Biogeography of Long-Jawed Spiders Reveals Multiple Colonization of the Caribbean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Material Acquisition
2.2. Molecular Procedures
2.3. Species Delimitation
2.4. Phylogenetic Analyses
2.4.1. Two Gene, Species Level Phylogeny
2.4.2. All-Terminal, Single Gene Phylogeny
2.4.3. Caribbean Tetragnatha Monophyly Testing
2.4.4. Time-Calibrated Phylogenetic Reconstruction
2.5. Ancestral Area Estimation and Biogeographic Stochastic Mapping
3. Results
3.1. Species Delimitation
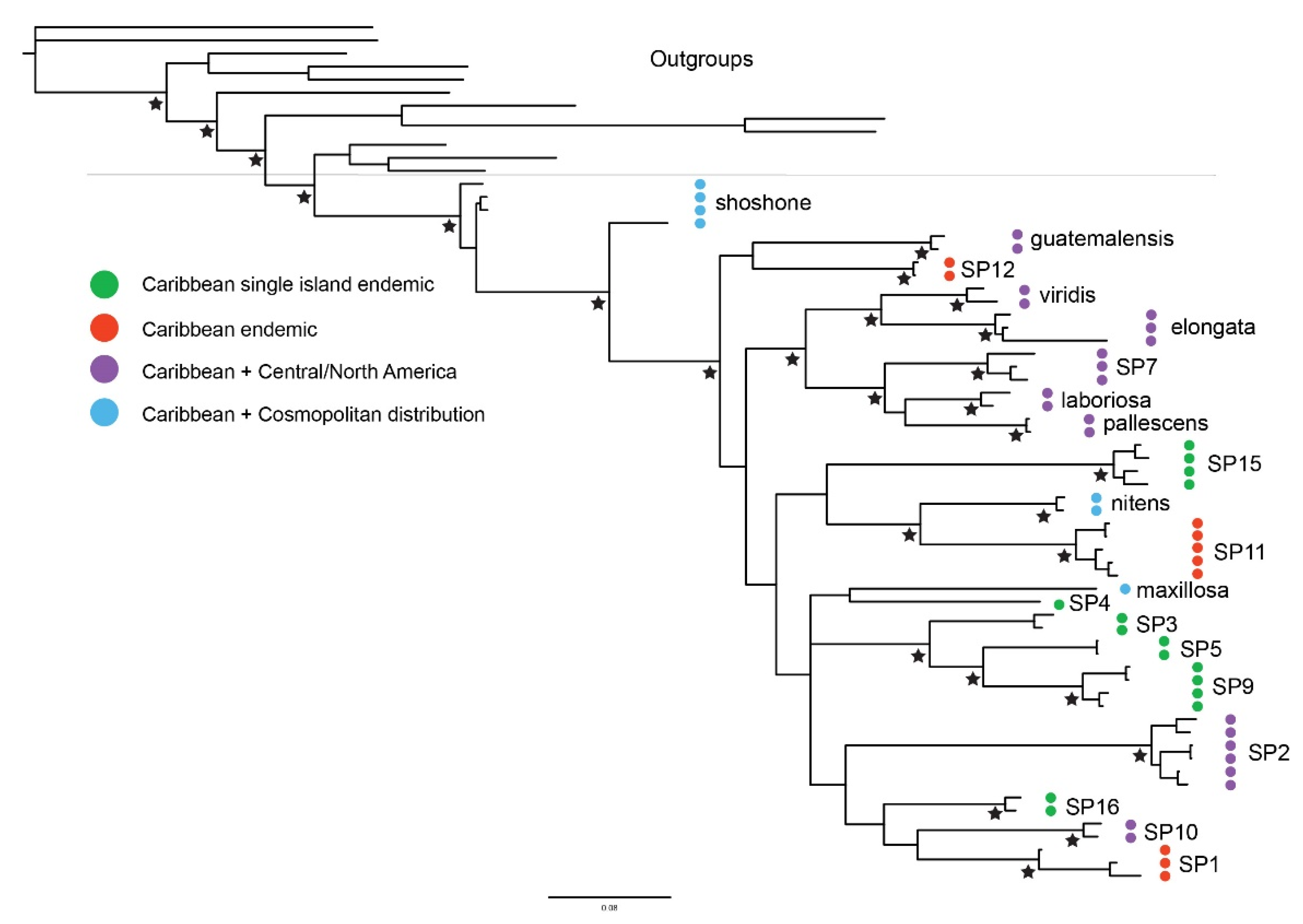
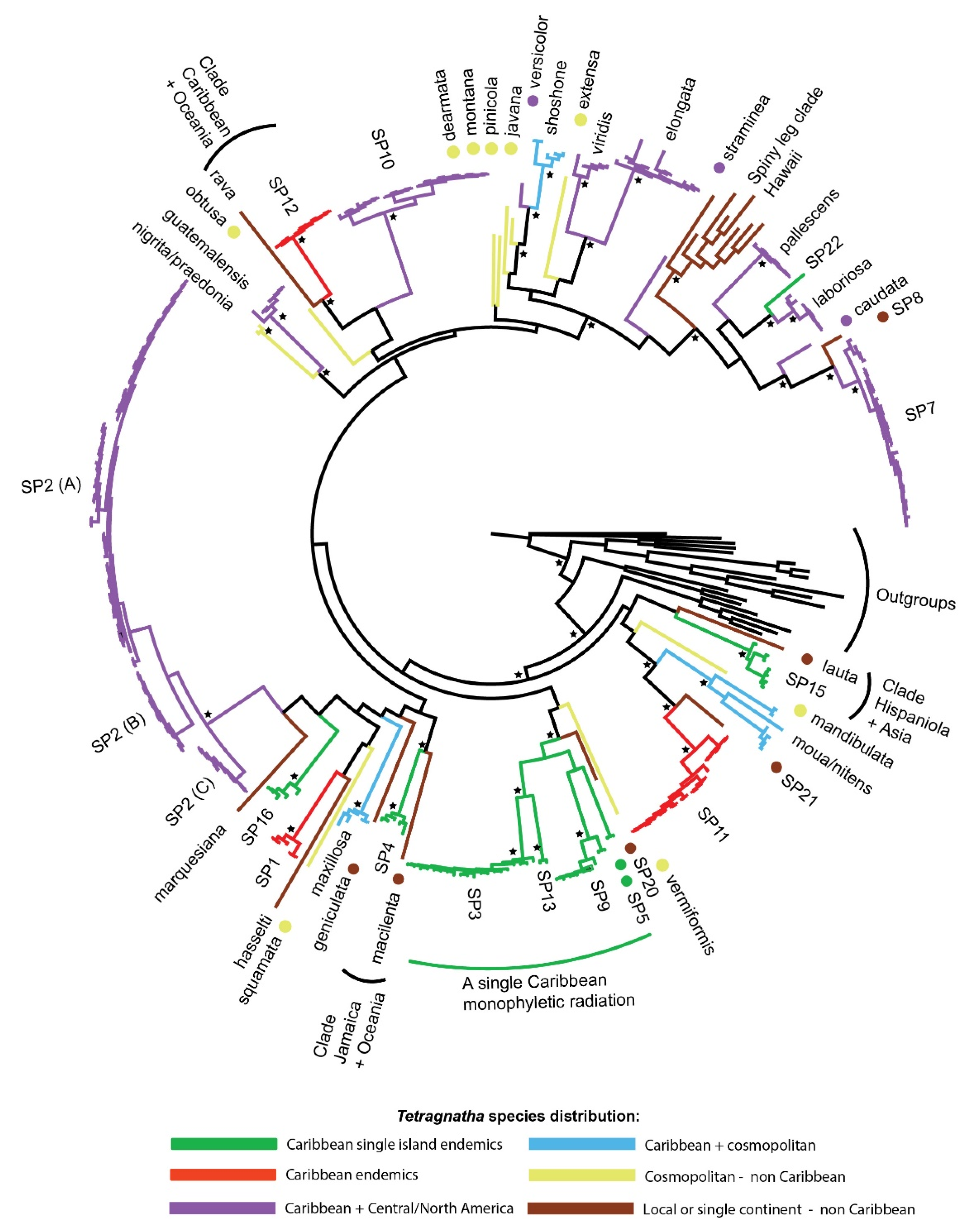
3.2. Molecular Phylogeny
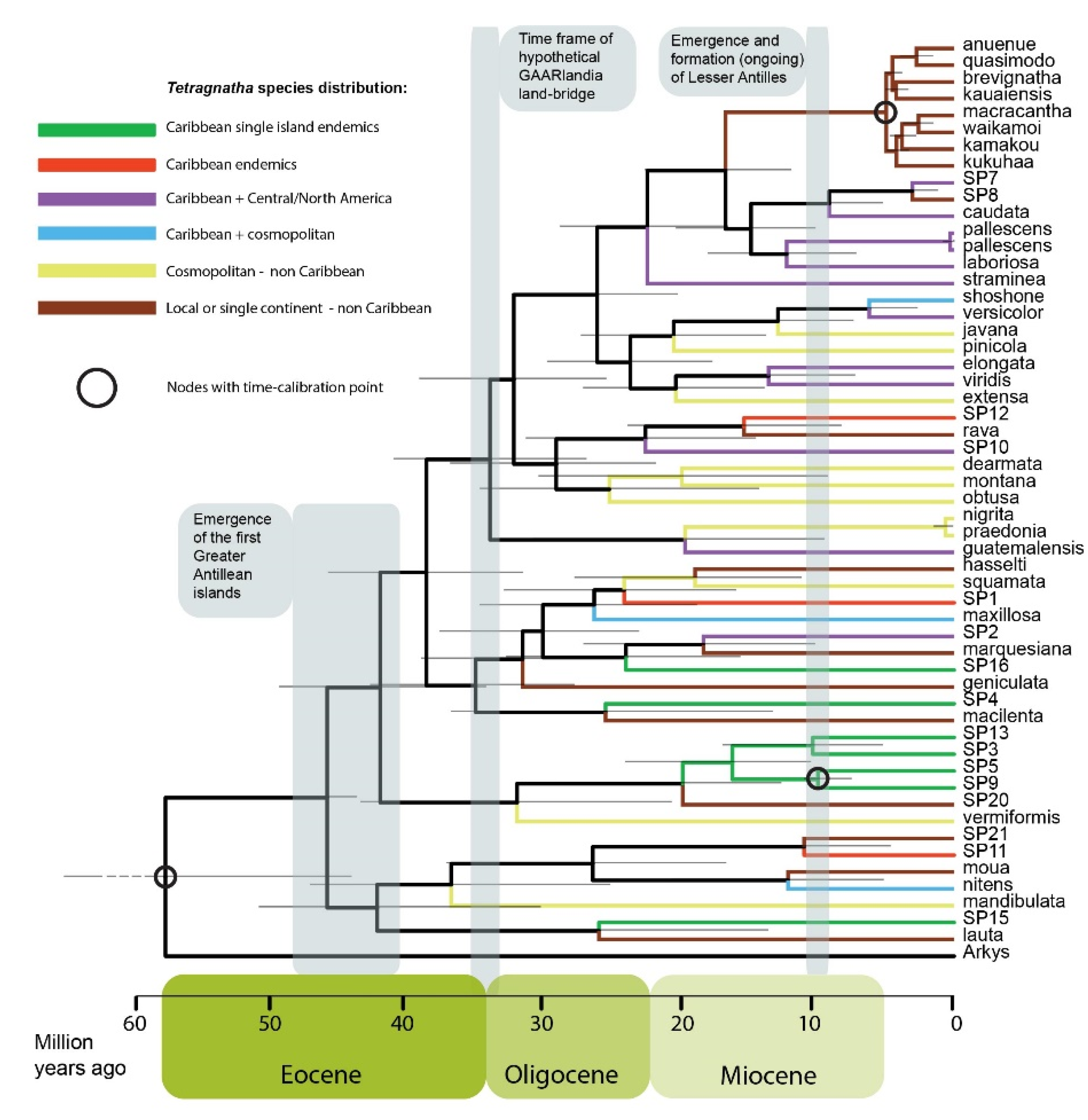
3.3. Time Calibrated Phylogeny
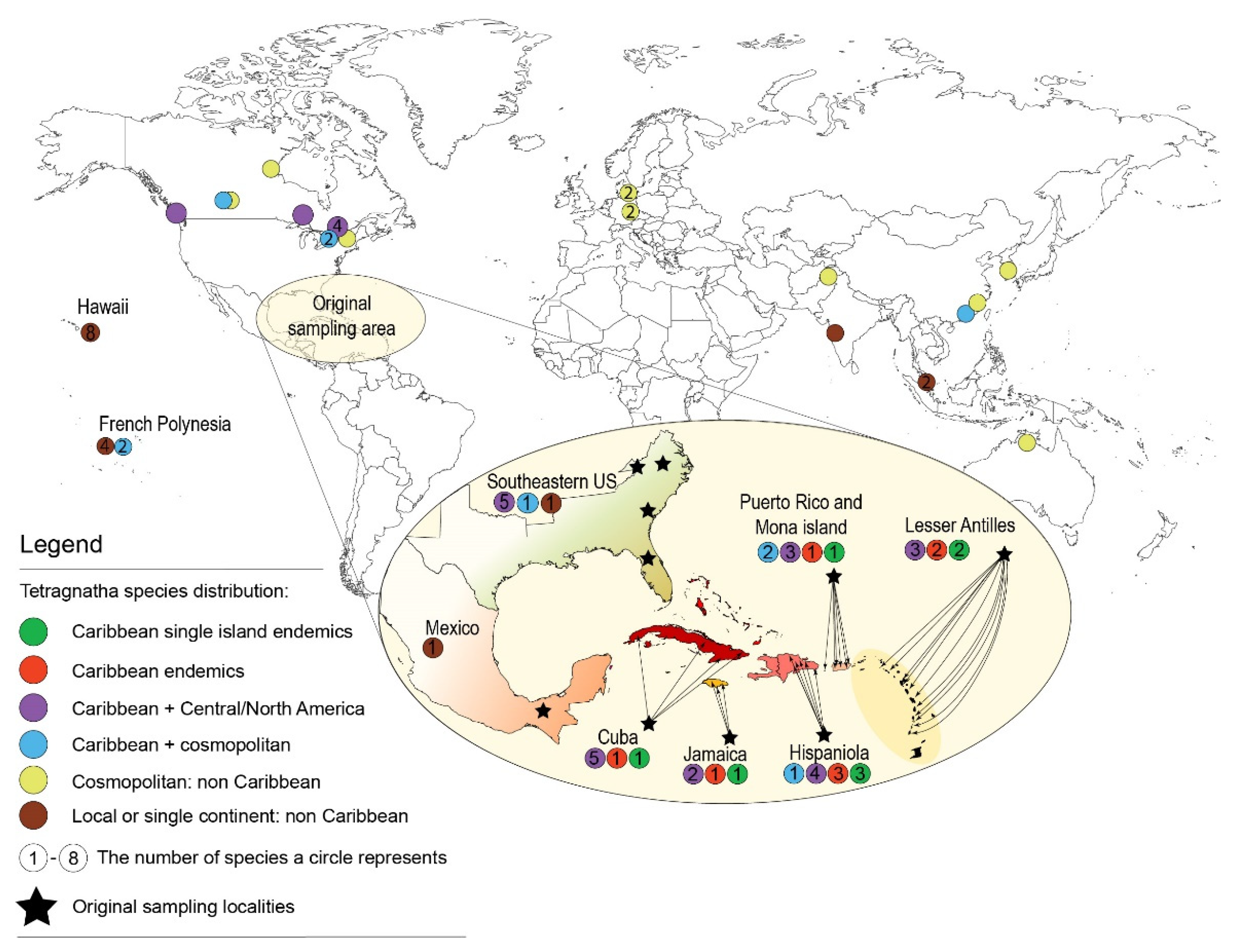
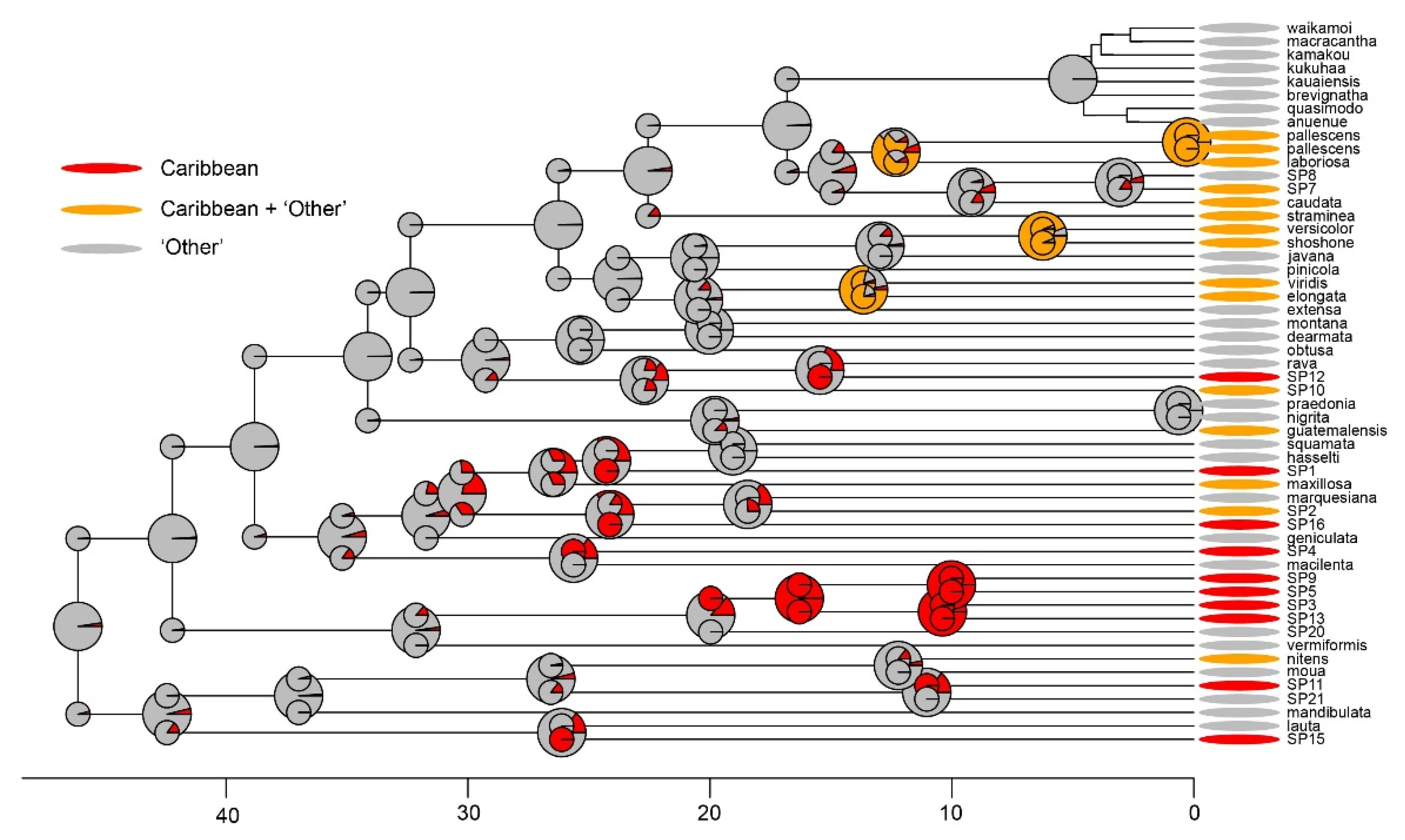
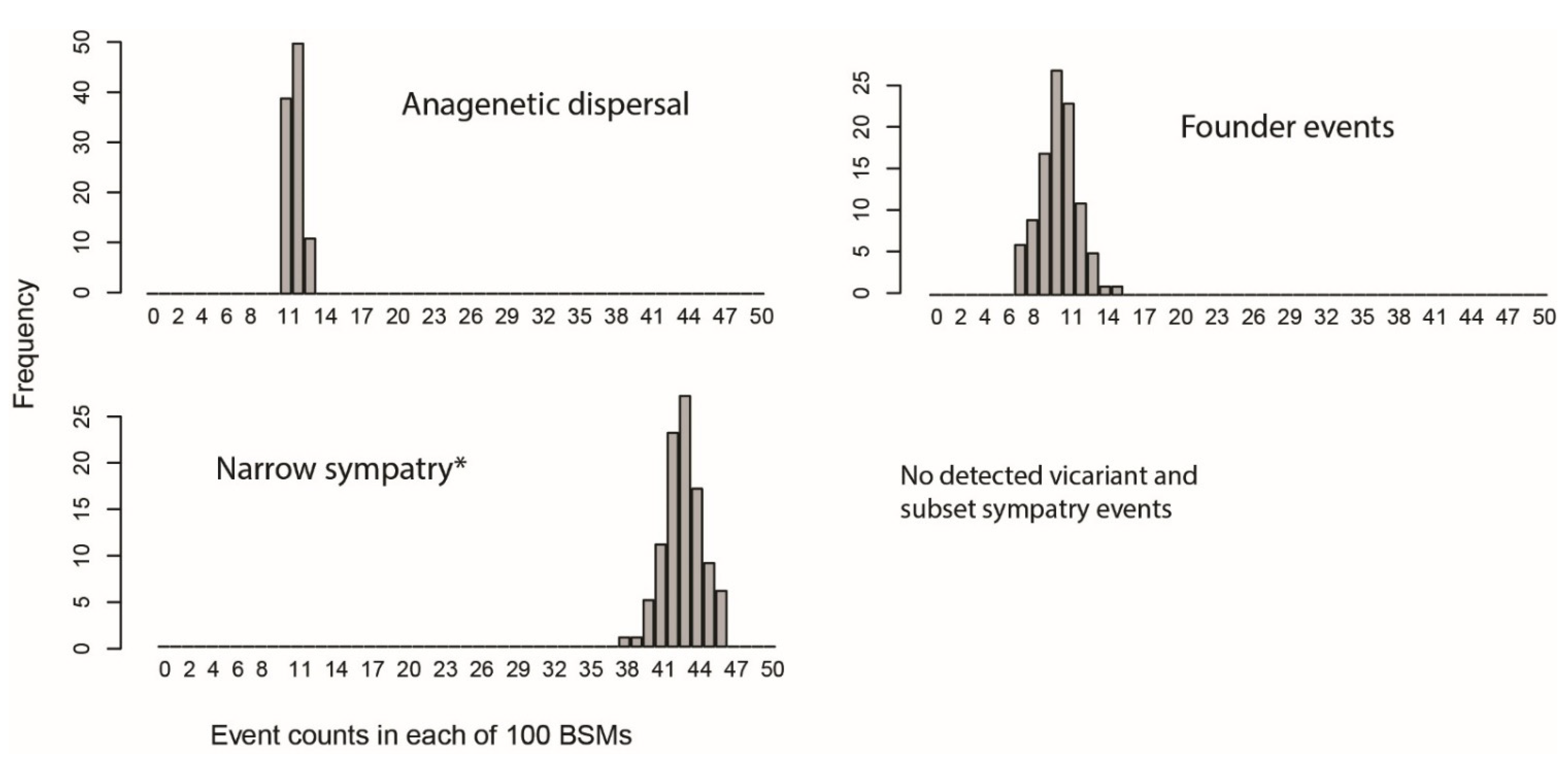
3.4. Biogeographic Analyses
4. Discussion
4.1. Caribbean Tetragnatha Are Not Monophyletic
4.2. Biogeographic History of Caribbean Tetragnatha
4.3. Tetragnatha Dispersal Abilities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozak, K.H.; Wiens, J.J. What explains patterns of species richness? The relative importance of climatic-niche evolution, morphological evolution, and ecological limits in salamanders. Ecol. Evol. 2016, 6, 5940–5949. [Google Scholar] [CrossRef] [Green Version]
- Kozak, K.H.; Wiens, J.J. Testing the Relationships between Diversification, Species Richness, and Trait Evolution. Syst. Biol. 2016, 65, 975–988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabosky, D.L.; Hurlbert, A.H. Species richness at continental scales is dominated by ecological limits. Am. Nat. 2015, 185, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Schluter, D.; Pennell, M.W. Speciation gradients and the distribution of biodiversity. Nature 2017, 546, 48–55. [Google Scholar] [CrossRef]
- Rajakaruna, H.; Lewis, M. Do yearly temperature cycles reduce species richness? Insights from calanoid copepods. Theor. Ecol. 2018, 11, 39–53. [Google Scholar] [CrossRef]
- Marin, J.; Hedges, S.B. Time best explains global variation in species richness of amphibians, birds and mammals. J. Biogeogr. 2016, 43, 1069–1079. [Google Scholar] [CrossRef]
- Borda-de-Água, L.; Whittaker, R.J.; Cardoso, P.; Rigal, F.; Santos, A.M.C.C.; Amorim, I.R.; Parmakelis, A.; Triantis, K.A.; Pereira, H.M.; Borges, P.A.V. Dispersal ability determines the scaling properties of species abundance distributions: A case study using arthropods from the Azores. Sci. Rep. 2017, 7, 3899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenoir, J.; Virtanen, R.; Oksanen, J.; Oksanen, L.; Luoto, M.; Grytnes, J.A.; Svenning, J.C. Dispersal ability links to cross-scale species diversity patterns across the Eurasian Arctic tundra. Glob. Ecol. Biogeogr. 2012, 21, 851–860. [Google Scholar] [CrossRef]
- Claramunt, S.; Derryberry, E.P.; Remsen, J.V.; Brumfield, R.T. High dispersal ability inhibits speciation in a continental radiation of passerine birds. Proc. R. Soc. B Biol. Sci. 2012, 279, 1567–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agnarsson, I.; Cheng, R.-C.; Kuntner, M. A multi-clade test supports the intermediate dispersal model of biogeography. PLoS ONE 2014, 9, e86780. [Google Scholar] [CrossRef]
- Opatova, V.; Arnedo, M.A. Spiders on a Hot Volcanic Roof: Colonisation Pathways and Phylogeography of the Canary Islands Endemic Trap-Door Spider Titanidiops canariensis (Araneae, Idiopidae). PLoS ONE 2014, 9, e115078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitta, E.; Kassara, C.; Tzanatos, E.; Giokas, S.; Sfenthourakis, S. Between-island compositional dissimilarity of avian communities. Ecol. Res. 2014, 29, 835–841. [Google Scholar] [CrossRef]
- Essl, F.; Steinbauer, K.; Dullinger, S.; Mang, T.; Moser, D. Telling a different story: A global assessment of bryophyte invasions. Biol. Invasions 2013, 15, 1933–1946. [Google Scholar] [CrossRef]
- Kuntner, M.; Agnarsson, I. Phylogeography of a successful aerial disperser: The golden orb spider Nephila on Indian Ocean islands. BMC Evol. Biol. 2011, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Turk, E.; Čandek, K.; Kralj-Fišer, S.; Kuntner, M. Biogeographical history of golden orbweavers: Chronology of a global conquest. J. Biogeogr. 2020, 47, 1333–1344. [Google Scholar] [CrossRef]
- Darwin, C. On the Origin of the Species; John Murray: London, UK, 1859; Volume 5, ISBN 1546622497. [Google Scholar]
- Warren, B.H.; Simberloff, D.; Ricklefs, R.E.; Aguilée, R.; Condamine, F.L.; Gravel, D.; Morlon, H.; Mouquet, N.; Rosindell, J.; Casquet, J.; et al. Islands as model systems in ecology and evolution: Prospects fifty years after MacArthur-Wilson. Ecol. Lett. 2015, 18, 200–217. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, R.G.; Collar, D.C.; Pasachnik, S.A.; Niemiller, M.L.; Puente-Rolón, A.R.; Revell, L.J. Ecological specialization and morphological diversification in Greater Antillean boas. Evolution 2016, 70, 1882–1895. [Google Scholar] [CrossRef]
- Ricklefs, R.E.; Bermingham, E. The West Indies as a laboratory of biogeography and evolution. Philos. Trans. R. Soc. B Biol. Sci. 2008, 363, 2393–2413. [Google Scholar] [CrossRef] [Green Version]
- Frankham, R. Inbreeding and Extinction: Island Populations. Conserv. Biol. 2008, 12, 665–675. [Google Scholar] [CrossRef]
- Meiri, S. Oceanic island biogeography: Nomothetic science of the anecdotal. Front. Biogeogr. 2017, 9, e32081. [Google Scholar] [CrossRef]
- Gillespie, R.G.; Croom, H.B.; Hasty, G.L. Phylogenetic Relationships and Adaptive Shifts among Major Clades of Tetragnatha Spiders (Araneae: Tetragnathidae) in Hawaí. Pacific Sci. 1997, 51, 380–394. [Google Scholar]
- Crews, S.C.; Esposito, L.A. Towards a synthesis of the Caribbean biogeography of terrestrial arthropods. BMC Evol. Biol. 2020, 20, 12. [Google Scholar] [CrossRef]
- Čandek, K.; Agnarsson, I.; Binford, G.J.; Kuntner, M. Biogeography of the Caribbean Cyrtognatha spiders. Sci. Rep. 2019, 9, 397. [Google Scholar] [CrossRef] [Green Version]
- Ali, J.R. Colonizing the Caribbean: Is the GAARlandia land-bridge hypothesis gaining a foothold? J. Biogeogr. 2012, 39, 431–433. [Google Scholar] [CrossRef]
- Iturralde-Vinent, M.A.; MacPhee, R.D.E. Paleogeography of the Caribbean Region: Implications for Cenozoic Biogeography. Bull. Am. Museum Nat. Hist. 1999, 238, 791. [Google Scholar] [CrossRef] [Green Version]
- Ali, J.R.; Hedges, S.B. Colonizing the Caribbean: New geological data and an updated land-vertebrate colonization record challenge the GAARlandia land-bridge hypothesis. J. Biogeogr. 2021, 48, jbi.14234. [Google Scholar] [CrossRef]
- Gillespie, R.G.; Benjamin, S.P.; Brewer, M.S.; Rivera, M.A.J.; Roderick, G.K. Repeated Diversification of Ecomorphs in Hawaiian Stick Spiders. Curr. Biol. 2018, 28, 941–947.e3. [Google Scholar] [CrossRef] [Green Version]
- Cotoras, D.D.; Bi, K.; Brewer, M.S.; Lindberg, D.R.; Prost, S.; Gillespie, R.G. Co-occurrence of ecologically similar species of Hawaiian spiders reveals critical early phase of adaptive radiation. BMC Evol. Biol. 2018, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Agnarsson, I.; van Patten, C.; Sargeant, L.; Chomitz, B.; Dziki, A.; Binford, G.J. A radiation of the ornate Caribbean ‘smiley-faced spiders’, with descriptions of 15 new species (Araneae: Theridiidae, Spintharus). Zool. J. Linn. Soc. 2018, 182, 758–790. [Google Scholar] [CrossRef]
- Agnarsson, I.; LeQuier, S.M.; Kuntner, M.; Cheng, R.-C.; Coddington, J.A.; Binford, G.J. Phylogeography of a good Caribbean disperser: Argiope argentata (Araneae, Araneidae) and a new ‘cryptic’ species from Cuba. Zookeys 2016, 2016, 25–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamberland, L.; McHugh, A.; Kechejian, S.; Binford, G.J.; Bond, J.E.; Coddington, J.; Dolman, G.; Hamilton, C.A.; Harvey, M.S.; Kuntner, M.; et al. From Gondwana to GAARlandia: Evolutionary history and biogeography of ogre-faced spiders (Deinopis). J. Biogeogr. 2018, 45, 2442–2457. [Google Scholar] [CrossRef] [Green Version]
- Čandek, K.; Agnarsson, I.; Binford, G.J.; Kuntner, M. Caribbean golden orbweaving spiders maintain gene flow with North America. Zool. Scr. 2020, 49, 210–221. [Google Scholar] [CrossRef]
- World Spider Catalog World Spider Catalog. 2021. Available online: https://wsc.nmbe.ch/ (accessed on 10 October 2021).
- Foelix, R. Biology of Spiders, 3rd ed.; Oxford University Press: Oxford, UK, 2011; ISBN 9780199734825. [Google Scholar]
- Čandek, K.; Pristovšek Čandek, U.; Kuntner, M. Machine learning approaches identify male body size as the most accurate predictor of species richness. BMC Biol. 2020, 18, 105. [Google Scholar] [CrossRef] [PubMed]
- Cotoras, D.D.; Murray, G.G.R.; Kapp, J.; Gillespie, R.G.; Griswold, C.E.; Simison, W.B.; Green, R.E.; Shapiro, B. Ancient DNA resolves the history of Tetragnatha (Araneae, Tetragnathidae) spiders on Rapa Nui. Genes 2017, 8, 403. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, R.G. Spiders of the Genus Tetragnatha) (Araneae, Tetragnathidae) in the Society Islands. J. Arachnol. 2003, 31, 157–172. [Google Scholar] [CrossRef]
- Gillespie, R.G. Geographical context of speciation in a radiation of Hawaiian Tetragnatha spiders (Araneae, Tetragnathidae). J. Arachnol. 2005, 33, 313–322. [Google Scholar] [CrossRef]
- Casquet, J.; Bourgeois, Y.X.C.; Cruaud, C.; Gavory, F.; Gillespie, R.G.; Thébaud, C. Community assembly on remote islands: A comparison of Hawaiian and Mascarene spiders. J. Biogeogr. 2015, 42, 39–50. [Google Scholar] [CrossRef]
- Okuma, C.; Kisimoto, R. Airborne spiders collected over the East China Sea. Jpn. J. Appl. Entomol. Zool. 1981, 25, 296–308. [Google Scholar] [CrossRef]
- Bell, J.R.; Bohan, D.A.; Shaw, E.M.; Weyman, G.S. Ballooning dispersal using silk: World fauna, phylogenies, genetics and models. Bull. Entomol. Res. 2005, 95, 69–114. [Google Scholar] [CrossRef]
- Hedges, S.B. Paleogeography of the Antilles and Origin of West Indian Terrestrial Vertebrates. Ann. Missouri Bot. Gard. 2006, 93, 231–244. [Google Scholar] [CrossRef]
- Shapiro, L.; Binford, G.J.; Agnarsson, I. Single island endemism despite repeated dispersal in Caribbean Micrathena (Araneae: Araneidae), an updated phylogeographic analysis. Diversity 2021, 13, 1–27, in press. [Google Scholar]
- Agnarsson, I.; Coddington, J.A.; Kuntner, M. Systematics: Progress in the study of spider diversity and evolution. In Spider Research in the 21st Century: Trends and Perspectives; Penney, D., Ed.; Siri Scientific Press: Manchester, UK, 2013; pp. 55–111. ISBN 0957453019. [Google Scholar]
- Coddington, J.A.; Griswold, C.E.; Silva, D.; Peqaranda, E.; Larcher, S.F.; Penaranda, E.; Larcher, S.F. Designing and testing sampling protocols to estimate biodiversity in tropical ecosystems. In The Unity of Evolutionary Biology: Proceedings of the Fourth International Congress of Systematic and Evolutionary Biology; Dioscorides Press: Portland, OR, USA, 1991; pp. 44–60. [Google Scholar]
- Vidergar, N.; Toplak, N.; Kuntner, M. Streamlining DNA barcoding protocols: Automated DNA extraction and a new cox1 primer in arachnid systematics. PLoS ONE 2014, 9, e113030. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [CrossRef]
- Hedin, M.C.; Maddison, W.P. A Combined Molecular Approach to Phylogeny of the Jumping Spider Subfamily Dendryphantinae (Araneae: Salticidae). Mol. Phylogenet. Evol. 2001, 18, 386–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunn, G.B.; Theisen, B.F.; Christensen, B.; Arctander, P. Simplicity-correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order isopoda. J. Mol. Evol. 1996, 42, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Whiting, M.F. Mecoptera is paraphyletic: Multiple genes and phylogeny of Mecoptera and Siphonaptera. Zool. Scr. 2002, 31, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Maddison, W.P.; Maddison, D.R. Mesquite: A modular system for evolutionary analysis. Evolution 2008, 62, 1103–1118. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Drummond, A.J. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017, 17, 42. [Google Scholar] [CrossRef] [Green Version]
- Bidegaray-Batista, L.; Arnedo, M.A. Gone with the plate: The opening of the Western Mediterranean basin drove the diversification of ground-dweller spiders. BMC Evol. Biol. 2011, 11, 317. [Google Scholar] [CrossRef] [Green Version]
- Peck, S.B. Diversity and distribution of beetles (Insecta: Coleoptera) of the northern Leeward Islands. Insecta Mundi 2011, 678, 159. [Google Scholar]
- Garrison, N.L.; Rodriguez, J.; Agnarsson, I.; Coddington, J.A.; Griswold, C.E.; Hamilton, C.A.; Hedin, M.C.; Kocot, K.M.; Ledford, J.M.; Bond, J.E. Spider phylogenomics: Untangling the spider tree of life. PeerJ 2016, 4, e1719. [Google Scholar] [CrossRef] [Green Version]
- Bond, J.E.; Garrison, N.L.; Hamilton, C.A.; Godwin, R.L.; Hedin, M.C.; Agnarsson, I. Phylogenomics resolves a spider backbone phylogeny and rejects a prevailing paradigm for orb web evolution. Curr. Biol. 2014, 24, 1765–1771. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, D.; Benavides, L.R.; Arnedo, M.A.; Giribet, G.; Griswold, C.E.; Scharff, N.; Hormiga, G. Rounding up the usual suspects: A standard target-gene approach for resolving the interfamilial phylogenetic relationships of ecribellate orb-weaving spiders with a new family-rank classification (Araneae, Araneoidea). Cladistics 2017, 33, 221–250. [Google Scholar] [CrossRef]
- Matzke, N.J. BioGeoBEARS: BioGeography with Bayesian (and Likelihood) Evolutionary Analysis in R Scripts. R Packag. Version 0.2 2013, 1. Available online: https://CRAN.R-project.org/package=BioGeoBEARS (accessed on 10 September 2021).
- R Core Team. R: A Language and Environment for Statistical Computing. 2018. Available online: https://www.r-project.org/ (accessed on 15 November 2021).
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Čandek, K.; Kuntner, M. DNA barcoding gap: Reliable species identification over morphological and geographical scales. Mol. Ecol. Resour. 2015, 15, 268–277. [Google Scholar] [CrossRef]
- Brandley, M.C.; Wang, Y.; Guo, X.; Montes de Oca, A.N.; Fería-Ortíz, M.; Hikida, T.; Ota, H. Accommodating heterogenous rates of evolution in molecular divergence dating methods: An example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Syst. Biol. 2011, 60, 3–15. [Google Scholar] [CrossRef]
- Sánchez-Ruiz, A.; Brescovit, A.D.; Alayón, G. Four new caponiids species (Araneae, Caponiidae) from the West Indies and redescription of Nops blandus (Bryant). Zootaxa 2015, 3972, 43–64. [Google Scholar] [CrossRef]
- Zhang, J.; Maddison, W.P. New euophryine jumping spiders from the Dominican Republic and Puerto Rico (Araneae: Salticidae: Euophryinae). Zootaxa 2012, 3476, 1–54. [Google Scholar] [CrossRef]
- Crews, S.C.; Puente-Rolón, A.R.; Rutstein, E.; Gillespie, R.G. A comparison of populations of island and adjacent mainland species of Caribbean Selenops (Araneae: Selenopidae) spiders. Mol. Phylogenet. Evol. 2010, 54, 970–983. [Google Scholar] [CrossRef]
- Dziki, A.; Binford, G.J.; Coddington, J.A.; Agnarsson, I. Spintharus flavidus in the Caribbean—A 30 million year biogeographical history and radiation of a ‘widespread species’. PeerJ 2015, 3, e1422. [Google Scholar] [CrossRef] [Green Version]
- McHugh, A.; Yablonsky, C.; Binford, G.J.; Agnarsson, I. Molecular phylogenetics of Caribbean Micrathena (Araneae: Araneidae) suggests multiple colonisation events and single island endemism. Invertebr. Syst. 2014, 28, 337–349. [Google Scholar] [CrossRef]
- Gillespie, R.G.; Baldwin, B.G.; Waters, J.M.; Fraser, C.I.; Nikula, R.; Roderick, G.K. Long-distance dispersal: A framework for hypothesis testing. Trends Ecol. Evol. 2012, 27, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.R.; Grant, B.R. Adaptive radiation of Darwin’s finches. Am. Sci. 2002, 90, 130. [Google Scholar] [CrossRef]
- Lovette, I.J.; Bermingham, E.; Ricklefs, R.E. Clade-specific morphological diversification and adaptive radiation in Hawaiian songbirds. Proc. R. Soc. B Biol. Sci. 2002, 269, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Hass, C.A.; Hedges, S.B.; Maxson, L.R. Molecular insights into the relationships and biogeography of West Indian anoline lizards. Biochem. Syst. Ecol. 1993, 21, 97–114. [Google Scholar] [CrossRef] [Green Version]
- Glor, R.E.; Gifford, M.E.; Larson, A.; Losos, J.B.; Schettino, L.R.; Lara, A.R.C.; Jackman, T.R. Partial island submergence and speciation in an adaptive radiation: A multilocus analysis of the Cuban green anoles. Proc. R. Soc. B Biol. Sci. 2004, 271, 2257–2265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Losos, J.B.; Schluter, D. Analysis of an evolutionary species-area relationship. Nature 2000, 408, 847–850. [Google Scholar] [CrossRef] [PubMed]
- Hedges, S.B. Biogeography of the West Indies: An overview. In Biogeography of the West Indies: Patterns and Perspectives; Woods, C.A., Sergile, F.E., Eds.; CRC Press: Boca Raton, FL, USA, 2001; pp. 15–33. ISBN 0849320011101. [Google Scholar]
- Říčan, O.; Piálek, L.; Zardoya, R.; Doadrio, I.; Zrzavý, J. Biogeography of the Mesoamerican Cichlidae (Teleostei: Heroini): Colonization through the GAARlandia land bridge and early diversification. J. Biogeogr. 2013, 40, 579–593. [Google Scholar] [CrossRef] [Green Version]
- Alonso, R.; Crawford, A.J.; Bermingham, E. Molecular phylogeny of an endemic radiation of Cuban toads (Bufonidae: Peltophryne) based on mitochondrial and nuclear genes. J. Biogeogr. 2012, 39, 434–451. [Google Scholar] [CrossRef]
- Dávalos, L.M. Phylogeny and biogeography of Caribbean mammals. Biol. J. Linn. Soc. 2004, 81, 373–394. [Google Scholar] [CrossRef]
- Binford, G.J.; Callahan, M.S.; Bodner, M.R.; Rynerson, M.R.; Núñez, P.B.; Ellison, C.E.; Duncan, R.P. Phylogenetic relationships of Loxosceles and Sicarius spiders are consistent with Western Gondwanan vicariance. Mol. Phylogenet. Evol. 2008, 49, 538–553. [Google Scholar] [CrossRef]
- Matos-Maraví, P.; Núñez Águila, R.; Peña, C.; Miller, J.Y.; Sourakov, A.; Wahlberg, N. Causes of endemic radiation in the Caribbean: Evidence from the historical biogeography and diversification of the butterfly genus Calisto (Nymphalidae: Satyrinae: Satyrini). BMC Evol. Biol. 2014, 14, 199. [Google Scholar] [CrossRef] [Green Version]
- Fritsch, P.W. Multiple Geographic Origins of Antillean Styrax. Syst. Bot. 2003, 28, 421–430. [Google Scholar] [CrossRef]
- van Ee, B.W.; Berry, P.E.; Riina, R.; Gutiérrez Amaro, J.E. Molecular Phylogenetics and Biogeography of the Caribbean-Centered Croton Subgenus Moacroton (Euphorbiaceae s.s.). Bot. Rev. 2008, 74, 132–165. [Google Scholar] [CrossRef]
- Nieto-Blázquez, M.E.; Antonelli, A.; Roncal, J. Historical Biogeography of endemic seed plant genera in the Caribbean: Did GAARlandia play a role? Ecol. Evol. 2017, 7, 10158–10174. [Google Scholar] [CrossRef]
- Graham, A. Geohistory models and Cenozoic paleoenvironments of the Caribbean region. Syst. Bot. 2003, 28, 378–386. [Google Scholar] [CrossRef]
- MacPhee, R.D.E.; Grimaldi, D.A. Mammal bones in Dominican amber. Nature 1996, 380, 489–490. [Google Scholar] [CrossRef]
- Shaw, K.L.; Gillespie, R.G. Comparative phylogeography of oceanic archipelagos: Hotspots for inferences of evolutionary process. Proc. Natl. Acad. Sci. USA 2016, 113, 7986–7993. [Google Scholar] [CrossRef] [Green Version]
- Dupin, J.; Matzke, N.J.; Särkinen, T.; Knapp, S.; Olmstead, R.G.; Bohs, L.; Smith, S.D. Bayesian estimation of the global biogeographical history of the Solanaceae. J. Biogeogr. 2017, 44, 887–899. [Google Scholar] [CrossRef]
- Magalhaes, I.L.F.; Santos, A.J.; Ramírez, M.J. Incorporating Topological and Age Uncertainty into Event-Based Biogeography of Sand Spiders Supports Paleo-Islands in Galapagos and Ancient Connections among Neotropical Dry Forests. Diversity 2021, 13, 418. [Google Scholar] [CrossRef]
- Gillespie, R.G. Biogeography of spiders on remote oceanic islands of the Pacific: Archipelagoes as stepping stones? J. Biogeogr. 2002, 29, 655–662. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, R.G.; Croom, H.B.; Palumbi, S.R. Multiple origins of a spider radiation in Hawaii. Proc. Natl. Acad. Sci. USA 1994, 91, 2290–2294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linck, E.; Schaack, S.; Dumbacher, J.P. Genetic differentiation within a widespread “supertramp” taxon: Molecular phylogenetics of the Louisiade White-eye (Zosterops griseotinctus). Mol. Phylogenet. Evol. 2016, 94, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.P.; Irestedt, M.; Joseph, L.; Rahbek, C.; Jønsson, K.A. Phylogeography of a ‘great speciator’ (Aves: Edolisoma tenuirostre) reveals complex dispersal and diversification dynamics across the Indo-Pacific. J. Biogeogr. 2018, 45, 826–837. [Google Scholar] [CrossRef]
- Crews, S.C.; Gillespie, R.G. Molecular systematics of Selenops spiders (Araneae: Selenopidae) from North and Central America: Implications for Caribbean biogeography. Biol. J. Linn. Soc. 2010, 101, 288–322. [Google Scholar] [CrossRef]
- Abel, C.; Schneider, J.M.; Kuntner, M.; Harms, D. Phylogeography of the ‘cosmopolitan’ orb-weaver Argiope trifasciata (Araneae: Araneidae). Biol. J. Linn. Soc. 2020, 131, 61–75. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čandek, K.; Agnarsson, I.; Binford, G.J.; Kuntner, M. Biogeography of Long-Jawed Spiders Reveals Multiple Colonization of the Caribbean. Diversity 2021, 13, 622. https://doi.org/10.3390/d13120622
Čandek K, Agnarsson I, Binford GJ, Kuntner M. Biogeography of Long-Jawed Spiders Reveals Multiple Colonization of the Caribbean. Diversity. 2021; 13(12):622. https://doi.org/10.3390/d13120622
Chicago/Turabian StyleČandek, Klemen, Ingi Agnarsson, Greta J. Binford, and Matjaž Kuntner. 2021. "Biogeography of Long-Jawed Spiders Reveals Multiple Colonization of the Caribbean" Diversity 13, no. 12: 622. https://doi.org/10.3390/d13120622
APA StyleČandek, K., Agnarsson, I., Binford, G. J., & Kuntner, M. (2021). Biogeography of Long-Jawed Spiders Reveals Multiple Colonization of the Caribbean. Diversity, 13(12), 622. https://doi.org/10.3390/d13120622







