siberian ancestor. The eastern supergroup comprises lineage I, comprising the populations from the eastern Khamar-Daban Ridge, the Eastern Sayan Mountains, and the Tannu-Ola Ridge, and lineage II, comprising the western Khamar-Daban populations. The western supergroup is represented by only lineage III, comprising the Western Sayan populations. We believe that the current phylogeographic structure indicates that the Eranthis range in South Siberia was split at least twice due to intense mountain building and progressive cooling and glaciations in the Late Cenozoic and the Quaternary. Our data also clearly show that E. tanhoensis is nested in the E. sibirica. Thus, describing E. tanhoensis as a separate species cannot be justified, as in this case, the monophyly of E. sibirica is compromised. Hence, until the monophyly issue is resolved, E. tanhoensis Erst should be considered a synonym of E. sibirica DC.
Author Contributions
Conceptualization, M.V.P.; methodology, M.V.P. and V.V.P.; validation, M.V.P.; formal analysis, M.V.P.; investigation, M.V.P. and V.V.P.; resources; M.V.P. and V.V.P.; data curation, M.V.P.; writing—original draft preparation, M.V.P.; writing—review and editing, M.V.P. and V.V.P.; visualization, M.V.P. and V.V.P.; supervision, M.V.P.; project administration, M.V.P.; funding acquisition, M.V.P. and V.V.P. All authors have read and agreed to the published version of the manuscript.
Funding
During 2017–2019 the research was funded by Russian Science Foundation, grant number 17-74-10074 (M.V.P.).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Original sequence data are available at GenBank (http://www.ncbi.nlm.nih.gov/genbank, accessed on 7 September 2022) by their accession numbers presented in Table 2. All data matrices and trees obtained in the study are available in TreeBASE by the following link: http://purl.org/phylo/treebase/phylows/study/TB2:S29659 (accessed on 1 August 2022).
Acknowledgments
The research was done using the equipment of the Core Facilities Center “Bioanalitika” and under the state assignments of Siberian Institute of Plant Physiology and Biochemistry, Siberian Branch of the Russian Academy of Sciences (No 121031300009-4). We sincerely thank Victor Chepinoga for inspiring us in our study, for his partial help with material collection (e.g., herbarium (NSK) materials from the Tannu-Ola Ridge), and accompanying us with some field work, Denis Sandanov for discussing some issues related to our study, Natalya Shvetsova and Polina Nelyubina for their help with plant sampling and for the lab assistance, Veronika Belova for her help with mounting of herbarium vouchers. We also thank the IRKU (especially Nadezhda Stepantsova), NSK herbaria for assistance and allowing us to deposit the vouchers (IRKU) and collect samples (NSK). Moreover, we are grateful to Taisya Protopopova for supporting and helping us during the expedition planning, Sergey and Anna Ivanovs for their assistance in accommodation and transportation during fieldwork in the Western Sayan Mts.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lee, C.S.; Yeau, S.H.; Lee, N.S. Taxonomic Status and Genetic Variation of Korean Endemic Plants, Eranthis byunsanensis and Eranthis pungdoensis (Ranunculaceae) based on nrDNA ITS and cpDNA Sequences. J. Plant Biol. 2012, 55, 165–177. [Google Scholar] [CrossRef]
- Oh, A.; Oh, B.-U. The speciation history of northern- and southern-sourced Eranthis (Ranunculaceae) species on the Korean peninsula and surrounding areas. Ecol. Evol. 2019, 9, 2907–2919. [Google Scholar] [CrossRef]
- Park, S.Y.; Jeon, M.J.; Ma, S.H.; Wahlsteen, E.; Amundsen, K.; Kim, J.H.; Suh, J.K.; Chang, J.S.; Joung, Y.H. Phylogeny and genetic variation in the genus Eranthis using NrITS and CpIS single nucleotide polymorphisms. Hortic. Environ. Biotechnol. 2019, 60, 239–252. [Google Scholar] [CrossRef]
- Xiang, K.-L.; Erst, A.S.; Yang, J.; Peng, H.-W.; Ortiz, R.D.C.; Jabbour, F.; Erst, T.V.; Wang, W. Biogeographic diversification of Eranthis (Ranunculaceae) reflects the geological history of the three great Asian plateaus. Proc. R. Soc. B Boil. Sci. 2021, 288, 20210281. [Google Scholar] [CrossRef] [PubMed]
- Erst, A.S.; Sukhorukov, A.P.; Mitrenina, E.Y.; Skaptsov, M.V.; Kostikova, V.A.; Chernisheva, O.A.; Troshkina, V.; Kushunina, M.; Krivenko, D.A.; Ikeda, H.; et al. An integrative taxonomic approach reveals a new species of Eranthis (Ranunculaceae) in North Asia. PhytoKeys 2020, 140, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Chepinoga, V.V.; Mishina, A.V.; Protopopova, M.V.; Pavlichenko, V.V.; Bystrov, S.O.; Vilor, M.A. New Data on Distribution of Several Nemoral Relict Plant Species on the Foothills of the Khamar-Daban Ridge (Southern Baikal). Bot. Zhurnal 2015, 100, 478–489. (In Russian) [Google Scholar]
- Timokhina, S.A.; Frizen, N.V.; Vlasova, N.V.; Zuev, V.V.; Kovtonyuk, N.K.; Baykov, K.S. Flora Sibiriae. Portulacaceae—Ranunculaceae; Malyshev, L.I., Peshkova, G.A., Eds.; Nauka: Novosibirsk, Russia, 1993; Volume 6, ISBN 5-02-030132-9. (In Russian) [Google Scholar]
- Chepinoga, V.V.; Protopopova, M.V.; Pavlichenko, V.V.; Dudov, S.V. Habitat Distribution Patterns of Nemoral Relict Plant Species on the Khamar-Daban Ridge (the South of Eastern Siberia) According to Grid Mapping Data. Russ. J. Ecol. 2021, 52, 212–222. [Google Scholar] [CrossRef]
- Shaulo, D.N.; Zykova, E.Y.; Shmakov, A.I.; Tupitsyna, N.N.; Molokova, N.I.; Artemov, I.A.; An’kova, T.V.; Sonnikova, A.E.; Shanmak, R.B.; Saak, N.V.; et al. Floristic fndings in south of Central Siberia: Krasnoyarsk Territory, Republics of Khakassia and Tuva. Turczaninowia 2019, 22, 80–93. (In Russian) [Google Scholar] [CrossRef]
- Protopopova, M.V.; Pavlichenko, V.V.; Gnutikov, A.A.; Adelshin, R.V.; Chepinoga, V.V. Application of Genetic Markers for Ecological Status Assessment of the Relict Plant Species of Baikal Siberia. RUDN J. Ecol. Life Safety 2015, 4, 28–36. (In Russian) [Google Scholar]
- Protopopova, M.V.; Pavlichenko, V.V.; Chepinoga, V.V. Some Aspects of Genetic Polymorphism in Two Relict Plant Species from Baikal Siberia. In Proceedings of the Modern Achivements in Population, Evolutionary, and Ecological Genetics: International Symposium, Vladivostoc—Vostok Marine Biological Station, Vladivostok, Russia, 1–10 September 2015; p. 57. [Google Scholar]
- Protopopova, M.V.; Pavlichenko, V.V.; Konovalov, A.D.; Chepinoga, V.V. The Study of the Historical Dynamics of Some Relic Plant Species in Baikal Siberia during Global Climatic Change Using Molecular Genetic Markers. In Proceedings of the All-Russian Conference with International Participation and School for Young Scientists: Factors of Plant and Microorganism Resistance in Extremal Nature Conditions and Technogenic Environment, Irkutsk, Russia, 12–15 September 2016; V.B. Sochava Institute of Geography SB RAS Publishing House: Irkutsk, Russia, 2016; pp. 250–251. (In Russian). [Google Scholar]
- Bobo-Pinilla, J.; Salmerón-Sánchez, E.; Mendoza-Fernández, A.J.; Mota, J.F.; Peñas, J. Conservation and Phylogeography of Plants: From the Mediterranean to the Rest of the World. Diversity 2022, 14, 78. [Google Scholar] [CrossRef]
- Shorthouse, D.P. SimpleMappr, an Online Tool to Produce Publication-Quality Point Maps. Available online: https://www.simplemappr.net (accessed on 27 July 2022).
- Polozhii, A.V.; Krapivkina, E.D. Relikty Tretichnyh Shirokolistvennyh Lesov vo Flore Sibiri [Relics of Tertiary Deciduous Forests in the Flora of Siberia]; Tomsk University Press: Tomsk, Russia, 1985. (In Russian) [Google Scholar]
- Krestov, P.V.; Barkalov, V.Y.; Omelko, A.M.; Yakubov, V.V.; Nakamura, Y.; Sato, K. Relic Vegetation Complexes in the Modern Refugia of Northeast Asia. Komarovskie Chtenia [V. L. Komarov Memorial Lectures] 2009, 56, 5–63. (In Russian) [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Utelli, A.; Roy, B.; Baltisberger, M. Molecular and morphological analyses of European Aconitum species (Ranunculaceae). Plant Syst. Evol. 2000, 224, 195–212. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Taberlet, P.; Gielly, L.; Pautou, G.; Bouvet, J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.A.; Simpson, B.B. Paraphyly of Tarasa (Malvaceae) and Diverse Origins of the Polyploid Species. Syst. Bot. 2003, 28, 723–737. [Google Scholar]
- Sang, T.; Crawford, D.J.; Stuessy, T.F. Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am. J. Bot. 1997, 84, 1120–1136. [Google Scholar] [CrossRef]
- Lahr, D.J.G.; Katz, L.A. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. BioTechniques 2009, 47, 857–866. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T.-A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C.R. Evolution of Protein Molecules; Academic Press: New York, NY, USA, 1969; Volume 3, pp. 21–132. [Google Scholar]
- Rambaut, A. FigTree: Tree Figure Drawing Tool, Version 1.4.3. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 July 2022).
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Ribosomal RNA genes in plants: Variability in copy number and in the intergenic spacer. Plant Mol. Biol. 1987, 9, 509–520. [Google Scholar] [CrossRef]
- Xu, B.; Zeng, X.-M.; Gao, X.-F.; Jin, D.-P.; Zhang, L.-B. ITS non-concerted evolution and rampant hybridization in the legume genus Lespedeza (Fabaceae). Sci. Rep. 2017, 7, 40057. [Google Scholar] [CrossRef]
- Belov, A.V.; Bezrukova, E.V.; Sokolova, L.P.; Abzayeva, A.A.; Letunova, P.P.; Fisher, E.E.; Orlova, L.A. Vegetation of the Baikal Region as an Indicator of Global and Regional Changes in Natural Conditions of North Asia in the Late Cainozoic. Geogr. Nat. Resour. 2006, 6, 5–18. (In Russian) [Google Scholar]
- Alter, S.P. K Istorii Formirovaniya Doliny Eniseya [to the History of the Yenisey Valley Formation]. Doklady instituta geografii Sibiri i Dal’nego Vostoka [Reports of the Institute of Geography of Siberia and the Far East]. 1965, 8, 38–44. (In Russian) [Google Scholar]
- Chepinoga, V.V.; Protopopova, M.V.; Pavlichenko, V.V. Detection of the most probable Pleistocene microrefugia on the northern macroslope of the Khamar-Daban Ridge (Southern Prebaikalia). Contemp. Probl. Ecol. 2017, 10, 38–42. [Google Scholar] [CrossRef]
- Protopopova, M.V.; Pavlichenko, V.V.; Orlova, D.A.; Chepinoga, V.V. Phylogeographic Structure of Anemone baicalensis (Ranunculaceae) Based on Plastid DNA Polymorphism (trnL-trnF) as an Evidence of Pleistocene Microrefugia Existence on the Khamar-Daban Ridge (Southern Baikal Region). Bull. Irkutsk State Univ. Ser. Biol. Ecol. 2019, 30, 3–15. [Google Scholar] [CrossRef]
- Malyschev, L.I. Flora Alpina Montium Sajanensium Orientalium; Nauka: Moscow, Russia; Leningrad, Russia, 1965. (In Russian) [Google Scholar]
- Krasnoborov, I.M. Flora Alpina Montium Sajanensium Occidentalium; Nauka: Moscow, Russia; Leningrad, Russia, 1976. (In Russian) [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).