Transcriptome Analyses Reveal Circadian-Related Expression Features in the Visual Systems of Two Snakes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequencing Samples Collection
2.2. cDNA Library Construction and mRNA Sequencing
2.3. De Novo Transcriptome Assembly and Transcription Abundance Statistic
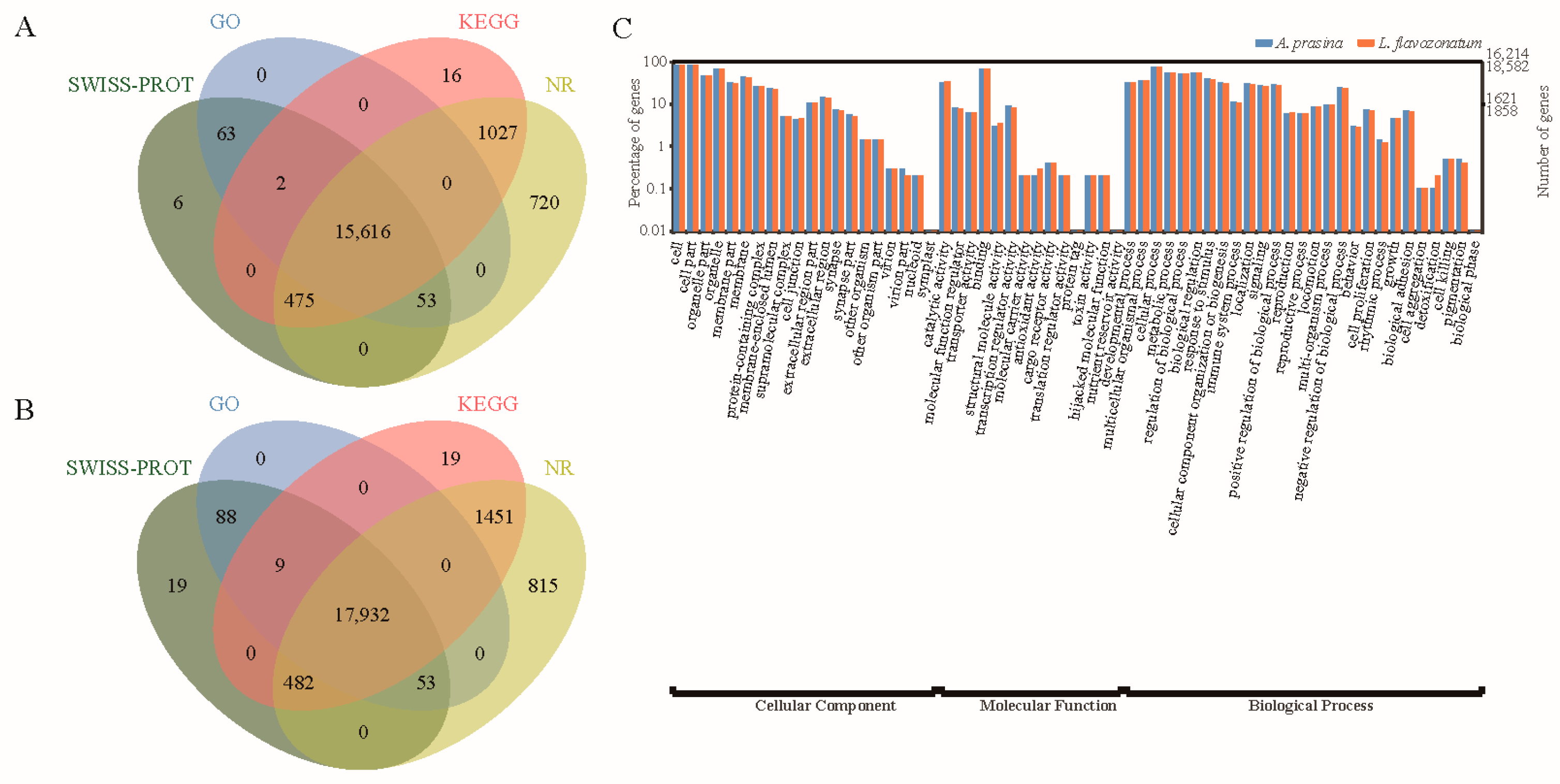
2.4. Transcriptome Assembly Statistics and Functional Annotation
2.5. Orthologous Genes Identification and Principal Component Analysis (PCA)
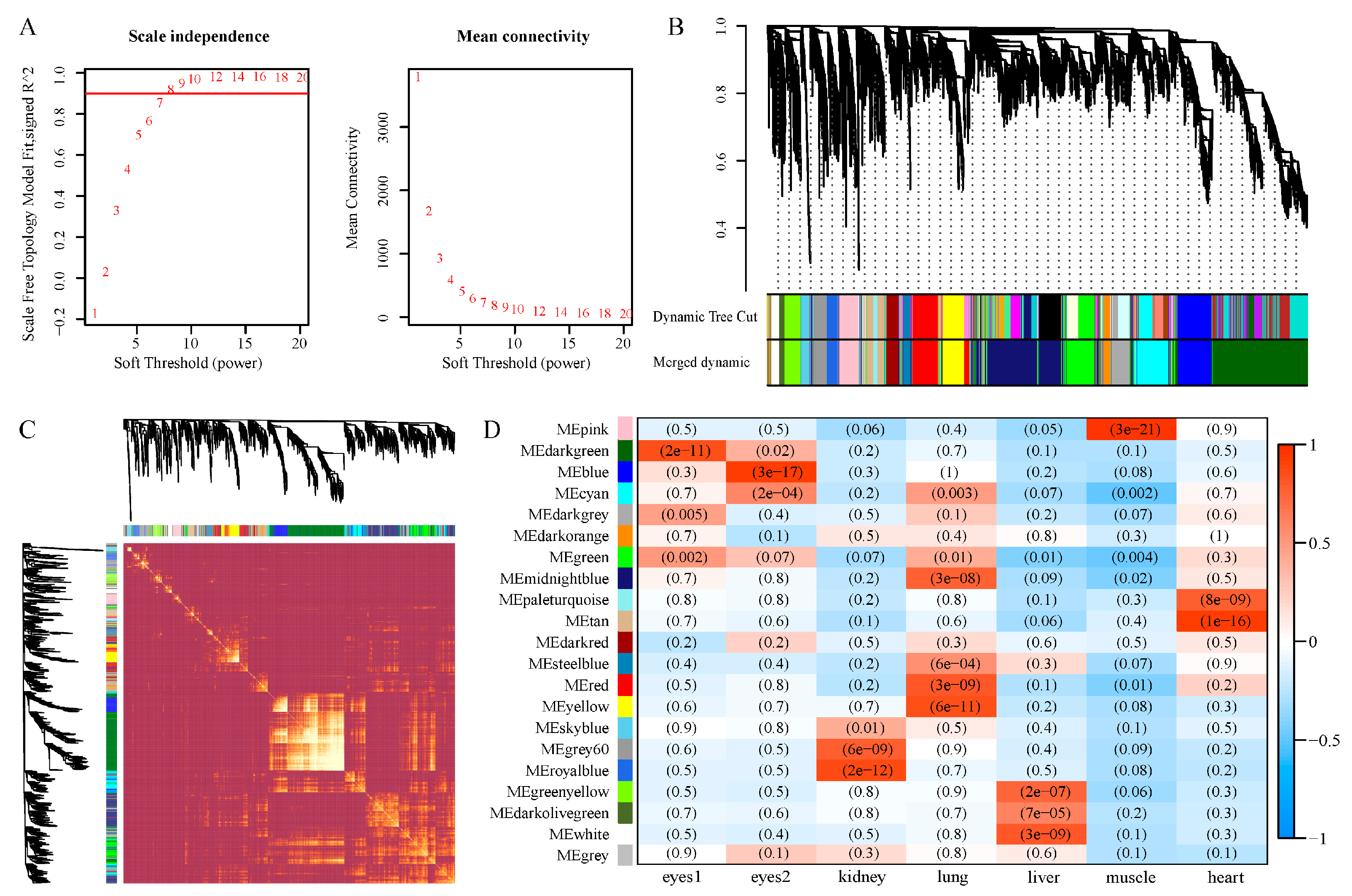
2.6. Weighted Gene Co-Expression Network Analysis (WGCNA) and Screening of Differential Expression Genes
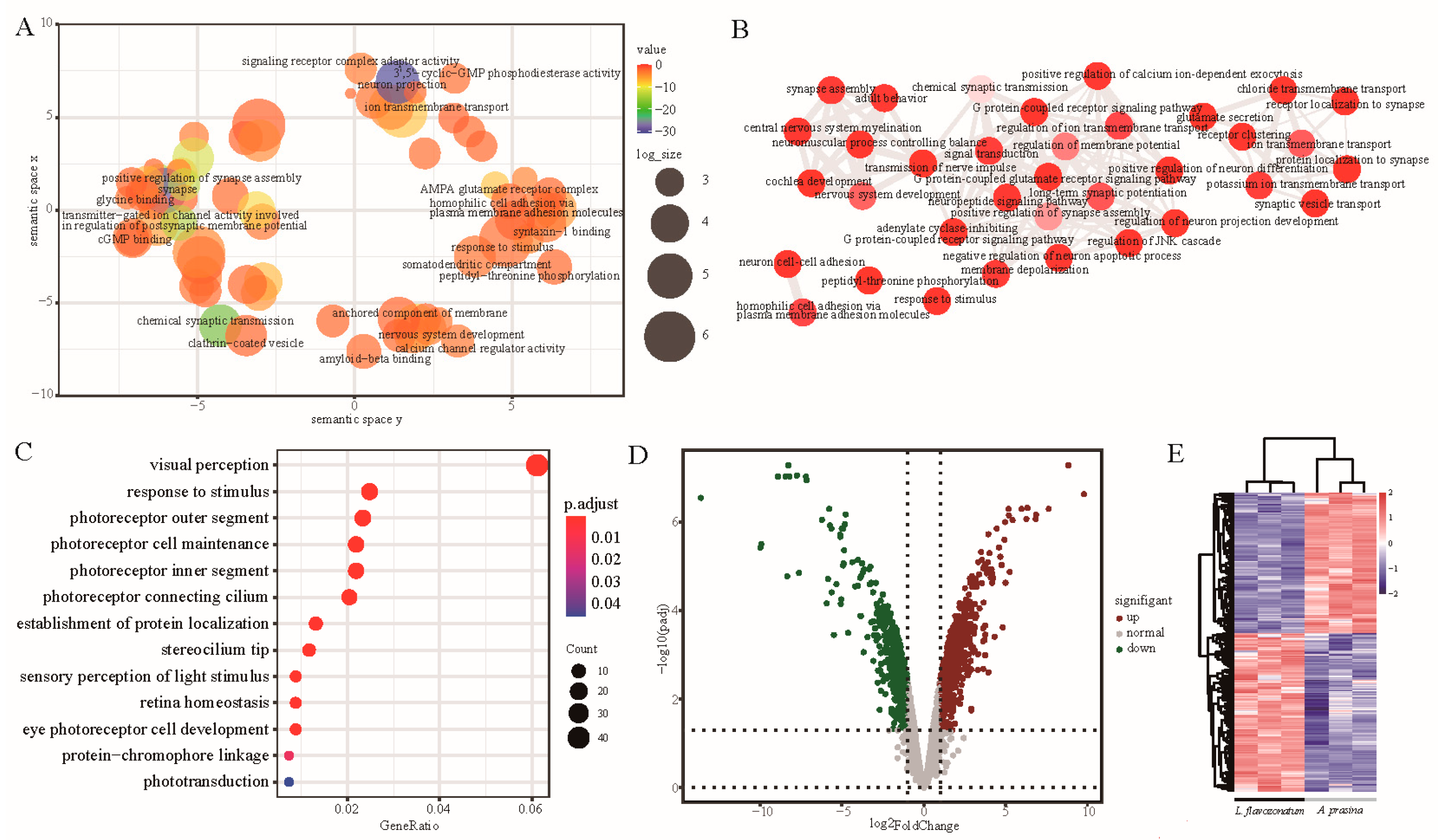
2.7. Gene Functional Enrichment and Pathways Analysis
3. Results
3.1. Transcriptome De Novo Assembly
3.2. Unigene Functional Annotation
3.3. Gene Expression and Cluster Analysis
3.4. Co-Expression Network Construction
3.5. Identification and GO Enrichment of Significant Eye Correlation Modules
3.6. Expression Pattern and Screening of Genes in Phototransduction Pathway
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Katti, C.; Stacey-Solis, M.; Coronel-Rojas, N.A.; Davies, W.I.L. Opsin-based photopigments expressed in the retina of a South American pit viper, Bothrops atrox (Viperidae). Vis. Neurosci. 2018, 35, E027. [Google Scholar] [CrossRef]
- Strader, C.D.; Fong, T.M.; Tota, M.R.; Underwood, D. Structure and Function of G Protein-Coupled Receptors. Annu. Rev. Biochem. 1994, 63, 101–132. [Google Scholar] [CrossRef]
- Kefalov, V.J. Rod and cone visual pigments and phototransduction through pharmacological, genetic, and physiological approaches. J. Biol. Chem. 2012, 287, 1635–1641. [Google Scholar] [CrossRef]
- Sakmar, T.P.; Fahmy, K. Properties and Photoactivity of Rhodopsin Mutants. Isr. J. Chem. 1995, 35, 325–337. [Google Scholar] [CrossRef]
- Yokoyama, S. Molecular evolution of vertebrate visual pigments. Prog. Retin. Eye Res. 2000, 19, 385–419. [Google Scholar] [CrossRef]
- Lamb, T.D. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 2013, 36, 52–119. [Google Scholar] [CrossRef]
- Rojasa, L.M.; McNeila, R.; Cabanaa, T.; Lachapelleb, P. Behavioral, Morphological and Physiological Correlates of Diurnal and Nocturnal Vision in Selected Wading Bird Species. Brain Behav. Evol. 1999, 53, 227–242. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, S.; Blow, N.S. Molecular evolution of the cone visual pigments in the pure rod-retina of the nocturnal gecko, Gekko gekko. Gene 2001, 276, 117–125. [Google Scholar] [CrossRef]
- Crescitelli, F. The Visual Cells and Visual Pigments of the Vertebrate Eye; Springer: Berlin, Germany, 1972; Volume VII/1, pp. 245–363. [Google Scholar]
- Jiang, D.; Klaus, S.; Zhang, Y.P.; Hillis, D.M.; Li, J.T. Asymmetric biotic interchange across the Bering land bridge between Eurasia and North America. Natl. Sci. Rev. 2019, 6, 739–745. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Fan, H.; Chen, Y.; Chang, J.; Zhan, X.; Wu, H.; Zhang, B.; Wang, M.; Zhang, W.; Yang, L.; et al. Spatial patterns and conservation of genetic and phylogenetic diversity of wildlife in China. Sci. Adv. 2021, 7, eabd5725. [Google Scholar] [CrossRef]
- Torello-Viera, N.F.; Marques, O.A.V. Daily Activity of Neotropical Dipsadid Snakes. South Am. J. Herpetol. 2017, 12, 128–135. [Google Scholar] [CrossRef]
- Mullin, S.J.; Cooper, R.J. The Foraging Ecology of the Gray Rat Snake (Elaphe obsoleta spiloides)—Visual Stimuli Facilitate Location of Arboreal Prey. Am. Midl. Nat. 1998, 140, 397–401. [Google Scholar] [CrossRef]
- Hauzman, E. Adaptations and evolutionary trajectories of the snake rod and cone photoreceptors. Semin. Cell Dev. Biol. 2020, 106, 86–93. [Google Scholar] [CrossRef]
- Schmitz, L.; Motani, R. Morphological differences between the eyeballs of nocturnal and diurnal amniotes revisited from optical perspectives of visual environments. Vis. Res. 2010, 50, 936–946. [Google Scholar] [CrossRef]
- Simoes, B.F.; Sampaio, F.L.; Douglas, R.H.; Kodandaramaiah, U.; Casewell, N.R.; Harrison, R.A.; Hart, N.S.; Partridge, J.C.; Hunt, D.M.; Gower, D.J. Visual Pigments, Ocular Filters and the Evolution of Snake Vision. Mol. Biol. Evol. 2016, 33, 2483–2495. [Google Scholar] [CrossRef]
- Cronin, T.W. Sensory Ecology: In Sea Snake Vision, One Plus One Makes Three. Curr. Biol. 2020, 30, R763–R766. [Google Scholar] [CrossRef]
- Simoes, B.F.; Sampaio, F.L.; Jared, C.; Antoniazzi, M.M.; Loew, E.R.; Bowmaker, J.K.; Rodriguez, A.; Hart, N.S.; Hunt, D.M.; Partridge, J.C.; et al. Visual system evolution and the nature of the ancestral snake. J. Evol. Biol. 2015, 28, 1309–1320. [Google Scholar] [CrossRef]
- Davies, W.L.; Cowing, J.A.; Bowmaker, J.K.; Carvalho, L.S.; Gower, D.J.; Hunt, D.M. Shedding light on serpent sight: The visual pigments of henophidian snakes. J. Neurosci. 2009, 29, 7519–7525. [Google Scholar] [CrossRef]
- Bhattacharyya, N.; Darren, B.; Schott, R.K.; Tropepe, V.; Chang, B.S.W. Cone-like rhodopsin expressed in the all-cone retina of the colubrid pine snake as a potential adaptation to diurnality. J. Exp. Biol. 2017, 220, 2418–2425. [Google Scholar] [CrossRef] [PubMed]
- Simoes, B.F.; Sampaio, F.L.; Loew, E.R.; Sanders, K.L.; Fisher, R.N.; Hart, N.S.; Hunt, D.M.; Partridge, J.C.; Gower, D.J. Multiple rod-cone and cone-rod photoreceptor transmutations in snakes: Evidence from visual opsin gene expression. Proc. Biol. Sci. 2016, 283, 20152624. [Google Scholar] [CrossRef]
- Amber, E.D.; Strine, C.T.; Suwanwaree, P.; Waengsothorn, S. Intra-Population Color Dimorphism of Ahaetulla prasina (Serpentes: Colubridae) in Northeastern Thailand. Curr. Herpetol. 2017, 36, 98–104. [Google Scholar] [CrossRef]
- Zhao, E. Snakes of China; Anhui Science Technology Publishing House: Hefei, China, 2006; p. 372. [Google Scholar]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Hauzman, E.; Bonci, D.M.O.; Suarez-Villota, E.Y.; Neitz, M.; Ventura, D.F. Daily activity patterns influence retinal morphology, signatures of selection, and spectral tuning of opsin genes in colubrid snakes. BMC Evol. Biol. 2017, 17, 249. [Google Scholar] [CrossRef]
- Hauzman, E.; Bonci, D.M.O.; Ventura, D.F. Visual ecology of snakes: Comparative study of the density and distribution of retinal neurons in diurnal and nocturnal species. Investig. Ophthalmol. Vis. Sci. 2016, 57, 4667. [Google Scholar]
- Krizaj, D.; Copenhagen, D.R. Calcium regulation in photoreceptors. Front. Biosci. 2002, 7, d2023–d2044. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.-W.; Stryer, L. Highly cooperative feedback control of retinal rod guanylate cyclase by calcium ions. Nature 1988, 334, 64–66. [Google Scholar] [CrossRef] [PubMed]
- Dizhoor, A.M.; Ray, S.; Kumar, S.; Niemi, G.; Spencer, M.; Brolley, D.; Walsh, K.A.; Philipov, P.P.; Hurley, J.B.; Stryer, L. Recoverin: A Calcium Sensitive Activator of Retinal Rod Guanylate Cyclase. Science 1991, 251, 915–918. [Google Scholar] [CrossRef] [PubMed]
- Ridge, K.D.; Palczewski, K. Visual rhodopsin sees the light: Structure and mechanism of G protein signaling. J. Biol. Chem. 2007, 282, 9297–9301. [Google Scholar] [CrossRef]
- Baylor, D.A.; Lamb, T.D.; Yau, K.W. Responses of retinal rods to single photons. J. Physiol. 1979, 288, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Fain, G.L.; Hardie, R.; Laughlin, S.B. Phototransduction and the evolution of photoreceptors. Curr. Biol. 2010, 20, R114–R124. [Google Scholar] [CrossRef]
- Bowmaker, J.K. Evolution of vertebrate visual pigments. Vision Res. 2008, 48, 2022–2041. [Google Scholar] [CrossRef] [PubMed]
- Wikler, K.C.; Rakic, P. Distribution of photoreceptor subtypes in the retina of diurnal and nocturnal primates. J. Neurosci. 1990, 10, 3390–3401. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Hadly, E.A.; Teng, W.; Hao, Y.; Liang, W.; Liu, Y.; Wang, H. Retinal transcriptome sequencing sheds light on the adaptation to nocturnal and diurnal lifestyles in raptors. Sci. Rep. 2016, 6, 33578. [Google Scholar] [CrossRef]
- Douglas, R.H.; Jeffery, G. The spectral transmission of ocular media suggests ultraviolet sensitivity is widespread among mammals. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132995. [Google Scholar] [CrossRef]
- Rasmussen, J.B. The retina of Psammodynastes pulverulentus (Boie, 1827) and Telescopus fallax (Fleischmann, 1831) with a discussion of their phylogenetic significance (Colubroidea, Serpentes). J. Zool. Syst. Evol. 1990, 28, 269–276. [Google Scholar] [CrossRef]
- Walls, G. The Vertebrate Eye and Its Adaptive Radiation; Fafner Publishing Company: New York, NY, USA, 1942. [Google Scholar]
- Schott, R.K.; Muller, J.; Yang, C.G.; Bhattacharyya, N.; Chan, N.; Xu, M.; Morrow, J.M.; Ghenu, A.H.; Loew, E.R.; Tropepe, V.; et al. Evolutionary transformation of rod photoreceptors in the all-cone retina of a diurnal garter snake. Proc. Natl. Acad. Sci. USA 2016, 113, 356–361. [Google Scholar] [CrossRef]
- Ridge, K.D.; Abdulaev, N.G.; Sousa, M.; Palczewski, K. Phototransduction: Crystal clear. Trends Biochem. Sci. 2003, 28, 479–487. [Google Scholar] [CrossRef]
- Peng, C.; Ren, J.L.; Deng, C.; Jiang, D.; Wang, J.; Qu, J.; Chang, J.; Yan, C.; Jiang, K.; Murphy, R.W.; et al. The Genome of Shaw’s Sea Snake (Hydrophis curtus) Reveals Secondary Adaptation to Its Marine Environment. Mol. Biol. Evol. 2020, 37, 1744–1760. [Google Scholar] [CrossRef]
- Osawa, S.; Weiss, E.R. A tale of two kinases in rods and cones. Adv. Exp. Med. Biol. 2012, 723, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Kohl, S.; Baumann, B.; Broghammer, M.; Jägle, H.; Sieving, P.; Kellner, U.; Spegal, R.; Anastasi, M.; Zrenner, E.; Sharpe, L.T.; et al. Mutations in the CNGB3 gene encoding the β-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 2000, 9, 2107–2116. [Google Scholar] [CrossRef]
- Gerstner, A.; Zong, X.; Hofmann, F.; Biel, M. Molecular Cloning and Functional Characterization of a New Modulatory Cyclic Nucleotide-Gated Channel Subunit from Mouse Retina. J. Neurosci. 2000, 20, 1324–1332. [Google Scholar] [CrossRef]
- Grau, T.; Artemyev, N.O.; Rosenberg, T.; Dollfus, H.; Haugen, O.H.; Cumhur Sener, E.; Jurklies, B.; Andreasson, S.; Kernstock, C.; Larsen, M.; et al. Decreased catalytic activity and altered activation properties of PDE6C mutants associated with autosomal recessive achromatopsia. Hum. Mol. Genet. 2011, 20, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Nishiguchi, K.M.; Sandberg, M.A.; Kooijman, A.C.; Martemyanov, K.A.; Pott, J.W.R.; Hagstrom, S.A.; Arshavsky, V.Y.; Berson, E.L.; Dryja, T.P. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature 2004, 427, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.-T.; Molday, R.S. Modulation of the cGMP-gated channel of rod photoreceptor cells by calmodulin. Nature 1993, 361, 76–79. [Google Scholar] [CrossRef]
- Burns, M.E.; Lamb, T.D. Visual Transduction by Rod and Cone Photoreceptors; MIT Press: Cambridge, MA, USA, 2004; Volume 1, pp. 215–233. [Google Scholar]
- Pignatelli, V.; Champ, C.; Marshall, J.; Vorobyev, M. Double cones are used for colour discrimination in the reef fish, Rhinecanthus aculeatus. Biol. Lett. 2010, 6, 537–539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cyriac, V.P.; Kodandaramaiah, U. Conspicuous colours reduce predation rates in fossorial uropeltid snakes. PeerJ 2019, 7, e7508. [Google Scholar] [CrossRef] [PubMed]
- Kaupp, U.B.; Niidome, T.; Tanabe, T.; Terada, S.; Bonigk, W.; Stiihmert, W.; Cooka, N.J.; Kangawa, K.; Matsuo, H.; Hirosell, T.; et al. Primary structure and functional expression from complementary DNA of the rod photoreceptor cyclic GMP.gated channel. Nature 1989, 342, 762–766. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Y.; Illing, M.; Molday, L.L.; Hsu, Y.T.; Yau, K.W.; Molday, R.S. Subunit 2 (or β) of retinal rod cGMP-gated cation channel is a component of the 240-kDa channel-associated protein and mediates Ca2+-calmodulin modulation. Proc. Natl. Acad. Sci. USA 1994, 91, 11757–11761. [Google Scholar] [CrossRef] [PubMed]
- Kitiratschky, V.B.D.; Behnen, P.; Kellner, U.; Heckenlively, J.R.; Zrenner, E.; Jägle, H.; Kohl, S.; Wissinger, B.; Koch, K.-W. Mutations in the GUCA1A Gene Involved in Hereditary Cone Dystrophies Impair Calciummediated Regulation of Guanylate Cyclase. Hum. Mutat. 2009, 30, E782–E796. [Google Scholar] [CrossRef] [PubMed]
- Baehr, W.; Karan, S.; Maeda, T.; Luo, D.-G.; Li, S.; Bronson, J.D.; Watt, C.B.; Yau, K.-W.; Frederick, J.M.; Palczewski, K. The Function of Guanylate Cyclase 1 and Guanylate Cyclase 2 in Rod and Cone Photoreceptors. J. Biol. Chem. 2007, 282, 8837–8847. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, C.-Y.; Song, M.-H.; Peng, Z.-L.; Wu, W.; Peng, C.; Yang, K.; Li, J.-T. Transcriptome Analyses Reveal Circadian-Related Expression Features in the Visual Systems of Two Snakes. Diversity 2021, 13, 621. https://doi.org/10.3390/d13120621
Tang C-Y, Song M-H, Peng Z-L, Wu W, Peng C, Yang K, Li J-T. Transcriptome Analyses Reveal Circadian-Related Expression Features in the Visual Systems of Two Snakes. Diversity. 2021; 13(12):621. https://doi.org/10.3390/d13120621
Chicago/Turabian StyleTang, Chen-Yang, Meng-Huan Song, Zhong-Liang Peng, Wei Wu, Changjun Peng, Kong Yang, and Jia-Tang Li. 2021. "Transcriptome Analyses Reveal Circadian-Related Expression Features in the Visual Systems of Two Snakes" Diversity 13, no. 12: 621. https://doi.org/10.3390/d13120621
APA StyleTang, C.-Y., Song, M.-H., Peng, Z.-L., Wu, W., Peng, C., Yang, K., & Li, J.-T. (2021). Transcriptome Analyses Reveal Circadian-Related Expression Features in the Visual Systems of Two Snakes. Diversity, 13(12), 621. https://doi.org/10.3390/d13120621






