Landscape Genetics of the Yellow-Bellied Toad (Bombina variegata) in the Northern Weser Hills of Germany
Abstract
:1. Introduction
2. Materials and Methods
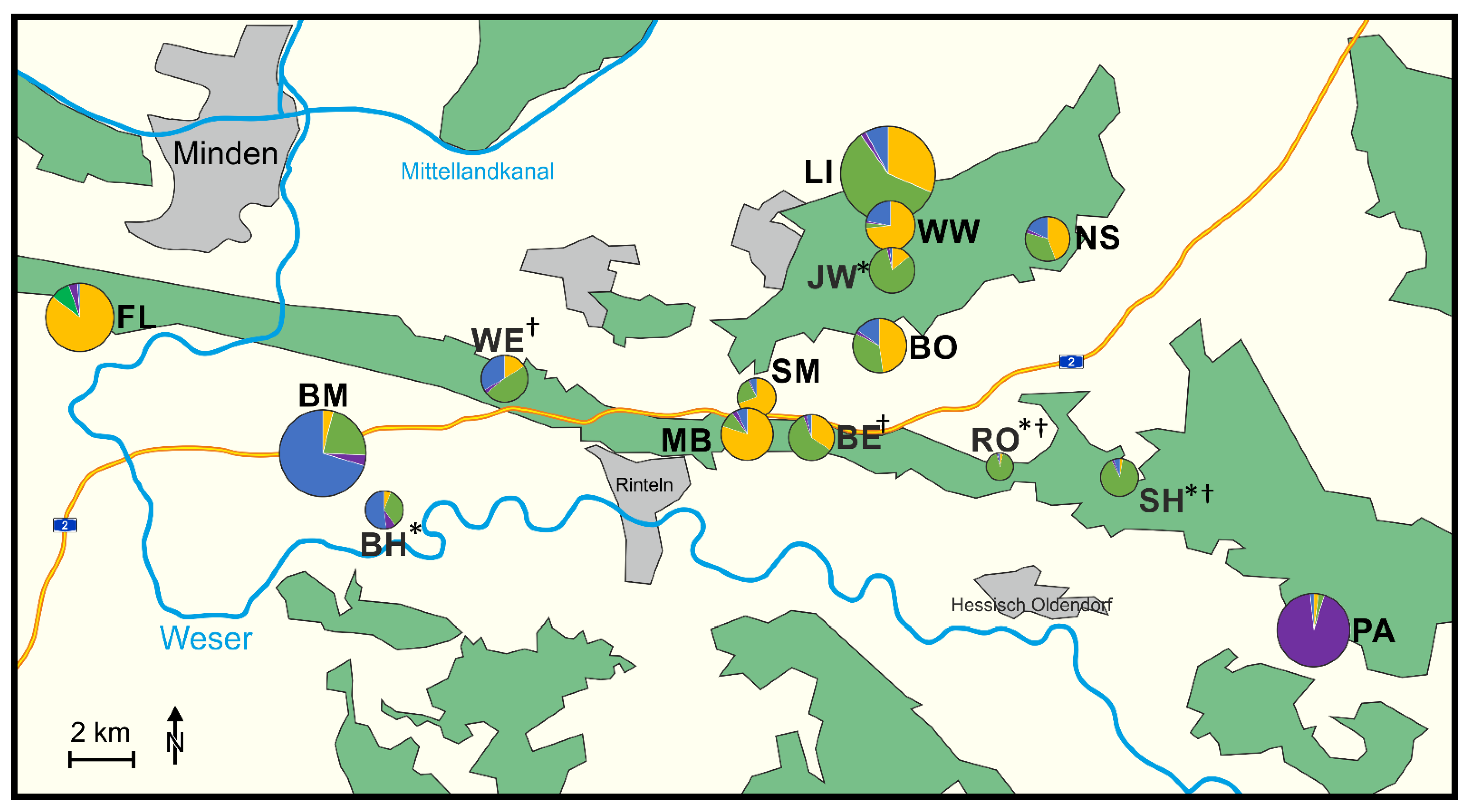
2.1. Study Area
2.2. Microsatellite Genotyping
2.3. Population Genetic Analysis
2.4. Landscape Genetic Analysis
3. Results
3.1. Population Genetic Diversity
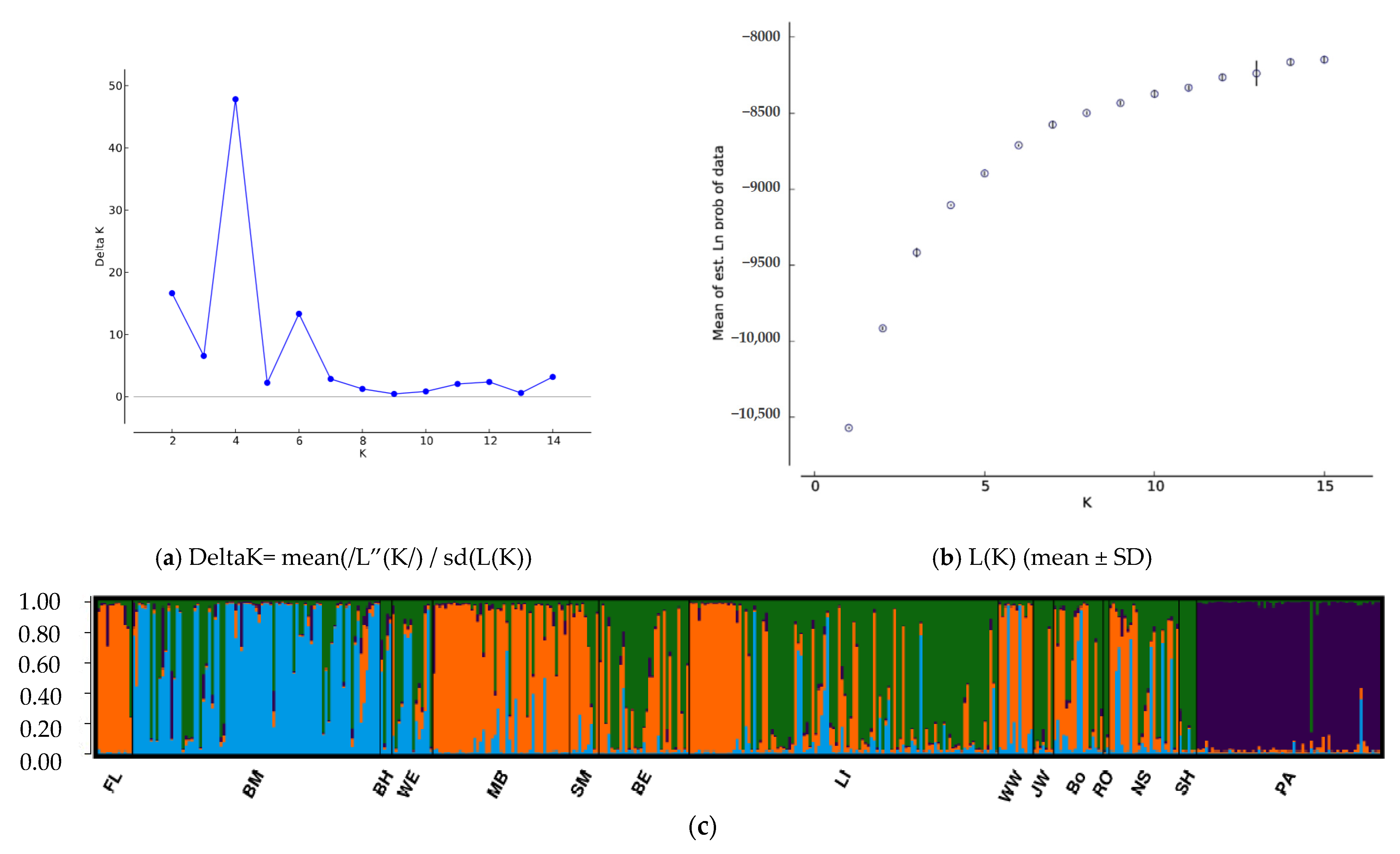
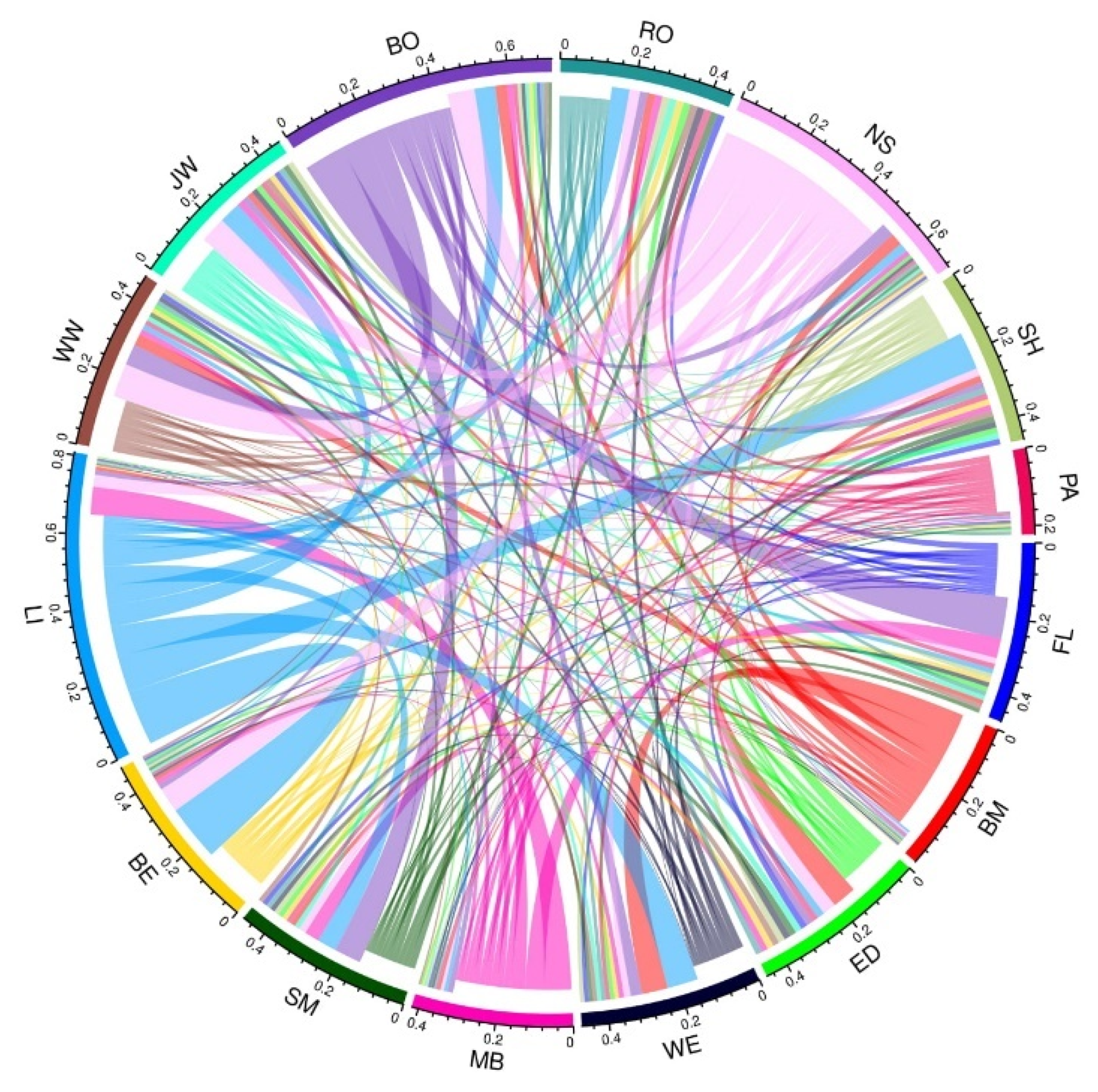
3.2. Population Structure and Migration
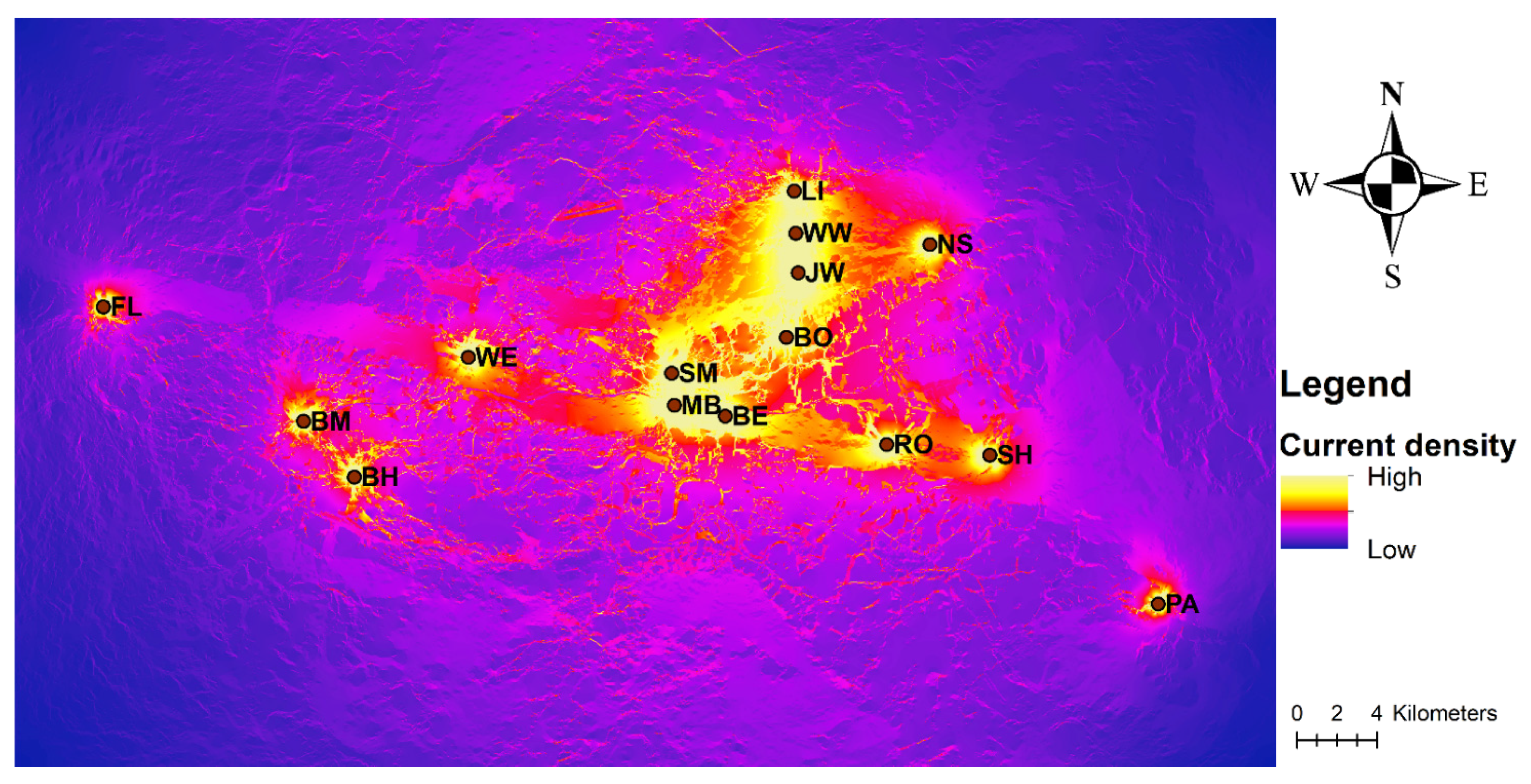
3.3. Landscape Genetic Analysis
4. Discussion
4.1. Genetic Diversity and Population Structure
4.2. Effects of Geographic and Resistance Distances on Gene Flow
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alford, R.A.; Richards, S.J. Global Amphibian Declines: A Problem in Applied Ecology. Annu. Rev. Ecol. Evol. Syst. 1999, 30, 133–165. [Google Scholar] [CrossRef] [Green Version]
- Dodd, C.K. Amphibian Ecology and Conservation: A Handbook of Techniques; Oxford University Press: Oxford, UK, 2010; p. 584. [Google Scholar]
- Holderegger, R.; Segelbacher, G. Naturschutzgenetik: Ein Handbuch Für Die Praxis; Haupt Verlag: Berlin, Germany, 2016; p. 248. [Google Scholar]
- Covarrubias, S.; Gonzalez, C.; Gutierrez-Rodríguez, C. Effects of natural anthopogenic Features on functional Connectivity of Anurans: A Review of Landscape Genetics Studies in temperate, subtropical and tropical species. J. Zool. 2021, 313, 159–171. [Google Scholar] [CrossRef]
- Spear, S.F.; Peterson, C.R.; Matocq, M.D.; Storfer, A. Landscape Genetics of the Blotched Tiger Salamander (Ambystoma Tigrinum Melanostictum). Mol. Ecol. 2005, 14, 2553–2564. [Google Scholar] [CrossRef]
- Churko, G. Evaluating the Landscape Connectivity of Five Amphibian Species Using Circuit Theory. Master’s Thesis, Swiss Federal Institute of Technology Zurich, Zurich, Switzerland, 2016. [Google Scholar]
- Website IUCN. IUCN Red List of threatened Species. Available online: https://www.iucnredlist.org/ (accessed on 22 November 2021).
- Kühnel, K.D.; Geiger, A.; Laufer, H.; Podloucky, R.; Schlüpmann, M. Rote Liste Und Gesamtartenliste Der Lurche (Amphibia) Deutschlands. Nat. Biol. Vielfalt 2009, 70, 259–288. [Google Scholar]
- Podloucky, R.; Fischer, C. Rote Liste Der Gefährdeten Amphibien Und Reptilien in Niedersachsen Und Bremen. Inf. Nat. Niedersachs. 1994, 14, 119–120. [Google Scholar]
- Schlüpmann, M.; Geiger, A. Rote Liste der gefährdeten Kriechtiere (Reptilia) und Lurche (Amphibia) in Nordrhein-Westfalen. In Rote Liste der Gefährdeten Pflanzen und Tiere in Nordrhein-Westfalen, 3rd ed.; Wasner, U., Wolff-Straub, R., Eds.; Landesanstalt für Ökologie, Bodenordnung u. Forsten/Landesamt f. Agrarordnung: Recklinghausen, Germany, 1999; Volume 17, pp. 375–404. [Google Scholar]
- Nöllert, A.; Scheidt, U.; Uthleb, H. Rote Liste Der Lurche (Amphibia) Thüringens—Rote Liste Thürigens. Jena. Nat. Heft 2001, 18, 43–46. [Google Scholar]
- Website BfN. Ergebnisse Nationaler FFH-Bericht 2019, Erhaltungszustande und Gesamttrends der Arten in der Atlantischen Biogeografischen Region. Available online: https://www.bfn.de/themen/natura2000/berichte-monitoring/nationaler-ffh-bericht/berichtsdaten.html (accessed on 5 June 2020).
- Günther-Diringer, D.; Berner, K.; Koenzen, U.; Kurth, A.; Modrak, P.; Ackermann, W.; Ehlert, T.; Heyden, J. Methodische Grundlagen Zum Auenzustandsbericht 2021: Erfassung, Bilanzierung Und Bewertung von Flussauen; Heft no. 591; BfN: Bonn, Germany, 2021; p. 57. [Google Scholar]
- NLWKN. Vollzugshinweise Zum Schutz von Amphibien-Und Reptilienarten in Niedersachsen, Amphibienarten Des Anhangs II Der FFH-Richtlinie Mit Priorität Für Erhaltungs-Und Entwicklungsmaßnahmen, Gelbbauchunke (Bombina Variegata)—Niedersächsische Strategie Zum Arten-Und Biotopschutz; NLWKN: Hannover, Germany, 2003; Unpublished work. [Google Scholar]
- Gollmann, B.; Gollmann, G. Die Gelbbauchunke: Von Der Suhle Zur Radspur; Laurenti: Bielefeld, Germany, 2012; p. 176. [Google Scholar]
- Hantzschmann, A.; Sinsch, U.; Göttlicher, C.; Pröhl, H. Conservation Genetics of Yellow-Bellied Toads (Bombina variegata): A Matter of Geographical Scale and Location. Conserv. Genet. 2020, 22, 83–96. [Google Scholar] [CrossRef]
- Gollmann, B.; Gollmann, G.; Miesler, M. Habitatnutzung Und Wanderungen in Einer Gelbbauchunken-Population (Bombina v. variegata). Z. Feldherpetol. 2000, 7, 1–16. [Google Scholar]
- Wright, S. Evolution and the Genetics of Populations, Volume 2, Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978; p. 520. [Google Scholar]
- McRae, B.H. Isolation by Resistance. Evolution 2006, 60, 1551–1561. [Google Scholar] [CrossRef] [PubMed]
- Oswald, P.; Rodríguez, A.; Bourke, J.; Wagner, N.; de Buhr, N.; Buschmann, H.; von Köckritz-Blickwede, M.; Pröhl, H. Locality, Time and Heterozygosity Affect Chytrid Infection in Yellow-Bellied Toads. Dis. Aquat. Org. 2020, 142, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Höppner, C.; Nadjafzadeh, M.; Buschmann, H. Wiederansiedlungsvorhaben für die Gelbbauchunke (Bombina variegata) Im Nördlichen Weserbergland. Z. Feldherpetol. 2017, 20, 1–36. [Google Scholar]
- Pröhl, H.; Auffarth, J.; Bergmann, T.; Buschmann, H.; Balkenhol, N. Conservation Genetics of the Yellow-Bellied Toad (Bombina variegata): Population Structure, Genetic Diversity and Landscape Effects in an Endangered Amphibian. Conserv. Genet. 2021, 22, 513–529. [Google Scholar] [CrossRef]
- Stuckas, H.; Tiedemann, R. Eight New Microsatellite Loci for the Critically Endangered Fire-bellied Toad Bombina and Their Cross-species Applicability among Anurans. Mol. Ecol. Notes 2006, 6, 150–152. [Google Scholar] [CrossRef]
- Hauswaldt, J.S.; Schröder, C.; Tiedemann, R. Nine New Tetranucleotide Microsatellite Markers for the Fire-Bellied Toad (Bombina bombina). Mol. Ecol. Notes 2007, 7, 49–52. [Google Scholar] [CrossRef]
- Alberto, F. MsatAllele_1.0: An R Package to Visualize the Binning of Microsatellite Alleles. J. Hered. 2009, 100, 394–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the Number of Clusters of Individuals Using the Software STRUCTURE: A Simulation Study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ersts, P.J. Geographic Distance Matrix Generator, version 1.2.3; AMNH: New York, NY, USA, 2020. [Google Scholar]
- Wilson, G.A.; Rannala, B. Bayesian Inference of Recent Migration Rates Using Multilocus Genotypes. Genetics 2003, 163, 1177–1191. [Google Scholar] [CrossRef] [PubMed]
- Rannala, B. BayesAss Edition 3.0 User´s Manual, Update 2015; University of California Davis: Davis, CA, USA, 2007; Volume 14, p. 12. [Google Scholar]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An Information Aesthetic for Comparative Genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Goudet, J. FSTAT, (Version 1.2): A Computer Program to Calculate F-Statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Goudet, J. Hierfstat a Package for R to Compute and Test Hierarchical F-Statistics. Mol. Ecol. Notes 2004, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population-Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [PubMed]
- Dᾳbrowski, M.J.; Bornelöv, S.; Kruczyk, M.; Baltzer, N.; Komorowski, J. ‘True’ Null Allele Detection in Microsatellite Loci: A Comparison of Methods, Assessment of Difficulties and Survey of Possible Improvements. Mol. Ecol. Resour. 2014, 15, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population Genetics Software for Exact Tests and Ecumenicism. J. Hered. 1995, 68, 248–249. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L. Maximum Likelihood Estimation of the Frequency of Null Alleles at Microsatellite Loci. Conserv. Genet. 2006, 7, 991–995. [Google Scholar] [CrossRef]
- ESRI. ArcGIS Desktop. Release 10; ESRI: Redlands, CA, USA, 2011. [Google Scholar]
- Peterman, W.; Pope, N.S. The Use and Misuse of Regression Models in Landscape Genetic Analyses. Mol. Ecol. 2020, 30, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Wyman, R.L. Soil Acidity and Moisture and the Distribution of Amphibians in Five Forests of Southcentral New York. Copeia 1988, 1988, 394–399. [Google Scholar] [CrossRef]
- Eckhardt, F.S.; (Amt für Umwelt und ländlicher Raum, Celle, Lower Saxony, Germany). Personal communication, 2020.
- Hall, K.R.; Anantharaman, R.; Landau, V.A.; Clark, M.; Dickson, B.G.; Jones, A.; Platt, J.; Edelman, A.; Shah, V.B. Circuitscape in Julia: High Performance Connectivity Modelling to Support Conservation Decisions. Land 2021, 10, 2–24. [Google Scholar]
- Peterman, W. ResistanceGA: An R Package for the Optimization of Resistance Surfaces Using Genetic Algorithms. Methods Ecol. Evol. 2018, 9, 1638–1647. [Google Scholar] [CrossRef] [Green Version]
- Row, J.R.; Knick, S.T.; Oyler-McCance, S.J.; Lougheed, S.C.; Fedy, B.C. Developing Approaches for Linear Mixed Modelling in Landscape Genetics through Landscape-Directed Dispersal Simulations. Ecol. Evol. 2017, 7, 3751–3761. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. Adegenet: A R Package for the Multivariate Analysis of Genetic Markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefcheck, J.S. PiecewiseSEM: Piecewise Structural Equation Modeling in R for Ecology, Evolution, and Systematics. Methods Ecol. Evol. 2016, 7, 573–579. [Google Scholar] [CrossRef]
- Shirk, A.J.; Landguth, E.L.; Cushman, S.A. A Comparison of Individual-Based Genetic Distance Metrics for Landscape Genetics. Mol. Ecol. Resour. 2017, 17, 1308–1317. [Google Scholar] [CrossRef] [PubMed]
- Zeileis, A.; Hothorn, T. Lmtest: Testing Linear Regression Models. R News 2002, 2, 7–10. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, version 2.5–7; 2020. [Google Scholar]
- Weihmann, F.; Weihmann, S.; Weihmann, T. Conservation Genetic Analysis of a Central-European Range-Margin Population of the Yellow-Bellied Toad (Bombina v. variegata). Conserv. Genet. 2019, 20, 557–569. [Google Scholar] [CrossRef]
- Jacob, A.; Scheel, B.; Buschmann, H. Raumnutzung in Einer Metapopulation Der Gelbbauchunke (Bombina variegata) an Ihrer Nördlichen Verbreitungsgrenze. Z. Feldherpetol. 2009, 16, 85–102. [Google Scholar]
- Schmidt, P. Kriterien zur Bewertung des Erhaltungszustandes der Populationen der Gelbbauchunke Bombina variegata (Linnaeus, 1758). In Berichte des Landesamtes für Umweltschutz Sachsen-Anhalt, Sonderheft 2; Schnitter, P., Eichen, C., Ellwanger, G., Neukrchen, M., Schröder, E., Eds.; Landesamt für Umweltschutz Sachsen Anhalt: Halle, Germany, 2006; pp. 243–244. [Google Scholar]
- Nadjafzadeh, M.; Kutter, T.; Höppner, C.; Jentschke, I.; Berkhan, R.; Scheel, B.; Oswald, P.; Pröhl, H. Stärkung und Vernetzung von Vorkommen der Gelbbauchunke als Leitart für dynamische Offenlandschaften—Ziele und Erfolge eines bundesweiten Projekts. Nat. Landsch. 2020, 95, 301–309. [Google Scholar]
- Lowe, W.H.; Allendorf, F.W. What Can Genetics Tell Us about Population Connectivity? Mol. Ecol. 2010, 19, 3038–3051. [Google Scholar] [CrossRef] [PubMed]
- Naumann, S.; Davis, M.; Kaphengst, T.; Pieters, M.; Rayment, M. Design, Implementation and Cost Elements of Green Infrastructure Projects; DG Environment 2011, Final report to the European Commission, 1–142, Contact no. 070307/2010/577182/ETU/F.1; DG Environments: Kopenhagen, Danmark, 2011. [Google Scholar]
- Khosravi, R.; Hemami, M.R.; Malekian, M.; Silva, T.L.; Rezaei, H.R.; Brito, J.C. Effect of Landscape Features on Genetic Structure of the Goitered Gazelle (Gazella subgutturosa) in Central Iran. Conserv. Genet. 2018, 19, 323–336. [Google Scholar] [CrossRef]
- Emel, S.L.; Olson, D.H.; Knowler, L.L.; Storfer, A. Comparative Landscape Genetics of Two Endemic Torrent Salamander Species, Rhyacotriton kezeri and R. variegatus: Implications for Forest Management and Species Conservation. Conserv. Genet. 2019, 20, 801–815. [Google Scholar] [CrossRef] [Green Version]
- Hails, R.S. Assessing the Risks Associated with new Agricultural Practices. Nature 2002, 418, 685–688. [Google Scholar] [CrossRef] [PubMed]
| Landscape Category | Resistance Tier | Assigned Resistance Value |
|---|---|---|
| Streams, Ponds & Moist Soil [6,41] | Habitat | 0 (reduces resistance value of Forest and Grassland to 0 when overlapping) |
| Forest (dry) [6,15] | Favorable matrix | 3 |
| Grassland (dry) [6,15] | Favourable matrix | 3 |
| Agricultural Land [14,15] | Less favorable matrix | 6 |
| A2 Underpasses | Less favorable matrix | 6 |
| Weser [6,42] | Strong barrier | 9 |
| A2 Motorway [6,14] | Strong barrier | 9 |
| Imperviousness [14] | Habitat to strong barrier | 0% to 100%, adjusted to fit within 0 to 10 range |
| Model ID | Model | Explanation |
|---|---|---|
| 1 | UNDIF | Resistance calculated using an undifferentiated landscape |
| 2 | REF | Reference resistance values listed in Table 1 |
| 3 | UNDER3 | Underpasses A2 with resistance value 3 |
| 4 | UNDER9 | Underpasses A2 with resistance value 9 |
| 5 | AGRI3 | Agricultural Land with resistance value 3 |
| 6 | AGRI9 | Agricultural Land with resistance value 9 |
| 7 | WET0 | Streams, Ponds & Moist Soil do not reduce resistance value of layers Grassland and Forest from 3 to 0 |
| 8 | WESER3 | Weser with resistance value 3 |
| 9 | WESER6 | Weser with resistance value 6 |
| Sample Site | Habitat | Distance to Next Population (km) | N | Gd | He | Ho | FIS | Np | Ar |
|---|---|---|---|---|---|---|---|---|---|
| FL | Former clay pit, nature reserve | 8.68 | 12 | 0.42 | 0.42 | 0.38 | 0.05 | 1 | 1.86 |
| BM | Sand/gravel pit | 2.33 | 85 | 0.48 | 0.48 | 0.35 | 0.26 | 5 | 2.08 |
| BH * | Former sand pit, nature reserve | 2.33 | 4 | 0.4 | 0.4 | 0.43 | −0.20 | 0 | 1.81 |
| WE † | Active quarry | 5.19 | 14 | 0.49 | 0.49 | 0.37 | 0.21 | 2 | 2.09 |
| MB | Active quarry | 0.68 | 47 | 0.51 | 0.50 | 0.43 | 0.15 | 2 | 2.11 |
| SM | Farm track/wheel tracks/stepping stone | 0.68 | 10 | 0.49 | 0.48 | 0.34 | 0.26 | 1 | 2.06 |
| BE † | Inactive quarry | 2.56 | 31 | 0.47 | 0.47 | 0.39 | 0.16 | 3 | 2.05 |
| LI | Nature reserve, inactive quarry | 1.06 | 106 | 0.54 | 0.54 | 0.42 | 0.21 | 5 | 2.23 |
| WW | Forest meadow | 1.06 | 12 | 0.59 | 0.65 | 0.49 | 0.12 | 2 | 2.3 |
| JW * | Forest meadow | 1.41 | 7 | 0.51 | 0.50 | 0.41 | 0.12 | 0 | 2.03 |
| BO | Former clay pit, nature reserve | 1.81 | 17 | 0.47 | 0.47 | 0.44 | 0.04 | 2 | 2.05 |
| RO *† | Inactive quarry | 2.94 | 2 | 0.58 | 0.52 | 0.4 | −0.03 | 0 | 2.1 |
| NS | Military training site | 5.69 | 24 | 0.53 | 0.53 | 0.48 | 0.07 | 3 | 2.21 |
| SH *† | Active quarry | 2.94 | 6 | 0.41 | 0.40 | 0.37 | 0.004 | 0 | 1.89 |
| PA | Inactive quarry | 7.65 | 63 | 0.39 | 0.39 | 0.33 | 0.15 | 10 | 1.85 |
| Mean/Total (± SD) | - | - | 440 | 0.49 | 0.48 | 0.4 | 0.1 | 2.4 | 2.05 |
| - | (±0.06) | (±0.06) | (±0.05) | (±0.12) | (±2.58) | (±0.14) |
| Model | Delta BIC | Marginal R2 | LogLik for Complex Model | Chisq | Pr (>Chisq) |
|---|---|---|---|---|---|
| FST values | |||||
| FST~UNDIF | 0 | 0.001 | 89.389 | ||
| FST~REF | −1.25 | 0.292 | 92.772 | 6.767 | 0.034 * |
| FST~UNDER3 | −1.07 | 0.299 | 92.862 | 6.947 | 0.031 * |
| FST~UNDER9 | −1.25 | 0.292 | 92.772 | 6.767 | 0.034 * |
| FST~AGRI3 | −4.18 | 0.032 | 91.305 | 3.832 | 0.147 |
| FST~AGRI9 | −2.24 | 0.324 | 92.276 | 5.774 | 0.056 |
| FST~WET0 | −1.17 | 0.294 | 92.811 | 6.844 | 0.033 * |
| FST~WESER3 | −1.49 | 0.283 | 92.652 | 6.526 | 0.038 * |
| FST~WESER6 | −1.34 | 0.288 | 92.726 | 6.675 | 0.036 * |
| Migration rates | |||||
| MIG~UNDIF | 0 | 0.08 | 122.08 | ||
| MIG~REF | −0.99 | 0.179 | 125.593 | 7.028 | 0.023 * |
| MIG~UNDER3 | −0.41 | 0.19 | 125.879 | 7.6 | 0.022 * |
| MIG~UNDER9 | −0.99 | 0.179 | 125.593 | 7.028 | 0.03 * |
| MIG~AGRI3 | −4.60 | 0.085 | 123.788 | 3.418 | 0.181 |
| MIG~AGRI9 | −0.42 | 0.19 | 125.876 | 7.593 | 0.022 * |
| MIG~WET0 | −1.02 | 0.178 | 125.576 | 6.994 | 0.03 * |
| MIG~WESER3 | −0.93 | 0.18 | 125.62 | 7.08 | 0.03 * |
| MIG~WESER6 | −0.97 | 0.179 | 125.604 | 7.05 | 0.03 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kleißen, J.; Balkenhol, N.; Pröhl, H. Landscape Genetics of the Yellow-Bellied Toad (Bombina variegata) in the Northern Weser Hills of Germany. Diversity 2021, 13, 623. https://doi.org/10.3390/d13120623
Kleißen J, Balkenhol N, Pröhl H. Landscape Genetics of the Yellow-Bellied Toad (Bombina variegata) in the Northern Weser Hills of Germany. Diversity. 2021; 13(12):623. https://doi.org/10.3390/d13120623
Chicago/Turabian StyleKleißen, Jasmin, Niko Balkenhol, and Heike Pröhl. 2021. "Landscape Genetics of the Yellow-Bellied Toad (Bombina variegata) in the Northern Weser Hills of Germany" Diversity 13, no. 12: 623. https://doi.org/10.3390/d13120623
APA StyleKleißen, J., Balkenhol, N., & Pröhl, H. (2021). Landscape Genetics of the Yellow-Bellied Toad (Bombina variegata) in the Northern Weser Hills of Germany. Diversity, 13(12), 623. https://doi.org/10.3390/d13120623





