Abstract
Planetary extinction of biodiversity underscores the need for taxonomy. Here, we scrutinize spider taxonomy over the last decade (2008–2018), compiling 2083 published accounts of newly described species. We evaluated what type of data were used to delineate species, whether data were made freely available, whether an explicit species hypothesis was stated, what types of media were used, the sample sizes, and the degree to which species constructs were integrative. The findings we report reveal that taxonomy remains largely descriptive, not integrative, and provides no explicit conceptual framework. Less than 4% of accounts explicitly stated a species concept and over one-third of all new species described were based on 1–2 specimens or only one sex. Only ~5% of studies made data freely available, and only ~14% of all newly described species employed more than one line of evidence, with molecular data used in ~6% of the studies. These same trends have been discovered in other animal groups, and therefore we find it logical that taxonomists face an uphill challenge when justifying the scientific rigor of their field and securing the needed resources. To move taxonomy forward, we make recommendations that, if implemented, will enhance its rigor, repeatability, and scientific standards.
1. Introduction
The biological field of taxonomy and systematics, the science of describing and classifying species, is often maligned as merely descriptive [1,2,3,4,5]. Despite this characterization, taxonomic products play a pivotal role by providing the underlying framework for every biological study [6]—rigorous and repeatable ecological, biochemical, comparative, evolutionary, and physiological studies would be impossible without accurate species delimitation. Nevertheless, taxonomy is typically regarded as a science in crisis [5,7,8]. Fewer students are being trained in organismal expertise, taxonomic works are under cited, funding for taxonomic research is limited, and the paucity of professional taxonomic positions at academic institutions portends unimportance among fellow researchers [7,8,9,10,11]. Arguments that the number of taxonomists has increased in recent years [12] are debatable [8] and likely reflect changes in scientific publication practices rather than increasing taxonomic expertise [13]. Recent reliance on citation metrics and journal impact factors for making hiring decisions, promotions, and other rewards [14] reinforces that taxonomic work is undervalued [9]. Citations and the correlated impact of taxonomic journals [15] mirrors the bias against proper full citation of scientific names [4,7,16,17,18]. The perceived diminished impact of taxonomy becomes self-perpetuating. The latest example of this dangerous trend was the proposed suppression (and subsequent reversal, due to backlash) in early 2020 of impact factors from journals with high instances of self-citations—oftentimes taxonomic journals, a direct consequence of the tradition not to cite taxonomic authorities—as expressed in an announcement from Clarivate (https://jcr.help.clarivate.com/Content/title-suppressions.htm (accessed on 28 June 2020)).
Contemporary taxonomists find themselves at the forefront of the battle lines drawn by human-driven climate change and mass extinction due to habitat loss and other factors [19,20]. As this large and rapid extinction event unfolds [21], taxonomists are thrust into the unsustainable position of documenting this monumental historical loss of biodiversity [22] and, in some cases, grimly identifying and naming new species already extinct or destined thusly. One of the authors of this paper (JEB) has first-hand experience describing new species (Californian trapdoor spiders) after their extinction [23]. Taxonomy alone stands between a species being lost to both extinction and obscurity. As such, one could argue that never before has the discipline been so important; it is impossible to “save,” conserve, and/or inventory undiscovered species. Yet, lag times between when a new species is collected and when it is described remain on average as high as 35 years or more with only 15% of all species described within five years of collection [13]. The implications of the Anthropocene epoch will never fully be understood without an exhaustive inventory of Earth’s biodiversity—a task where taxonomists will play fundamental roles. Taxonomic work is an undeniably critical component to saving our planet.
The arguments above notwithstanding, it would be fair to say that the taxonomic literature, from nearly any organismal field, represents works that span a very broad range, from those that are very descriptive (e.g., taxonomic descriptions of a single species that lack broader context) to large-scale phylogenetically-informed monographs that explicitly contain many elements of hypothesis testing and experimentation. Both types of work have utility and value. Moreover, considerable variation across many taxonomic groups exists as a function of species diversity and taxonomic “maturity”. For example, taxonomically-biased groups such as birds and mammals [24], in which the majority of species have likely been described, can afford now to be broadly integrative. In contrast, megadiverse arthropod groups with many undescribed species remaining [25,26] likely lag behind collectively in terms of highly sophisticated approaches to species delimitation, in part because taxonomists must find the balance between the urgent need to rapidly describe species and the production of more integrative works evaluating species using other approaches (e.g., molecular taxonomy). That said, we probably know very little about the progress in most taxonomic fields of study, and what we do know is based largely on anecdotal observations of individual works with which we are most familiar.
To our knowledge, there are few, if any, examples of multi-year surveys within any one taxonomic field evaluating the data and rigor of the species hypotheses being formulated (but see Liu et al. 2019). As such, we aimed to evaluate the data being collected and the species hypotheses being proposed across the large taxonomic field of spider systematics (Order: Araneae). Spiders are a hyper-diverse group comprising approximately 50,000 species parceled among >4000 genera and 129 families (World Spider Catalog 2021; WSC [27]). By some estimates, there are likely well over 120,000–200,000 species [28], an estimate supported by non-asymptotic new species discovery (WSC). Progress in spider taxonomy is documented via the WSC where each year hundreds of species descriptions are catalogued with the accompanying literature. The WSC is an information-rich resource, a rare database when compared against most other major lineages on the Tree of Life, that facilitates questions such as the ones we pose herein. Using this unique dataset, we surveyed the spider taxonomic literature from 2008 to 2018 (11 years). As part of our survey, we tabulated 22 parameters that included the type of data used in species delimitation, the species concept employed (if stated), and the number of specimens available for each species described. The results we outline below present a sobering overview of spider taxonomy (and perhaps of taxonomy in general): few papers describe an explicit species hypothesis, very few studies are integrative with the vast majority relying on one morphological data point, and a large proportion of species are based on two or fewer specimens.
We, like nearly all biologists, agree that taxonomy is important and more is needed, but it does not seem like anyone is really willing to ask hard questions about why it has a less than desirable reputation. If it is important, relevant, hypothesis-driven work, we see no reason why it should garner less respect now, or in the past, than any other field of biological science. As a model system that likely reflects the state of the science, we aim to take a critical look at the field of spider taxonomy by asking some difficult questions about the nature of the work we are doing with the hope of provoking change. We feel it is best that this criticism be honest and direct, and we acknowledge openly that it certainly applies to some of the works of this paper’s authors.
2. Methods
We downloaded nearly all the taxonomic works documented in the WSC during the time period of 2008–2018. Each investigator documented authorship and the number of new species described per publication; our review focused exclusively on newly described taxa during the study time period. Only a few non-English works were omitted from this study for which we were unable to find a translation that allowed confident data scoring. Table 1 below lists the parameters reviewed and how they were scored. Binary scorings were based on interrogative NO/YES (0/1) responses, whereas others were quantitative (documenting absolute numbers of observations or counts). Generally, we assessed the following: (1) the type of data used to establish species constructs; (2) how species were illustrated; (3) whether raw specimen data were downloadable; (4) how many specimens were examined for each species; and (5) what sexes were available for each species. The number of specimens available was tabulated as (1), (2), or (>2); >2 is somewhat arbitrary and underestimates the paucity of data associated with some species, but objectively captures the variation in the dataset without documenting the absolute number of specimens for all species. A study was classified as ‘integrative’ if morphological data (i.e., genitalic or other) was used in combination with at least one other data source. For species concept, we assessed each paper to determine whether the author stated explicitly what species concept they used to delineate taxa. The data were tabulated in a MS Excel spreadsheet; summary statistics and bar graphs were produced using the base R statistics packages and carried out in R-Studio [29].

Table 1.
Parameters evaluated and how they were assessed/scored.
Although most of the parameters evaluated can be objectively scored, the role that a particular data type played in species delimitation might be viewed as a matter of opinion sans asking the author directly. Generally, the team erred on the side of inclusivity—for example, if a researcher documented having collected ecological or behavioral data, we assumed those data played a role in delimiting species. For molecular studies where multiple populations were sequenced, if not stated it was considered implicit that the researcher employed a phylogenetic lineage species concept (indeed in nearly every case monophyly or exclusivity was stated as the criterion), otherwise a species concept was only noted if explicitly stated.
3. Results
Spanning an 11 year time period (2008–2018), we evaluated 2083 spider taxonomic works that described 8433 new species. The data are summarized in Table 2, Table 3 and Table 4 and Figure 1; the raw data can be downloaded as a .csv (comma separated) file at https://doi.org/10.6084/m9.figshare.17263835.v1. Nearly half the papers surveyed (>990) described a single new species with a median and average of two and four new species per publication, respectively. Note that some of these papers describing new species were part of a larger work or revision that also redescribed existing nominal taxa. Because we were primarily interested in the data used to describe new species, per se, we did not evaluate the context. In retrospect, this would have been a worthwhile parameter to assess and as such may extend the data set to include this at some point in the future.

Table 2.
The 2008–2018 summary data showing numbers of species described, numbers of papers examined, types of data considered, numbers of studies scored as integrative, and the number of papers that communicated an explicit species concept.

Table 3.
The 2008–2018 summary data showing the type of imaging used to illustrate species and number of papers with electronic data available in a downloaded format.

Table 4.
The 2008–2018 summary data showing numbers of species for which males and females were described and the number of specimens associated with each new species description. Percentages (of Totals) are the percentage of each column relative to the total number of species evaluated during the project time period.
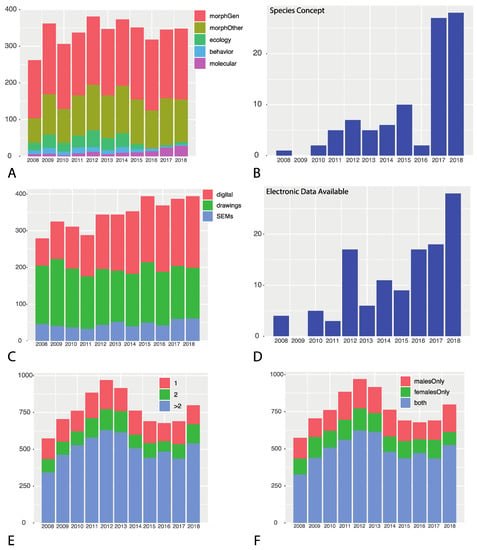
Figure 1.
Summary of data per year 2008–2018. (A) Number of species based on each data type; (B) number of publications explicitly stating a species concept; (C) illustration type used in each publication; (D) prevalence of downloadable electronic data; (E) number of specimens examined per new species; (F) prevalence of both male and female specimens per new species described.
3.1. How Integrative Is Spider Taxonomy?
Most surveyed studies rely on morphology alone (Figure 1A), with few including additional types of evidence. Only 14% of taxonomic works were classified as integrative using the criterion requiring two or more data types, whereas all studies classified as integrative included morphology as one of the data sources. Our evaluation of the degree to which taxonomy is integrative is liberal because most instances of “integrative taxonomy” we documented did not invoke an explicit statement of a predefined integrative approach, e.g., [29,30,31], by the author(s). Consequently, our results report a best-case scenario for integrative taxonomy. As shown in Figure 1A, genitalic morphology is the dominant data type followed by ‘morphology—other’, the latter representing morphological differences such as coloration, patterning, setal differences, and measurements. Ecological and behavioral data were included to a modest degree from 2008 to 2014, though diminish in prevalence thereafter.
At this time, molecular data have not been heavily involved in spider taxonomy (Figure 1A), though of the molecular studies we surveyed, almost all were integrative; we defined 98% as integrative with only two ostensibly not including other lines of evidence. Studies were considered to have a molecular component if they included any molecular data and thus spanned a wide range of data types from COI barcoding to multi-locus genomic studies. Molecular data were used in ~43% of all studies we considered to be integrative. Despite the increased access to cheaper molecular tools over the last decade, e.g., [32], molecules remain surprisingly scarce in spider taxonomic studies (see below for potential explanations). We document 130 occurrences of molecular usage over the 11 years, accounting for ~6% of the total publications surveyed, with nearly 2/3s of the molecular-based studies published within the last three surveyed years (2016–2018).
3.2. Are Species Constructs Defined Conceptually?
The vast majority of spider taxonomic publications fail to articulate any defined species concept (Figure 1B); only 93 of 2083 (4%) publications defined an operational species criterion or concept. From our survey, approximately five concepts are employed—morphological, phylogenetic, cohesion, general lineage, and the unified species concept.
3.3. What Data Are Being Used? What Is Available, and How Much?
Modern day spider species descriptions are primarily illustrated via drawings, scanning electron microscopy (SEM), and digital images (Figure 1C). From 2008 to 2012, drawings accounted for the greatest proportion of illustrations, whereas in subsequent years digital images using light microscopy and other techniques (e.g., image stacking) have become more prevalent [33]. The proportion of SEM illustration has remained relatively constant and likely reflects the need for fine-scale detail not possible using light microscopy. We did not collect data regarding the deposition of images in online databases; however, anecdotally it seems that very few authors archive images in any online repositories such as Morphbank (http://www.morphbank.net/). We observe a transition to high resolution light photographic images, a positive trend, given the recent push to digitize collections. Although drawings certainly have value as interpretive images, high quality photographs of actual specimens (e.g., holotypes) are potentially data rich and in some cases may abrogate the need for borrowing material from collections [34].
Raw measurement data, included in every spider species description, is seldom available as searchable supplementary data (Figure 1D). We found only 118 (~5%) documented occurrences where raw measurement data were available in a downloadable format (e.g., as a spreadsheet or database). Our survey shows a minor sustained upward trend in increasing data availability after 2015. For 2018, 28 of 207 papers, approximately 14%, provided data available for download. As discussed by Bik [6], availability of traditional taxonomic data would “fundamentally improve database resources for all scientific disciplines”.
Although taxonomic works are potentially data rich, a non-trivial number of spider species are based on little material and thus sample sizes are small. During the time period reviewed, 34% were based on two or fewer specimens (Figure 1E) and 35% of all species were based on one sex (Figure 1F). Twenty percent (20%) of all spider species were based on a single specimen (i.e., the sample size for a new species is n = 1). In numerous instances, a publication consisted entirely of the description of only one new species, based on only one specimen (185 in total, 8.5%).
4. Discussion
Current trends in spider taxonomy are predominantly of non-revisionary nature and non-integrative approaches, findings that seem consistent with trends reported in other groups (e.g., insects—[10]). This suggests that taxonomy has been slow to improve upon the traditional morphology-only approach despite rapid development and easy availability of a vast number of other data sources—a finding likely shared within taxonomic groups across the Tree of Life. Such a slow pace of transition to more inclusive taxonomy seems to characterize not only spiders, but also other hyper-diverse taxa, with calls to devise somewhat controversial methods [35] to drastically shortcut species descriptions for groups containing many undescribed species (e.g., barcode and rapid digital imaging of ichneumonoid wasps, [36].
4.1. How Integrative Is Taxonomy?
The spider taxonomic literature, like that of many other hyper-diverse taxa and ‘non-charismatic microbiota’, appears to be largely dominated by traditional morphological taxonomy. If relying on a single source of data, it can be argued that morphology is more important than other data sources, for example, by providing essential information on taxa and the necessary link to taxonomy going back to its beginnings [37]. Nevertheless, the field overwhelmingly lacks an explicit integrative perspective and the sole reliance on morphology restricts the field. From the typical number of new species described per paper (1–2 species), one can probably infer the field is not generally comparative (note that this was not an evaluative aim of our study), and one might also infer anecdotally that the literature is dominated by alpha taxonomy, with the majority of papers being non-monographic [7]. This is a trend that has been countered by the creation of taxonomic initiatives, for example the USA National Science Foundation PEET program (Partnership for Enhancing Expertise in Taxonomy [11]. However, more such efforts, especially international and global initiatives, are necessary.
The scarcity of molecular data is likely the consequence of a number of factors. First, many species are known from only one to a few specimens and thus taxonomists may hesitate damaging type specimens for DNA extraction. Likewise, while it is possible to extract DNA from older museum specimens using techniques that allow for non-destructive sampling [38], the same reluctance to damage or destroy precious material likely prevails. Further, morphological taxonomy can be produced on a salary alone, whereas molecular data often require additional funding.
All things considered, why have molecular and/or integrative approaches not become more common in spider taxonomy, particularly the former? All other fields in the life sciences have been methodologically transformed over the past decade by the major technological innovation of high-throughput sequencing technologies. Yet, this does not seem to be true for the taxonomy of spiders and other megadiverse organisms see [39] for a summary of how taxonomy could be advanced using genomics, likely in part due to the challenges in striking a balance between the needs for rapid discovery on one hand and taxonomic rigorousness on the other. That notwithstanding, rapid technological advances have essentially rejuvenated molecular phylogenetic systematics and have an amazing potential to invigorate taxonomy as well [40]. One likely general explanation for the slow adoption of molecular methods is that the majority of traditional taxonomists, in particular those from countries with little financial aid for scientific research available, are resource limited—a point that directly links with the problematic impact-factor decision by Clarivate (mentioned above) and has significant potential to harm our colleagues in these countries. However, the cost of obtaining molecular data is ever decreasing. The argument could further be made that morphological data are sufficient, yet numerous molecular studies at multiple hierarchical levels consistently show instances where morphology underestimates diversity and/or fails to accurately delimit species [41,42,43,44]; discussed in more detail below.
4.2. Are Species Constructs Defined Conceptually?
As we discussed above, taxonomy provides a potentially rich hypothetic-deductive framework for testing species boundaries [2,5,7]. Yet, spider taxonomists generally fail to convey any information on how they conceptualized the species they are describing, including some authors of this paper. Additionally, while we are certainly a part of this past, we argue, however, that the well-worn argument that taxonomic species constructs are rigorous species boundary hypotheses is vastly strengthened by placing the work in the context of an operational species construct. Otherwise, we believe taxonomists have played a role in diminishing their own work’s rigor. Arguably, taxonomic works that lack an explicit hypothetical framework could be branded as purely descriptive, and essentially unfalsifiable. We do believe that good ‘diagnoses’ sections found in many papers potentially provide some testable hypotheses, as do papers that clearly place species in a phylogenetic context and those using molecular data (e.g., ability to demonstrate monophyly, and lack of gene flow, important elements of most species concepts, at least intrinsically). However, the explicit use of species concepts as the underlying hypothetical framework of taxonomy, would vastly improve and clearly place taxonomic works on par with any other hypothesis-based sciences.
The absence of a stated conceptual species framework may provide some insight into why the value of taxonomy is diminished as a largely “descriptive science”; that is, why taxonomy is viewed as non-experimental observation and thus not as intellectually informed as other scientific disciplines perceived to be more hypothesis-driven. Although descriptive science should have merit in its capacity to discover and illuminate novel phenomena, a number of authors posit that taxonomy is just as much a hypothesis-driven science as any other [2]. Nicely articulated by Haszpruner [45], taxonomic works formulate and implicitly convey tests of three or more hypotheses: (1) species delimitation; (2) species classification; and (3) homology statements (often multiple hierarchical hypotheses) regarding organismal form and function. In short, a taxonomist with a collection of specimens postulates tests of what constitutes the limits of a species, where in the hierarchy of life that species is placed, and what synapomorphies support their hypotheses. The latter constitutes a complex nested set of homology statements that represent hypotheses based on, for example, anatomy, function, and ontogeny. Moreover, broad-scale studies (e.g., a family-level taxonomic revision, monograph with good diagnoses, and morphological studies in a phylogenetic framework) could be characterized as carefully designed experiments that include informed taxon and character selection used to construct phylogenetic inferences specifically aimed to test species, genera, and family-level limits (e.g., tests of monophyly). Taxonomy is potentially both descriptive and experimental, by employing levels of experimentation, sophistication, and knowledge that are equal among its counterpart disciplines in the life sciences. “That a taxonomic study is hypothesis-driven and analytical from its very beginning is not obvious to the uninitiated” [5]—for taxonomy to increase potential to receive its due credit, this needs to become explicit and obvious.
4.3. Species and Data: What Is Being Used? What Is Available, and How Much?
In general, we find that during the surveyed period, studies only employ a small portion of the available relevant data used to formulate and test the taxonomic hypotheses. In a large proportion of studies, the data presented barely meet an acceptable minimum; indeed we can think of no other field of biology (or other scientific discipline, in general) where such a paucity of data would be the acceptable basis for scientific inference.
Regardless, the historical taxonomic literature is (potentially) incredibly data rich, containing valuable information related to geographic distributions, temporal occurrence patterns, and morphology that captures a vast wealth of quantitative and qualitative observations. Because these data are generally inaccessible, one could argue that they simply do not exist—“If it isn’t online, it might as well not exist” [1,6] p. 2. Although some taxonomic journals such as Zookeys (Magnolia Press) use XML markup to facilitate downstream data extraction, there seems little reason for not making taxonomic data available in electronic form. For example, with the recent advances to spider phylogeny [46,47,48,49], accessible morphological data could be used to enhance comparative evolutionary studies that are de rigueur in other more exhaustively studied groups such as mammals, birds, and amphibians and reptiles.
Additionally, rare, singleton species (species known from only one specimen) are not uncommon in biodiversity samples, representing over 30% of all species found in tropical arthropod inventories [50]. In a 10 year survey (2000–2010) of American Museum of Natural History publications, Lim et al. [51] found that 17.7% of new species were known from only one specimen. Although very rare species are expected in collections and biodiversity surveys, some authors have proposed that new species should never be described on the basis of a single specimen. Species hypotheses using so little data are prone to anatomical mistakes and lack thorough evaluation of infra- and interspecific variation [29]. Lim et al. [51] suggest that these problems are not germane to only morphological species delimitation because molecular approaches such as GMYC and PAA only accommodate singleton species to some degree; many species are indeed rare and current molecular analytical methods are severely limited in treating these situations. In general, we believe such species descriptions in isolation, and absent a context should cease to exist, but clearly species based on single specimens may be very appropriate for inclusion in broader revisions. That is, single species and/or single specimen descriptions may be justifiable in cases given a very clear and well-defined context. These could include biological (e.g., unique species biology, part of a thorough biodiversity inventory), revisionary (e.g., adding a new species to a recent revision), or phylogenetic (e.g., critical phylogenetic taxon) contexts where their description serves a clear purpose despite the limitations detailed here.
We would also like to explore here the notion that many singleton species are unfalsifiable and represent a particularly poor inductive argument. As discussed by Wheeler [2], “species hypotheses are not efficiently tested in isolation” and “to critically test the distribution of attributes defining one species it is necessary to examine variation within that and all nearly related species”. The singleton specimen species problem often conveys no such test or distribution of attributes described by Wheeler; it is not a hypothesis that can be falsified based on a broader context character diagnosis—the essence of the species hypothesis. Alternatively, in the absence of any description of variation, the species is potentially typological and thus conceivably falsified upon discovery of any additional material showing variation. Second, the inductive assumptions of species based on one specimen are severe; it essentially assumes the uniformity of all individuals based on a single observation. It is not that we believe that the diagnosis of all species based on a single specimen are false, they may very well be “good species”, but they are certainly weak hypotheses, trivially falsifiable by a single additional datum. This merits acknowledging the flaws inherent in such logic and why the quality of such science might be viewed as wanting.
4.4. Recommendations
Below, we make four recommendations we think need to be quickly adopted by the taxonomic community. The first three are pragmatic changes to taxonomic publications that entail how species hypotheses are stated, data are made available, and provide minimum data standard guidelines. The fourth recommendation relates to an aspirational goal of ultimately achieving a more integrative taxonomy in the coming years. The International Code of Zoological Nomenclature (ICZN) establishes the rules for naming species but is silent with respect to what constitutes good scientific practice. Codes governing the naming of other taxa such as plants, fungi, and viruses similarly focus on the naming rules rather than scientific practice. If the field of taxonomy is going to gain the respect it deserves in the scientific community, then we need to move past simple compliance with the ICZN and other naming rules and adopt practices that are more in line with other scientific disciplines. Our recommendations are as follows:
(1) All taxonomic works should clearly state the species concept being applied. Failing to state what epistemological and/or conceptual-based criteria are being used to distinguish one specimen/species from the next should become an important factor for reviewers and editors in assessing manuscript quality. We emphasize that some of our own past works would have greatly benefited from doing so and we fail to see any valid reason for future taxonomic work not doing so. The notion that some underlying yet unstated species concept/hypothesis is implicit in every taxonomic work [sensu 5] should be no more acceptable to practicing taxonomists than would an experimental ecologist failing to describe experimental design, define hypotheses tested, and elaborate a statistical alpha level. Taxonomic journals, subject editors, and reviewers must start demanding that authors include such a statement. Such a change would benefit taxonomists in a number of ways including requiring authors to more carefully consider the criteria they are applying to differentiate taxa, by further enhancing the objectivity of species determination with explicit criteria, and by emphasizing to others that species descriptions are conceptually formulated and hypothesis driven.
(2) With increasing databank availability and ease of use, all data should be made electronically accessible. The demand for electronic data should include all quantitative, qualitative, specimen (e.g., geographic and collection event), and original (unaltered) image data. Nearly every journal that publishes papers containing genetic data have required authors for decades to deposit data in databases such as NCBI or EMBL. Although morphological databases with similar governmental support do not currently exist (they should!), archives such as Dryad (http://dryad.org), Symbiota (https://symbiota.org/), figshare (https://figshare.com/) are available and easy to use, and GBIF is online and free for archiving locality data (https://www.gbif.org/). Because some databases such as Dryad are fee based, for-profit journals capitalizing on the hard work of taxonomist authors, editors, and reviewers must work to ensure that free or minimal cost options are available to investigators that may not have adequate funding. The mandate to electronically archive all taxonomic associated data should begin immediately. Digitally archived morphological data should conform to field/organismal anatomy ontologies whenever possible, e.g., [52], the spider anatomy ontology (SPD) [53]. It follows that systematics and taxonomy need to also focus attention and computational resources to begin harvesting critical legacy morphological data but, in the meantime, cease accumulating data that are not electronically accessible. Efforts to employ computational approaches such as natural language and machine learning, coupled with well-developed ontologies, should be further exploited to facilitate recovery of 200+ years of data embedded in publications. As noted by Godfray [1], “the quantity of taxonomic information available on the web is pitiful”—a statement that remains as true today as it did over 18 years ago—that needs to change.
(3) Descriptions of new species based on singleton specimen data that are described outside a very clear context (see above) should stop. This practice strikes a balance between the positive aspects of describing rare new taxa with Dayrat’s [29] recommendation to preclude these altogether. Such data-deficient hypotheses should be as critically scrutinized in taxonomy as they would be in any other field of science. This should not necessarily stymie efforts describing rare taxa, but simply require that the taxonomic description thoroughly document interspecific variation and demonstrate sufficient evidence that a new, rare species is warranted. To be fair, some (but not all) singleton species descriptions typically examine types of related species, other material, or digital images of congeners, but we would argue that the bar needs to be higher. Unfortunately, this may leave some new candidate species waiting description (although it should motivate larger-scale studies, particularly international collaboration). As such, these specimens could be documented as candidate species in published taxonomic notes or using online data narratives. Nevertheless, the practice of describing a new taxon on the basis of very little data needs critical reevaluation. In general, it is not good scientific practice because it is a weakly formulated hypothesis (at best), logically flawed, and caters to the notion that taxonomy lacks robust data and rigorous publication standards.
(4) Finally, taxonomy must aspire to become more integrative and do so quickly [31]. Other fields in the life sciences have been transformed over the past decade via quantum leaps in genomics, proteomics, and biological imaging (to be fair most other fields do not have the task of describing hundreds of thousands of new entities, many of which are rare and difficult to discover, in a very short timeline). Taxonomy does not appear to have overwhelmingly capitalized on these advances. That said, it is important to recognize that not all taxonomists have access to technology and sufficient funding, thus the aspiration of a fully integrative taxonomy that includes genomics, for example, is primarily targeted at those labs able to do that sort of work (but see below)—with an emphatic call for global collaboration and an end to the colonialist mindset of “parachuting” into a region or country, collecting specimens, and describing that diversity without the help (and co-authorship) of local colleagues. We follow Dayrat’s [29] definition of integrative taxonomy “as the science that aims to delimit the units of life’s diversity from multiple and complementary perspectives (phylogeography, comparative morphology, population genetics, ecology, development, behaviour, etc.)”; therefore, integrative taxonomy is not limited to technology per se but fundamentally incorporates multiple data types. Whenever possible, future generations of taxonomists need to be trained in morphological taxonomy as well as modern techniques that capitalize on next generation technologies, thereby extending their potential to gain “complementary perspectives”—training only in the former severely limits marketability for jobs. Adopting an integrative approach is not just good practice because more modern techniques are in vogue, it is good practice because it is good science. Evolutionary biologists have long acknowledged the limitations of morphology in species delimitation. Phenomena such as convergent and parallel evolution, phenotypic plasticity, and morphological stasis (species crypsis) can obscure species boundaries requiring multiple lines of evidence to accurately resolve. If taxonomists are interested in “getting it right”, we need to consider other data related to the species origin and evolutionary trajectory [31]. The majority of spider taxonomic studies employing molecular data were ironically integrative for exactly these reasons; that is, molecular systematists have long acknowledged that one data point, one gene, one data type, is not sufficient to confidently assign populations to species. Species diagnosed using a single character system should be just as questionable as a species or phylogenetic hypothesis based on one gene, yet they are not. Taxonomy needs to genuinely transform itself as a collaborative, integrative information science [1]. Finally, we will also add that a recent review of the taxonomic literature characterized as “integrative” [54] seems to indicate that these works may “open doors” to top ranked journals and enhance citation performance.
The first three of the recommendations can easily be implemented through enhanced editorial practices and vigilant peer review that enforces a set of community standards. Recommendation #4 is a little more complicated in its implementation and should not be confused as a decree that a new integrative taxonomy be prohibitively costly and only technology driven. Although we do see an aspirational integrative taxonomy as taking full advantage of many of the methodological innovations available in a modern life sciences tool kit, this does not preclude integration of traditional morphology with other types of natural history data that can be collected at much lower or no cost (oftentimes just as valuable, e.g., differences in mating rituals and temporally different mating activity patterns). Moreover, such an aspiration should encourage partnerships and collaborations among scientists with access to those tools. There is a lot of quality taxonomy being done in parts of the world that must continue; however, if the field of taxonomy is going to advance it cannot remain stagnant for the purpose of holding to a standard that every practicing taxonomist can easily achieve. We certainly do not see other fields of biology only adhering to older practices as a means of accommodating all researchers.
5. Conclusions
The data we have presented here document a rather sobering depiction of the state of spider taxonomy, consistent with the sparse available evidence on other hyper-diverse taxonomic groups, e.g., [10]. We find that much of the work over the last decade is generally not integrative, not accessible electronically, and based on very few specimens and consequently little data. Spider taxonomists seldom state a species hypothesis or concept. Although there are certainly exceptional works that are broad in scope (monographic), integrative, and data rich/accessible, unfortunately such studies are the minority. As such, it would seem that many of the criticisms leveled at taxonomists including the stereotype that the work we do is largely descriptive, are justified. These wounds are to some extent self-inflicted. Despite the importance and relevance of the work we do, these data either bely the sophistication and intellectual underpinnings of taxonomy or, accurately depict the work being done. We believe that for many professional taxonomists, it is the former rather than the latter. Nevertheless, the majority of taxonomists do a very poor job emphasizing the tremendous intellectual contributions of the work they are doing within the context of those publications. As discussed recently by Wheeler [3], it is time that taxonomy got an “image makeover” and recognized for the “incomparable benefits to other sciences and society”. The recommendations we make above are aimed toward exactly such a makeover because it is simply not enough to just extol the virtues and intellectual content implicit in every taxonomic work—the intellectual content must be made explicit in every work, otherwise taxonomy will continue to dwindle in perception as a bona fide discipline along with its funding and academic positions. As a world community, we must heavily invest now in training modern integrative taxonomic specialists who take full advantage of all the tools modern biology has to offer. Currently, the resources needed to effectively discover and describe new species are seriously lacking, despite the fact that understanding the true diversity on our planet and finding answers to how that diversity has evolved is perhaps one of the most important endeavors we can answer as biologists. The resources needed for taxonomists to effectively do their jobs require immense person power and funding for field work, identification, data collection (e.g., morphological and DNA), specimen storage, and training the next generation. There is clear evidence that efforts such as the USA National Science Foundation’s PEET program, can dramatically advance the field. We strongly advocate that the NSF urgently consider reinstating the PEET program, and that similar efforts be initiated and/or sustained through other national, and especially international programs. It is not surprising that quick-fix approaches such as DNA barcoding [55] that propose to “democratize” taxonomy [56], garner funding and attention because their proponents have seemingly done a better job advocating for their science. DNA barcoding outwardly seems modern, rigorous and objective, and hypothesis driven. Consequently, governments continue to invest millions of dollars into this methodology—with little regard for the fact that for this approach to be truly effective, the species (and their boundaries) it seeks to identify need to be identified and diagnosed by trained taxonomists.
In closing, our planet is facing an extinction crisis. Taxonomy is integral to solving that crisis. It is time for taxonomists to stop complaining about being disrespected, underfunded, and not cited. Instead, we must acknowledge the problems with how we have been working on and presenting our science, as well as the role that we have played in fomenting negative perceptions. We must change some of our practices and demand the respect and resources that our noble field of taxonomy requires and deserves.
Author Contributions
Conceptualization, J.E.B.; methodology, J.E.B.; validation, all authors; formal analysis, J.E.B.; investigation, all authors; data curation, J.E.B.; writing—original draft preparation, J.E.B., I.A., M.K.; writing—review and editing, all authors; visualization, J.E.B.; supervision, J.E.B.; project administration, J.E.B.; funding acquisition, J.E.B., M.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by USA National Science Foundation grants AF 1401176 and DEB 1937604 to JEB and the Evert and Marion Schlinger Foundation. MK was supported by the Slovenian Research Agency, grants P1-0236, J1-9163.
Data Availability Statement
The raw data can be downloaded at https://doi.org/10.6084/m9.figshare.17263835.v1.
Acknowledgments
We thank Joel Ledford, Lynn Kimsey, Vera Opatova, Laura Montes de Oca, and two reviewers for critical comments on an earlier version of this manuscript (having commented does not imply agreement with all of the content!).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Godfray, H.C.J. Challenges for Taxonomy. Nature 2002, 417, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, Q.D. Taxonomic Triage and the Poverty of Phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, Q. A Taxonomic Renaissance in Three Acts. Megataxa 2020, 1, 4–8. [Google Scholar] [CrossRef][Green Version]
- Wägele, H.; Klussmann-Kolb, A.; Kuhlmann, M.; Haszprunar, G.; Lindberg, D.; Koch, A.; Wägele, J.W. The Taxonomist—An Endangered Race. A Practical Proposal for Its Survival. Front. Zool. 2011, 8, 25. [Google Scholar] [CrossRef]
- Sluys, R. The Unappreciated, Fundamentally Analytical Nature of Taxonomy and the Implications for the Inventory of Biodiversity. Biodivers. Conserv. 2013, 22, 1095–1105. [Google Scholar] [CrossRef]
- Bik, H.M. Let’s Rise up to Unite Taxonomy and Technology. PLoS Biol. 2017, 15, e2002231. [Google Scholar] [CrossRef]
- Agnarsson, I.; Kuntner, M. Taxonomy in a Changing World: Seeking Solutions for a Science in Crisis. Syst. Biol. 2007, 56, 531–539. [Google Scholar] [CrossRef]
- Bacher, S. Still Not Enough Taxonomists: Reply to Joppa et al. Trends Ecol. Evol. 2012, 27, 65–66. [Google Scholar] [CrossRef]
- Drew, L.W. Are We Losing the Science of Taxonomy? BioScience 2011, 61, 942–946. [Google Scholar] [CrossRef]
- Liu, Y.; Dietrich, C.; Braxton, S.; Wang, Y. Publishing Trends and Productivity in Insect Taxonomy from 1946 through 2012 Based on an Analysis of the Zoological Record for Four Species-Rich Families. Eur. J. Taxon. 2019, 504, 1–24. [Google Scholar] [CrossRef]
- Rodman, J.E.; Cody, J.H. The Taxonomic Impediment Overcome: NSF’s Partnerships for Enhancing Expertise in Taxonomy (PEET) as a Model. Syst. Biol. 2003, 52, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Joppa, L.N.; Roberts, D.L.; Pimm, S.L. The Population Ecology and Social Behaviour of Taxonomists. Trends Ecol. Evol. 2011, 26, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Bebber, D.P.; Wood, J.R.I.; Barker, C.; Scotland, R.W. Author Inflation Masks Global Capacity for Species Discovery in Flowering Plants. New Phytol. 2014, 201, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Gruber, T. Academic Sell-out: How an Obsession with Metrics and Rankings Is Damaging Academia. J. Mark. High. Educ. 2014, 24, 165–177. [Google Scholar] [CrossRef]
- Bilton, D.T. What’s in a Name? What Have Taxonomy and Systematics Ever Done for Us? J. Biol. Educ. 2014, 48, 116–118. [Google Scholar] [CrossRef]
- Vink, C.; Paquin, P.; Cruickshank, R. Taxonomy and Irreproducible Biological Science. BioScience 2012, 62, 451–452. [Google Scholar] [CrossRef]
- Packer, L.; Monckton, S.K.; Onuferko, T.M.; Ferrari, R.R. Validating Taxonomic Identifications in Entomological Research. Insect Conserv. Divers. 2018, 11, 1–12. [Google Scholar] [CrossRef]
- Monckton, S.K.; Johal, S.; Packer, L. Inadequate Treatment of Taxonomic Information Prevents Replicability of Most Zoological Research. Can. J. Zool. 2020, 633–642. [Google Scholar] [CrossRef]
- Ceballos, G.; Ehrlich, P.R.; Dirzo, R. Biological Annihilation via the Ongoing Sixth Mass Extinction Signaled by Vertebrate Population Losses and Declines. Proc. Natl. Acad. Sci. USA 2017, 114, E6089–E6096. [Google Scholar] [CrossRef]
- Young, H.S.; McCauley, D.J.; Galetti, M.; Dirzo, R. Patterns, Causes, and Consequences of Anthropocene Defaunation. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 333–358. [Google Scholar] [CrossRef]
- Régnier, C.; Achaz, G.; Lambert, A.; Cowie, R.H.; Bouchet, P.; Fontaine, B. Mass Extinction in Poorly Known Taxa. Proc. Natl. Acad. Sci. USA 2015, 112, 7761–7766. [Google Scholar] [CrossRef]
- Dubois, A. Zoological Nomenclature in the Century of Extinctions: Priority vs. ‘Usage’. Org. Divers. Evol. 2010, 10, 259–274. [Google Scholar] [CrossRef]
- Bond, J. Phylogenetic Treatment and Taxonomic Revision of the Trapdoor Spider Genus Aptostichus Simon (Araneae, Mygalomorphae, Euctenizidae). ZooKeys 2012, 252, 1–209. [Google Scholar] [CrossRef]
- Troudet, J.; Vignes-Lebbe, R.; Grandcolas, P.; Legendre, F. The Increasing Disconnection of Primary Biodiversity Data from Specimens: How Does It Happen and How to Handle It? Syst. Biol. 2018, 67, 1110–1119. [Google Scholar] [CrossRef] [PubMed]
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.B.; Worm, B. How Many Species Are There on Earth and in the Ocean? PLoS Biol. 2011, 9, e1001127. [Google Scholar] [CrossRef]
- Stork, N.E. How Many Species of Insects and Other Terrestrial Arthropods Are There on Earth? Annu. Rev. Entomol. 2018, 63, 31–45. [Google Scholar] [CrossRef] [PubMed]
- World Spider Catalog. Version 20.5. Natural History Museum Bern. Available online: http://wsc.nmbe.ch (accessed on 3 November 2021).
- Agnarsson, I.; Coddington, J.A.; Kuntner, M. Systematics: Progress in the Study of Spiders and Evolution. In Spider Research in the 21st Century: Trends and Perspectives; Siri Scientific Press: Manchester, UK, 2013; pp. 58–1111. ISBN 978-0-9574530-1-2. [Google Scholar]
- Dayrat, B. Towards Integrative Taxonomy: Integrative Taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–415. [Google Scholar] [CrossRef]
- DeSalle, R.; Egan, M.G.; Siddall, M. The Unholy Trinity: Taxonomy, Species Delimitation and DNA Barcoding. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1905–1916. [Google Scholar] [CrossRef]
- Padial, J.M.; Miralles, A. TRehvieew Integrative Future of Taxonomy. Front. Zool. 2010, 7, 1–4. [Google Scholar] [CrossRef]
- Pomerantz, A.; Peñafiel, N.; Arteaga, A.; Bustamante, L.; Pichardo, F.; Coloma, L.A.; Barrio-Amorós, C.L.; Salazar-Valenzuela, D.; Prost, S. Real-Time DNA Barcoding in a Rainforest Using Nanopore Sequencing: Opportunities for Rapid Biodiversity Assessments and Local Capacity Building. GigaScience 2018, 7, giy033. [Google Scholar] [CrossRef]
- Mertens, J.; Van Roie, M.; Merckx, J.; Dekoninck, W. The Use of Low Cost Compact Cameras with Focus Stacking Functionality in Entomological Digitization Projects. ZooKeys 2017, 712, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Brecko, J.; Mathys, A.; Dekoninck, W.; Leponce, M.; VandenSpiegel, D.; Semal, P. Focus Stacking: Comparing Commercial Top-End Set-Ups with a Semi-Automatic Low Budget Approach. A Possible Solution for Mass Digitization of Type Specimens. ZooKeys 2014, 464, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Zamani, A.; Vahtera, V.; Sääksjärvi, I.E.; Scherz, M.D. The Omission of Critical Data in the Pursuit of ‘Revolutionary’ Methods to Accelerate the Description of Species. Syst. Entomol. 2020, 46, 1–4. [Google Scholar] [CrossRef]
- Meierotto, S.; Sharkey, M.J.; Janzen, D.H.; Hallwachs, W.; Hebert, P.D.N.; Chapman, E.G.; Smith, M.A. A Revolutionary Protocol to Describe Understudied Hyperdiverse Taxa and Overcome the Taxonomic Impediment. Dtsch. Entomol. Z. 2019, 66, 119–145. [Google Scholar] [CrossRef]
- Clerck, C. Svenska Spindlar, Uti Sina Hufvud-Slågter Indelte Samt under Några Och Sextio Särskildte Arter Beskrefne Och Med Illuminerade Figurer Uplyste; Stockholmiae: Stockholm, Sweden, 1757; pp. 1–154. [Google Scholar]
- Patzold, F.; Zilli, A.; Hundsdoerfer, A.K. Advantages of an Easy-to-Use DNA Extraction Method for Minimal-Destructive Analysis of Collection Specimens. PLoS ONE 2020, 15, e0235222. [Google Scholar] [CrossRef]
- Coates, D.J.; Byrne, M.; Moritz, C. Genetic Diversity and Conservation Units: Dealing With the Species-Population Continuum in the Age of Genomics. Front. Ecol. Evol. 2018, 6, 165. [Google Scholar] [CrossRef]
- Gómez Daglio, L.; Dawson, M.N. Integrative Taxonomy: Ghosts of Past, Present and Future. J. Mar. Biol. Assoc. UK 2019, 99, 1237–1246. [Google Scholar] [CrossRef]
- Hamilton, C.A.; Formanowicz, D.R.; Bond, J.E. Species Delimitation and Phylogeography of Aphonopelma Hentzi (Araneae, Mygalomorphae, Theraphosidae): Cryptic Diversity in North American Tarantulas. PLoS ONE 2011, 6, e26207. [Google Scholar] [CrossRef]
- Hedin, M. High-Stakes Species Delimitation in Eyeless Cave Spiders (Cicurina, Dictynidae, Araneae) from Central Texas. Mol. Ecol. 2015, 24, 346–361. [Google Scholar] [CrossRef]
- Opatova, V.; Bond, J.E.; Arnedo, M.A. Ancient Origins of the Mediterranean Trap-Door Spiders of the Family Ctenizidae (Araneae, Mygalomorphae). Mol. Phylogenet. Evol. 2013, 69, 1135–1145. [Google Scholar] [CrossRef]
- Thomas, S.M.; Hedin, M. Multigenic Phylogeographic Divergence in the Paleoendemic Southern Appalachian Opilionid Fumontana Deprehendor Shear (Opiliones, Laniatores, Triaenonychidae). Mol. Phylogenet. Evol. 2008, 46, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Haszprunar, G. Species Delimitations—Not ‘Only Descriptive’. Org. Divers. Evol. 2011, 11, 249–252. [Google Scholar] [CrossRef]
- Bond, J.E.; Garrison, N.L.; Hamilton, C.A.; Godwin, R.L.; Hedin, M.; Agnarsson, I. Phylogenomics Resolves a Spider Backbone Phylogeny and Rejects a Prevailing Paradigm for Orb Web Evolution. Curr. Biol. 2014, 24, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Hormiga, G.; Giribet, G. Phylogenomic Analysis of Spiders Reveals Nonmonophyly of Orb Weavers. Curr. Biol. 2014, 24, 1772–1777. [Google Scholar] [CrossRef]
- Garrison, N.L.; Rodriguez, J.; Agnarsson, I.; Coddington, J.A.; Griswold, C.E.; Hamilton, C.A.; Hedin, M.; Kocot, K.M.; Ledford, J.M.; Bond, J.E. Spider Phylogenomics: Untangling the Spider Tree of Life. PeerJ 2016, 4, e1719. [Google Scholar] [CrossRef]
- Opatova, V.; Hamilton, C.A.; Hedin, M.; De Oca, L.M.; Král, J.; Bond, J.E. Phylogenetic Systematics and Evolution of the Spider Infraorder Mygalomorphae Using Genomic Scale Data. Syst. Biol. 2020, 69, 671–707. [Google Scholar] [CrossRef]
- Coddington, J.A.; Agnarsson, I.; Miller, J.A.; Kuntner, M.; Hormiga, G. Undersampling Bias: The Null Hypothesis for Singleton Species in Tropical Arthropod Surveys. J. Anim. Ecol. 2009, 78, 573–584. [Google Scholar] [CrossRef]
- Lim, G.S.; Balke, M.; Meier, R. Determining Species Boundaries in a World Full of Rarity: Singletons, Species Delimitation Methods. Syst. Biol. 2012, 61, 165–169. [Google Scholar] [CrossRef]
- Vogt, L.; Nickel, M.; Jenner, R.A.; Deans, A.R. The Need for Data Standards in Zoomorphology. J. Morphol. 2013, 274, 793–808. [Google Scholar] [CrossRef]
- Ramírez, M.J.; Michalik, P. The Spider Anatomy Ontology (SPD)—A Versatile Tool to Link Anatomy with Cross-Disciplinary Data. Diversity 2019, 11, 202. [Google Scholar] [CrossRef]
- Vinarski, M.V. Roots of the Taxonomic Impediment: Is the “Integrativeness” a Remedy? Integr. Zool. 2020, 15, 2–15. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.; Waterton, C.; Wynne, B. Taxonomy, Biodiversity and Their Publics in Twenty-First-Century DNA Barcoding. Public Underst. Sci. 2010, 19, 497–512. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).