Diversity of Pod Shape in Pisum
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Material
2.2. Pod Measurements
2.3. Statistical Analysis
3. Results
3.1. General Description
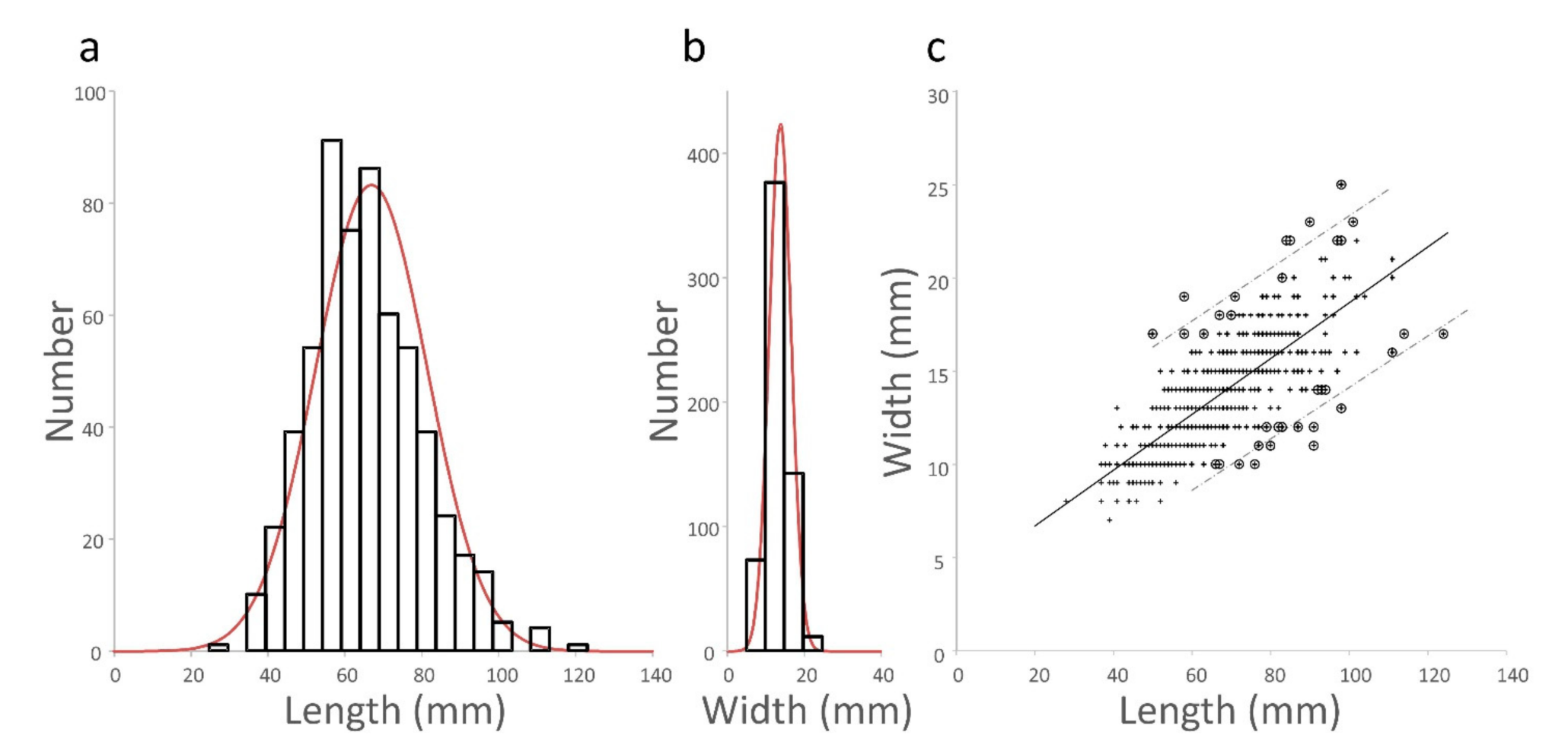
3.1.1. Distribution of Pod Length and Width Measurements
3.1.2. Length and Width Are Correlated
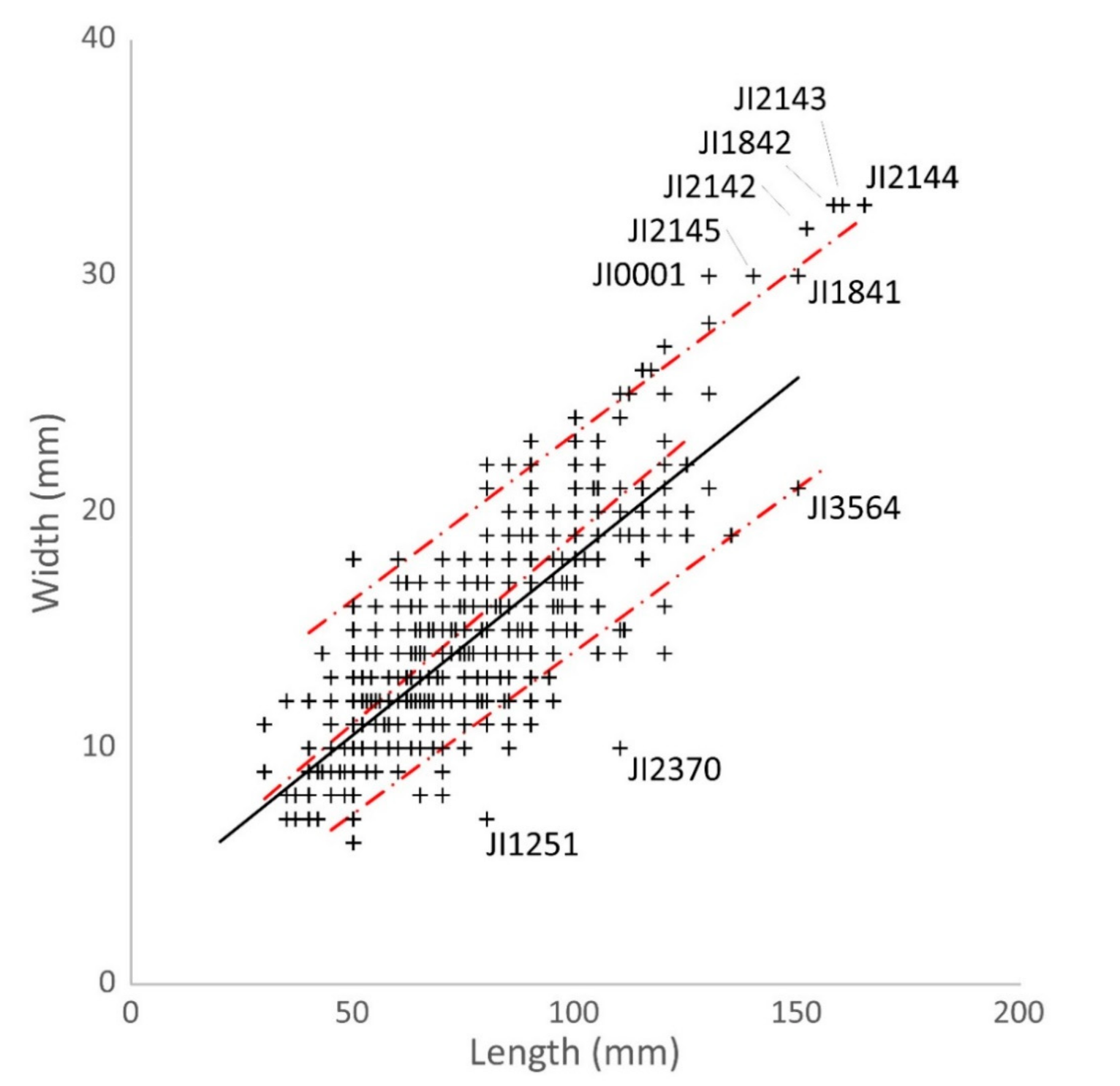
3.2. Historical Data
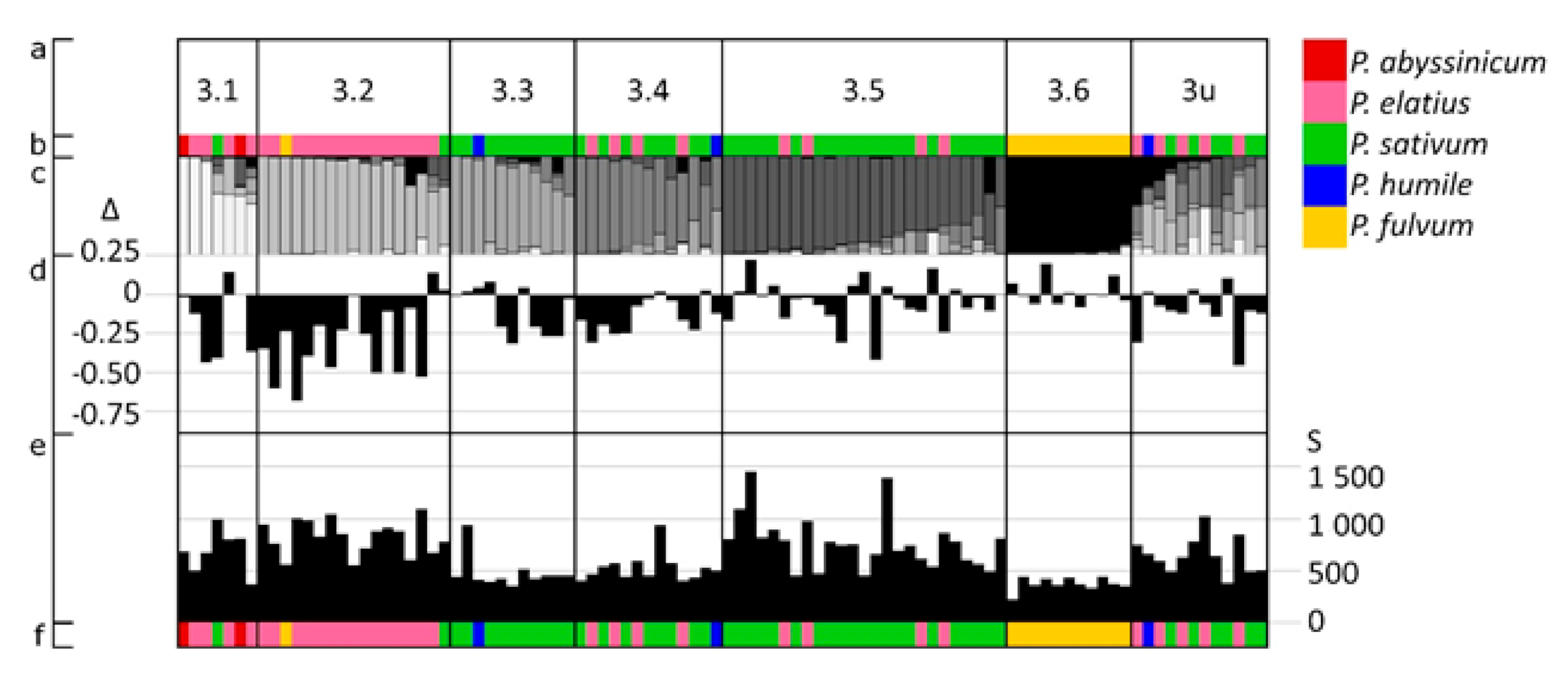
3.3. The Distribution of Pod Size, Length and Width Variation with Respect to Structure Groups
3.3.1. Pod Size
3.3.2. Pod Length: Width Ratio
3.3.3. Ovule Number, Pod Length and Pod Width
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Lewis, G.P.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the World; Kew Publishing: Kew, UK, 2005. [Google Scholar]
- Pirie, M.D.; Klitgaard, B.B.; Pennington, R.T. Revision and Biogeography of Centrolobium (Leguminosae-Papilionoideae). Syst. Bot. 2009, 34, 345–359. [Google Scholar] [CrossRef]
- Krapovickas, A.; Gregory, W.C.; Williams, D.E.; Simpson, C.E. The taxonomy of the genus Arachis (Leguminoseae). Bonplandia 2007, 16, 1–205. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of Species, 2nd ed.; Appleton & Coy: New York, NY, USA, 1861; p. 154. [Google Scholar]
- Literature.org. Available online: https://www.literature.org/authors/darwin-charles/the-origin-of-species/chapter-05.html (accessed on 19 January 2021).
- Aviezer, I.; Lev-Yadun, S. Pod and seed defensive coloration (camouflage and mimicry) in the genus Pisum. Is. J. Plant Sci. 2015, 62, 39–51. [Google Scholar] [CrossRef]
- Mejri, F.; Ben Khoud, H.; Njim, L.; Baati, T.; Selmi, S.; Martins, A.; Serralheiro, M.L.M.; Rauter, A.P.; Hosni, K. In vitro and in vivo biological properties of pea pods (Pisum sativum L.). Food Biosci. 2019, 32, 100482. [Google Scholar] [CrossRef]
- Sadras, V.O.; Lake, L.; Kaur, S.; Rosewarne, G. Phenotypic and genetic analysis of pod wall ratio, phenology and yield components in field pea. Field Crops Res. 2019, 241, 107551. [Google Scholar] [CrossRef]
- Gautam, K.K.; Syamal, M.M.; Singh, A.K.; Gupta, N. Variability, character association and path coefficient analysis of green pod yield and its related traits in pea (Pisum sativum L.). Legume Res. 2017, 40, 818–823. [Google Scholar] [CrossRef][Green Version]
- Bhuvaneswari, S.; Sharma, S.K.; Punitha, P.; Shashidhari, K.S.; Naveenkumar, K.L.; Prakash, N. Evaluation of morphological diversty of field pea (Pisum sativum subsp. arvense L.) germplasm under sub-tropical climate of Manipur. Legume Res. 2017, 40, 215–223. [Google Scholar]
- Blixt, S. Mutation genetics in Pisum. Agri. Hort. Genet. 1972, 30, 1–293. [Google Scholar]
- Jing, R.; Vershinin, A.; Grzebyta, J.; Shaw, P.; Smýkal, P.; Marshall, D.; Ambrose, M.J.; Ellis, T.H.N.; Flavell, A. The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- De Beukelaer, H.; Davenport, G.F.; Fack, V. Core Hunter 3: Flexible core subset selection. BMC Bioinform. 2018, 19, 203. [Google Scholar] [CrossRef] [PubMed]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef] [PubMed]
- Young, J.P.W.; Matthews, P. A distinct class of peas (Pisum sativum L.) from Afghanistan that show strain specificity for symbiotic rhizobium. Heredity 1982, 48, 203–210. [Google Scholar] [CrossRef]
- Li, X.; Liu, W.; Zhuang, L.; Zhu, Y.; Wang, F.; Chen, T.; Yang, J.; Ambrose, M.; Hu, Z.; Weller, J.L.; et al. BIGGER ORGANS and ELEPHANT EAR-LIKE LEAF1 control organ size and floral organ internal asymmetry in pea. J. Exp. Bot. 2019, 70, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Hradilová, I.; Trněný, O.; Brus, J.; Rathore, A.; Bariotakis, M.; Das, R.R.; Bhattacharyya, D.; Richards, C.; Coyne, C.J.; et al. Genomic diversity and macroecology of the crop wild relatives of domesticated pea. Sci. Rep. 2017, 7, 17384. [Google Scholar] [CrossRef] [PubMed]
- Trněný, O.; Brus, J.; Hradilová, I.; Rathore, A.; Das, R.R.; Kopecký, P.; Coyne, C.J.; Reeves, P.; Richards, C.; Smýkal, P. Molecular Evidence for Two Domestication Events in the Pea Crop. Genes 2018, 9, 535. [Google Scholar] [CrossRef] [PubMed]


| Gene Name | Symbol 1 | Phenotypic Description |
|---|---|---|
| acutilegumen | bt | pod apex pointed |
| convexum | co con | pod curved convex with respect to the seed side of attachment |
| concavum | cp cpa | pod curved concavely with respect to the seed side of attachment |
| contractivus | cotr curt | pod length reduced by ca. 20% |
| tenuis | te teu | pod breadth reduced by ca. 25% |
| latus | lt | increased pod width |
| Laitor | Laf | broader pod |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ellis, T.H.N.; Hofer, J.M.I.; Vikeli, E.; Ambrose, M.J.; Higuera-Poveda, P.; Wingen, L.U.; Chayut, N. Diversity of Pod Shape in Pisum. Diversity 2021, 13, 203. https://doi.org/10.3390/d13050203
Ellis THN, Hofer JMI, Vikeli E, Ambrose MJ, Higuera-Poveda P, Wingen LU, Chayut N. Diversity of Pod Shape in Pisum. Diversity. 2021; 13(5):203. https://doi.org/10.3390/d13050203
Chicago/Turabian StyleEllis, Thomas Henry Noel, Julie M. I. Hofer, Eleni Vikeli, Michael J. Ambrose, Paola Higuera-Poveda, Luzie U. Wingen, and Noam Chayut. 2021. "Diversity of Pod Shape in Pisum" Diversity 13, no. 5: 203. https://doi.org/10.3390/d13050203
APA StyleEllis, T. H. N., Hofer, J. M. I., Vikeli, E., Ambrose, M. J., Higuera-Poveda, P., Wingen, L. U., & Chayut, N. (2021). Diversity of Pod Shape in Pisum. Diversity, 13(5), 203. https://doi.org/10.3390/d13050203






