The Hidden Wood-Decaying Fungal Diversity: Rhizochaete from East Asia
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Herbarium Specimen Preparation
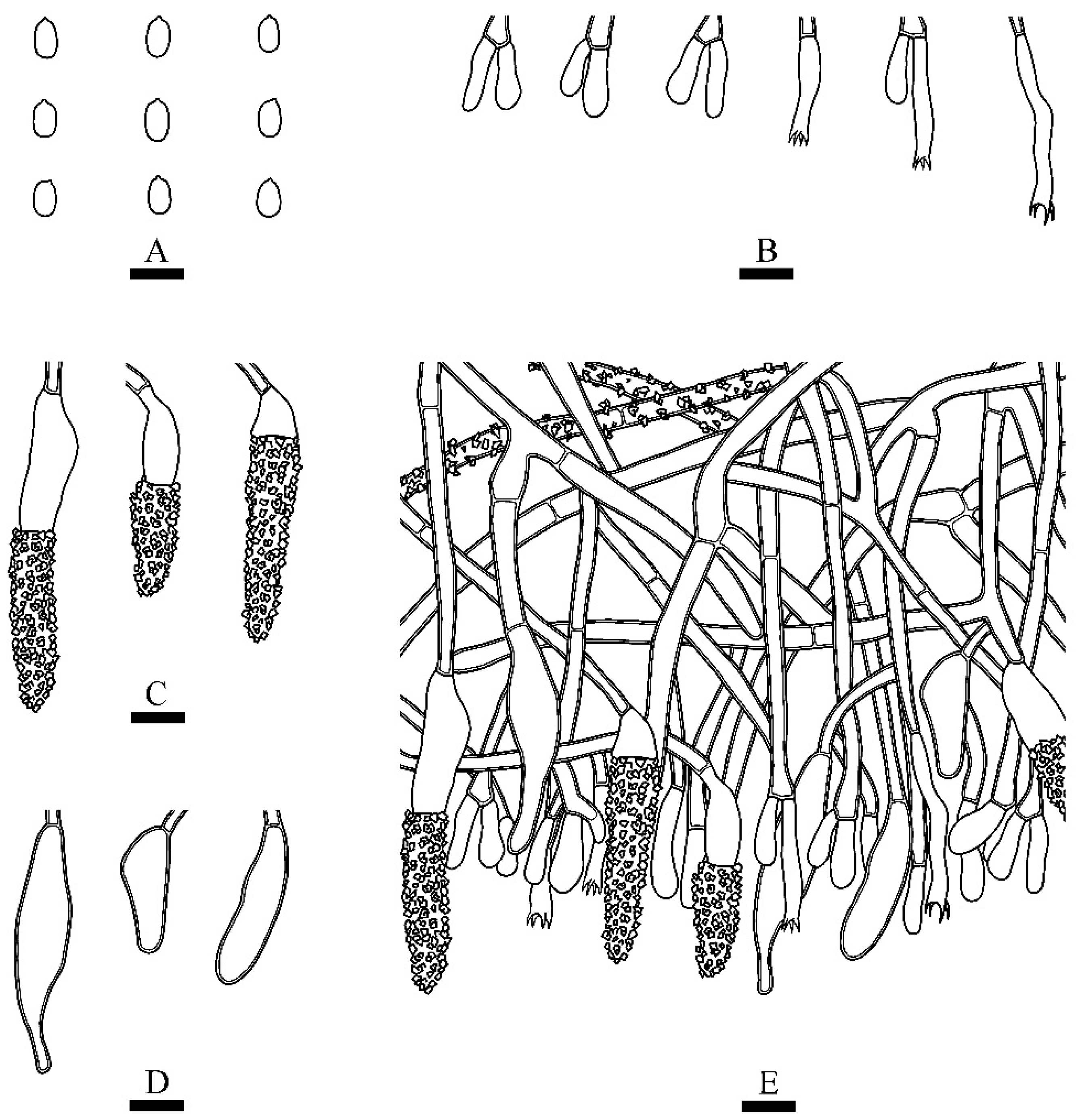
2.2. Morphology
2.3. Molecular Phylogeny
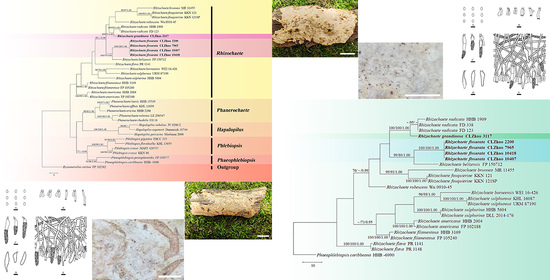
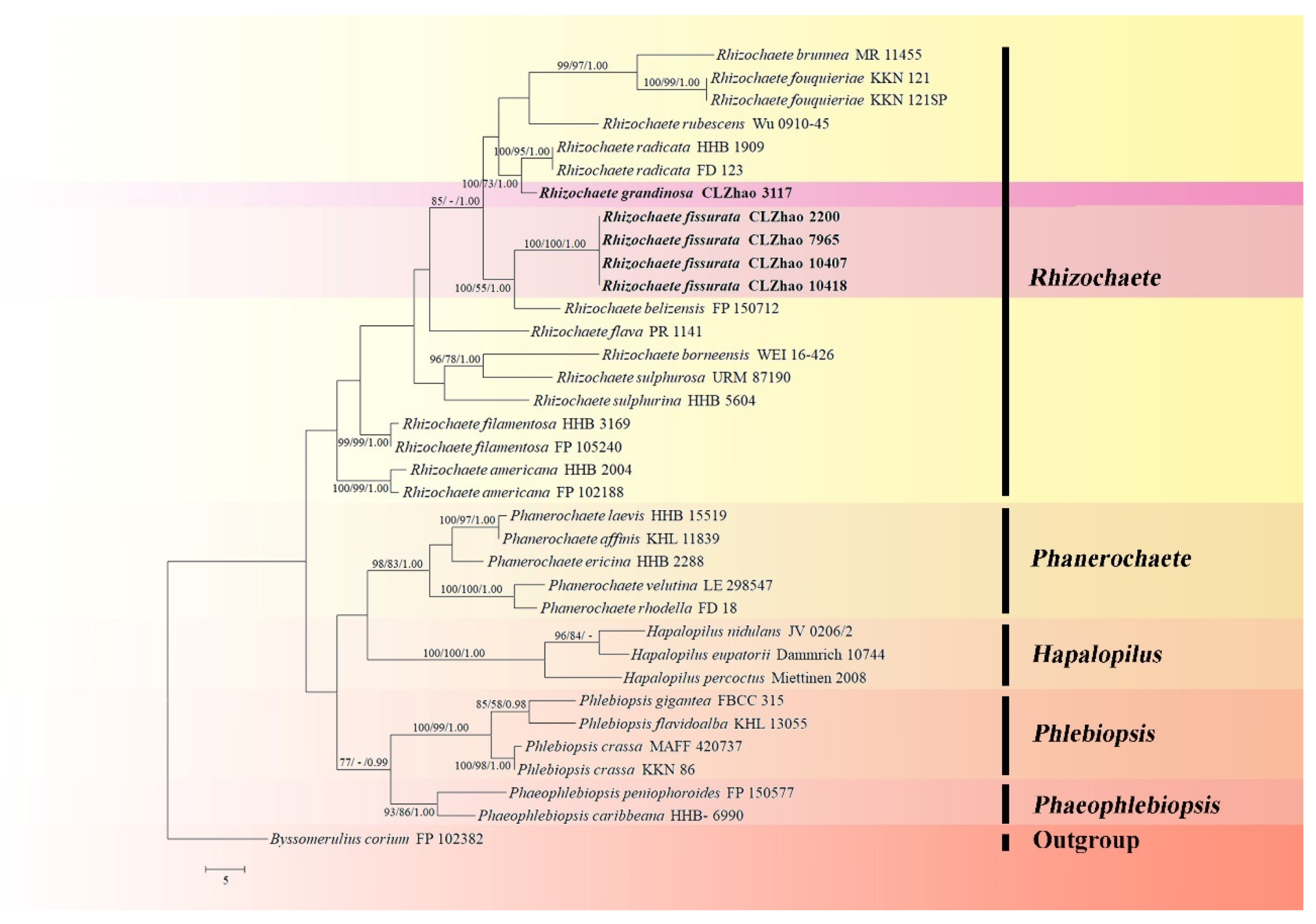
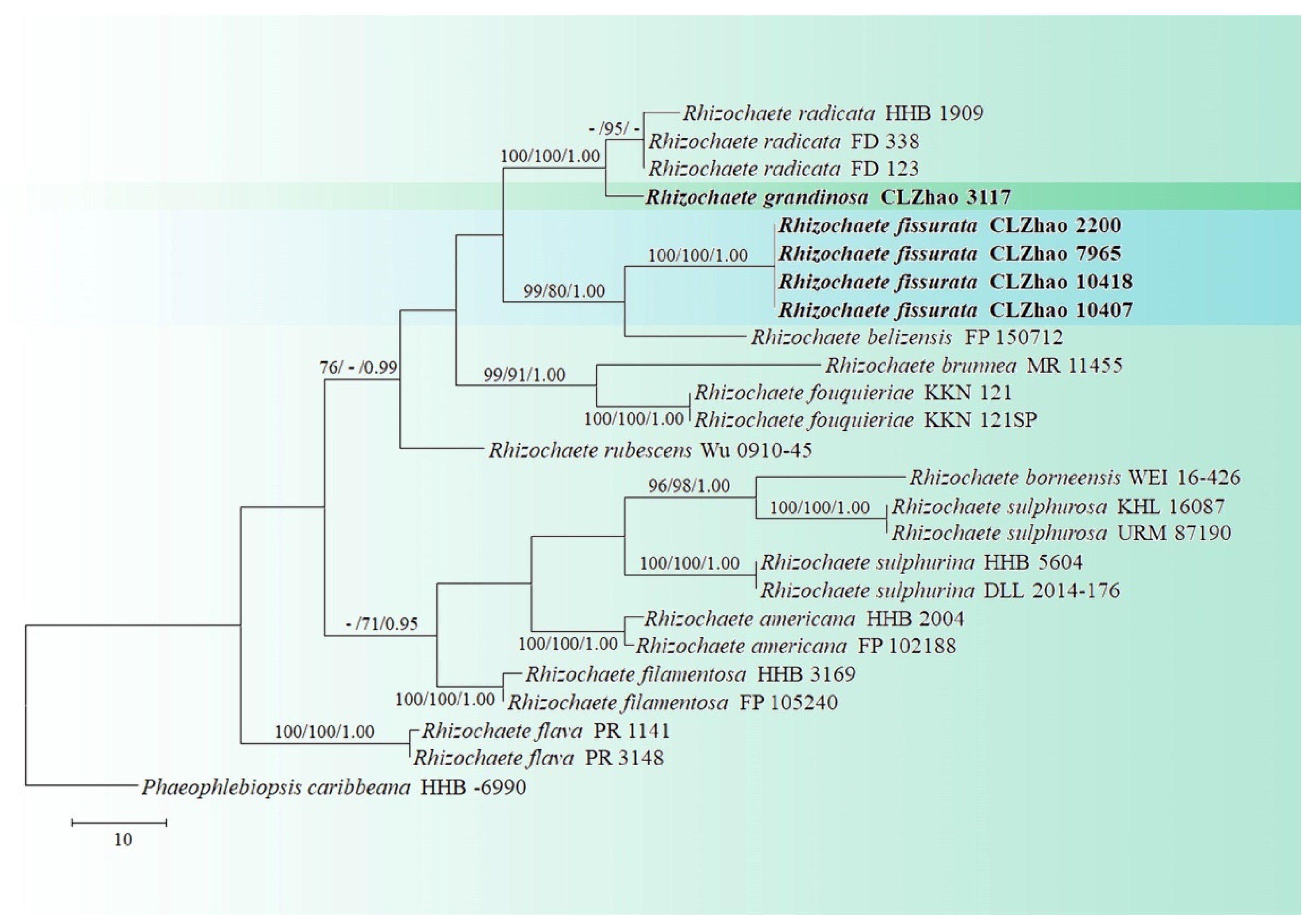
3. Results
3.1. Molecular Phylogeny
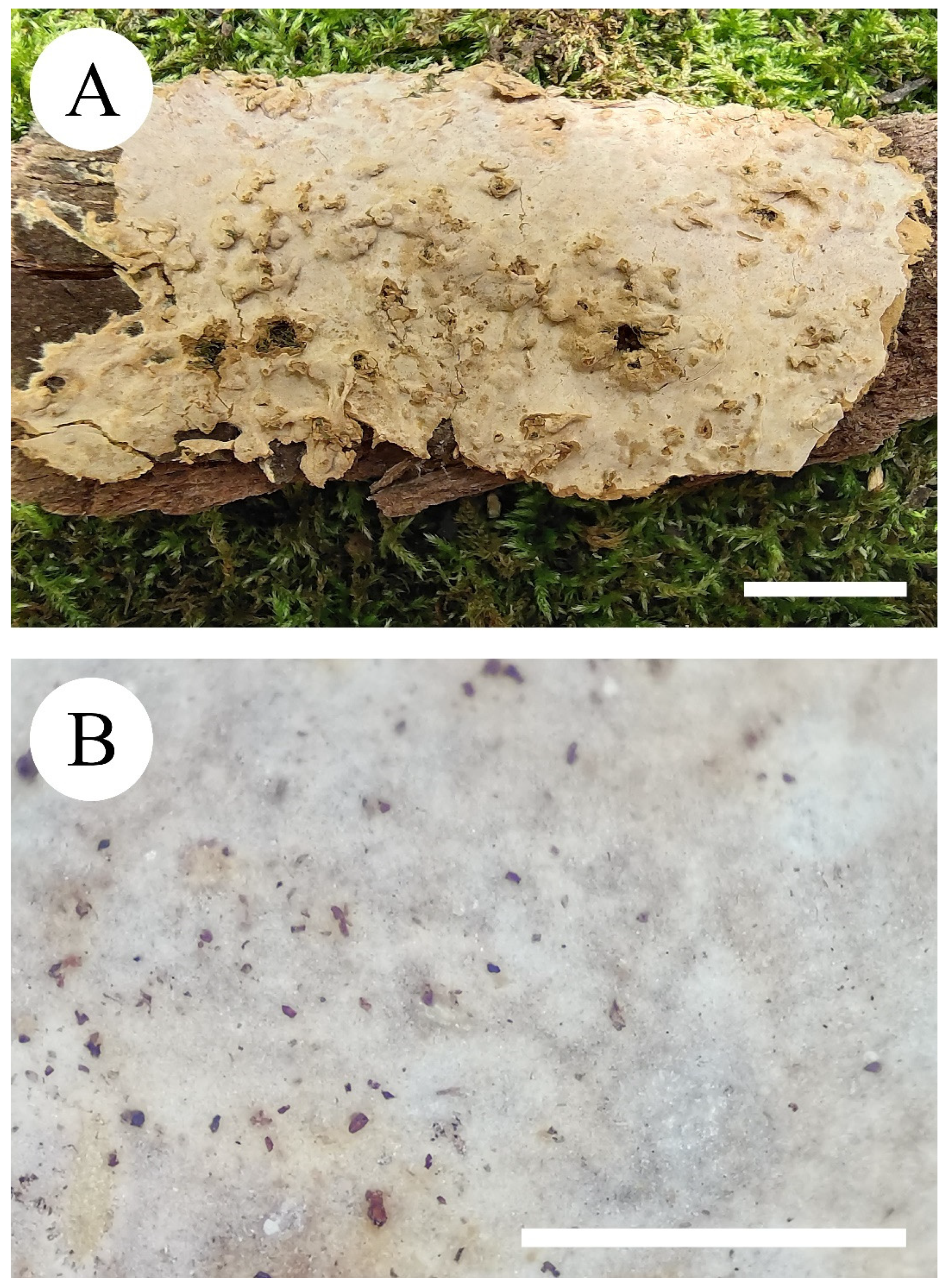
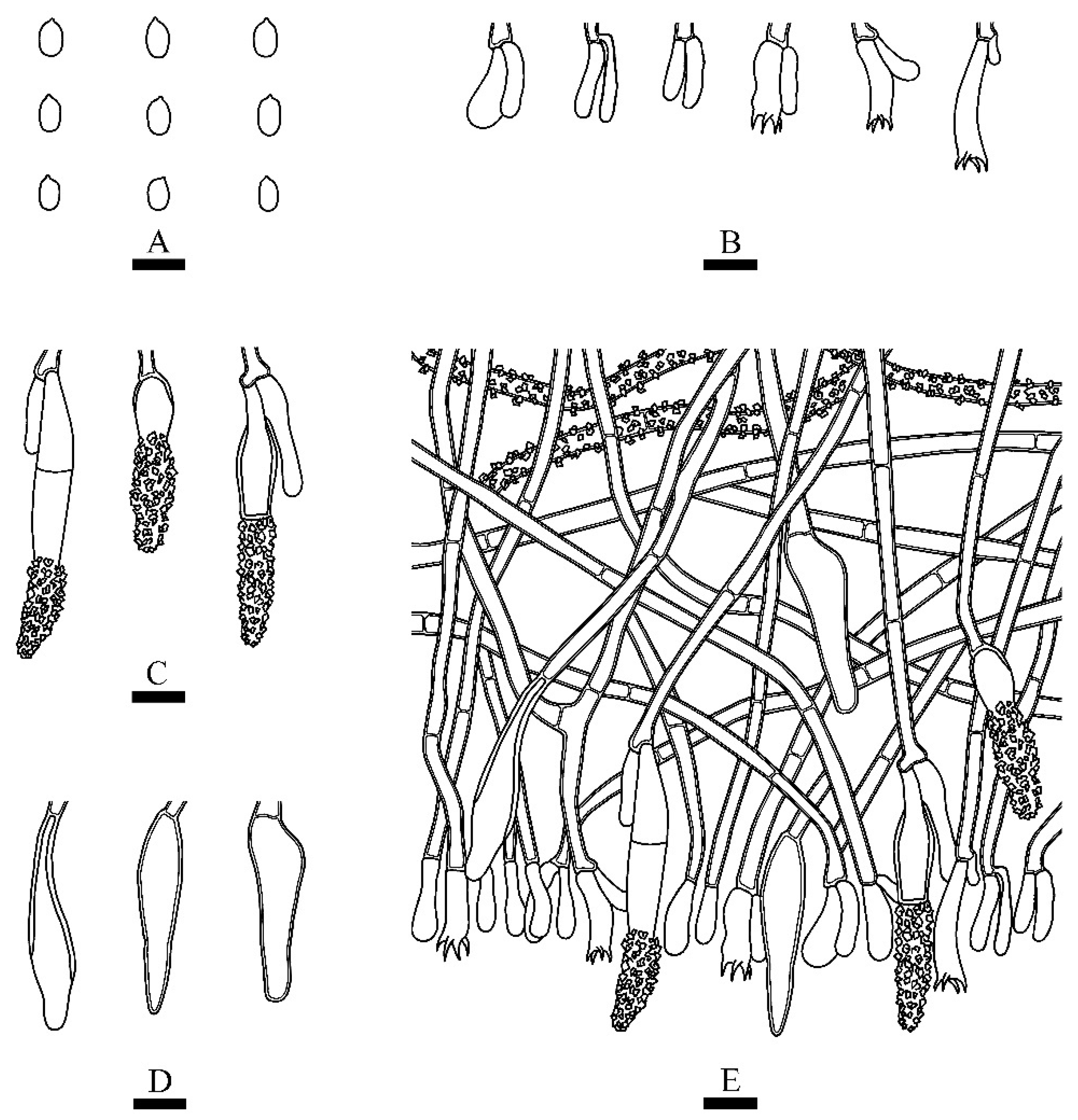
3.2. Taxonomy
4. Discussion
| Key to all species of Rhizochaete worldwide |
| 1. Generative hyphae regularly clamped....................................................................................................................................................................................................2 |
| 1. Generative hyphae primarily simple septa.............................................................................................................................................................................................6 |
| 2. Cystidia absent........................................................................................................................................................................................................................R. violascens |
| 2. Cystidia abundant......................................................................................................................................................................................................................................3 |
| 3. Cystidia up to 250 µm long with thick walls, basidia > 40 µm in length...............................................................................................................................................................................................................................................R. brunnea |
| 3. Cystidia up to 100 µm long with thin walls, basidia < 40 µm in length.............................................................................................................................................4 |
| 4. Basidiospores > 3 µm in width...........................................................................................................................................................................................R. fouquieriae |
| 4. Basidiospores < 3 µm in width................................................................................................................................................................................................................5 |
| 5. Basidiomes olive brown to yellowish brown, cystidia < 60 µm in length........................................................................................................................R. americana |
| 5. Basidiomes bright to dull yellow, cystidia > 60 µm in length..........................................................................................................................................R. sulphurina |
| 6. Cystidia with thin or slightly thickened walls, < 1 µm thick...............................................................................................................................................................7 |
| 6. Cystidia with distinctly thick walls, > 1 µm thick...............................................................................................................................................................................14 |
| 7. Hymenium turning violet or red in KOH..............................................................................................................................................................................................8 |
| 7. Hymenium not reacting or changing to orange or brown in KOH....................................................................................................................................................12 |
| 8. Hymenium turning red in KOH..............................................................................................................................................................................................................9 |
| 8. Hymenium turning violet in KOH........................................................................................................................................................................................................10 |
| 9. Subiculum brown..................................................................................................................................................................................................................R. filamentosa |
| 9. Subiculum colorless.................................................................................................................................................................................................................R. rubescens |
| 10. Subiculum yellow................................................................................................................................................................................................................R. sulphurosa |
| 10. Subiculum colorless...............................................................................................................................................................................................................................11 |
| 11. Hymenial surface smooth, cracking......................................................................................................................................................................................R. fissurata |
| 11. Hymenial surface grandinioid, not cracking....................................................................................................................................................................R. grandinosa |
| 12. Basidiomes bright yellow, unchanged in KOH.................................................................................................................................................................R. percitrina |
| 12. Basidiomes yellow to brownish orange, darkening in KOH............................................................................................................................................................13 |
| 13. Basidia > 30 µm in length.................................................................................................................................................................................R. rhizomorphosulphurea |
| 13. Basidia < 30 µm in length.............................................................................................................................................................................................................R. flava |
| 14. Cystidia < 50 µm in length..................................................................................................................................................................................................R. borneensis |
| 14. Cystidia > 50 µm in length...................................................................................................................................................................................................................15 |
| 15. Subiculum mustard yellow to brown, cystidia > 60 µm in length, basidiospores > 4 µm in length..............................................................................R. radicata |
| 15. Subiculum yellow, cystidia < 60 µm in length, basidiospores < 4 µm in length............................................................................................................R. belizensis |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hibbett, D.; Abarenkov, A.; Kõljalg, U.; Öpik, M.; Chai, B.; Cole, J.; Crous, P.; Robert, V.; Helgason, T.; Herr, J.R.; et al. Sequence-based classification and identification of Fungi. Mycologia 2016, 108, 1049–1068. [Google Scholar] [CrossRef]
- Hawksworth, D. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Blackwell, M. The fungi: 1, 2, 3…5.1 million species? Botany 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D. Global species numbers of fungi: Are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers. Conserv. 2012, 21, 2425–2433. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, O.; Spirin, V.; Vlasak, J.; Rivoire, B.; Stenroos, S.; Hibbett, D.S. Polypores and genus concepts in Phanerochaetaceae (Polyporales, Basidiomycota). Mycokeys 2016, 17, 1–46. [Google Scholar] [CrossRef]
- James, T.Y.; Stajich, J.E.; Hittinger, C.T.; Rokas, A. Toward a fully resolved fungal tree of life. Annu. Rev. Microbiol. 2020, 74, 291–313. [Google Scholar] [CrossRef]
- Nakasone, K.K.; Draeger, K.R.; Ortiz-Santana, B. A contribution to the taxonomy of Rhizochaete (Polyporales, Basidiomycota). Cryptogam. Mycol. 2017, 38, 81–99. [Google Scholar] [CrossRef]
- Greslebin, A.; Nakasone, K.K.; Rajchenberg, M. Rhizochaete, a new genus of phanerochaetoid fungi. Mycologia 2004, 96, 260–271. [Google Scholar] [CrossRef][Green Version]
- Chikowski, R.S.; Larsson, K.H.; Gibertoni, T.B. Three new combinations in Rhizochaete (Agaricomycetes, Fungi) and a new record to the Brazilian Amazonia. Nova Hedwig. 2016, 102, 185–196. [Google Scholar] [CrossRef]
- Floudas, D.; Hibbett, D.S. Revisiting the taxonomy of Phanerochaete (Polyporales, Basidiomycota) using a four gene dataset and extensive ITS sampling. Fungal Biol. 2015, 119, 679–719. [Google Scholar] [CrossRef]
- Bianchinotti, M.V.; Rajchenberg, M.; Greslebin, A.G. Parenthesome structure of some corticioid fungi. Mycol. Res. 2005, 109, 923–926. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.N.; He, S.H.; Nakasone, K.K.; Kumara, K.L.W.; Chen, C.C.; Liu, S.L.; Ma, H.X.; Huang, M.R. Global phylogeny and taxonomy of the wood-decaying fungal genus Phlebiopsis (Polyporales, Basidiomycota). Front. Microbiol. 2021, 12, 622460. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.H. Farvekort. The Danish Mycological Society’s Colour-Chart; Foreningen til Svampekundskabens Fremme: Greve, Danmark, 1996. [Google Scholar]
- Dai, Y.C. Polypore diversity in China with an annotated checklist of Chinese polypores. Mycoscience 2012, 53, 49–80. [Google Scholar] [CrossRef]
- Zhao, C.L.; Wu, Z.Q. Ceriporiopsis kunmingensis sp. nov. (Polyporales, Basidiomycota) evidenced by morphological characters and phylogenetic analysis. Mycol. Prog. 2017, 16, 93–100. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Bacteriology 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Larsson, K.H. Re-thinking the classification of corticioid fungi. Mycol. Res. 2007, 111, 1040–1063. [Google Scholar] [CrossRef]
- Volobuev, S.; Okun, M.; Ordynets, A.; Spirin, V. The Phanerochaete sordida group (Polyporales, Basidiomycota) in temperate Eurasia, with a note on Phanerochaete pallida. Mycol. Prog. 2015, 14, 80. [Google Scholar] [CrossRef]
- Takano, M.; Abe, H.; Hayashi, N. Extracellular peroxidase activity at the hyphal tips of the white-rot fungus Phanerochaete crassa WD1694. Wood Sci. 2006, 52, 429–435. [Google Scholar] [CrossRef]
- Kuuskeri, J.; Mäkelä, M.R.; Isotalo, J.; Oksanen, L.; Lundell, T. Lignocellulose-converting enzyme activity profiles correlate with molecular systematics and phylogeny grouping in the incoherent genus Phlebia (Polyporales, Basidiomycota). BMC Microbiol. 2015, 15, 217. [Google Scholar] [CrossRef] [PubMed]
- Justo, A.; Miettinen, O.; Floudas, D.; Ortiz-Santana, B.; Sjokvist, E.; Lindner, D.; Nakasone, K.K.; Niemela, T.; Larsson, K.H.; Ryvarden, L.; et al. A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 2017, 121, 798–824. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Wu, S.H.; Chen, C.Y. Hydnophanerochaete and Odontoefibula, two new genera of phanerochaetoid fungi (Polyporales, Basidiomycota) from East Asia. MycoKeys 2018, 39, 75–96. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Felsenstein, J. Confidence intervals on phylogenetics: An approach using bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Science Gateway: Enabling High-Impact Science for Phylogenetics Researchers with Limited Resources. Assoc. Comput. Mach. 2012, 39, 1–8. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2, Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nakasone, K.K.; Bergman, C.R.; Burdsall Jr, H.H. Phanerochaete filamentosa-Corticium radicatum Species Complex in North America. Sydowia 1994, 46, 44–62. [Google Scholar]
- Burdsall Jr, H.H. Taxonomic and distributional notes on Corticiaceae (Homobasidiomycetes, Aphyllophorales) of the southern appalachians. Algae Fungi 1975, 265–286. [Google Scholar]
- Wu, S.H. Nine new species of Phanerochaete from Taiwan. Mycol. Res. 1998, 102, 1126–1132. [Google Scholar] [CrossRef]
- Griffin, E.A.; Harrison, J.G.; McCormick, M.K.; Burghardt, K.T.; Parker, J.D. Tree diversity reduces fungal endophyte richness and diversity in a large-scale temperate forest experiment. Diversity 2019, 11, 234. [Google Scholar] [CrossRef]
- Girometta, C.E.; Bernicchia, A.; Baiguera, R.M.; Bracco, F.; Buratti, S.; Cartabia, M.; Picco, A.M.; Savino, E. An italian research culture collection of wood decay fungi. Diversity 2020, 12, 58. [Google Scholar] [CrossRef]
- Gargano, M.L.; Zervakis, G.I.; Isikhuemhen, O.S.; Venturella, G.; Calvo, R.; Giammanco, A.; Fasciana, T.; Ferraro, V. Ecology, phylogeny, and potential nutritional and medicinal value of a rare white “maitake” collected in a mediterranean Forest. Diversity 2020, 12, 230. [Google Scholar] [CrossRef]
- Polemis, E.; Fryssouli, V.; Daskalopoulos, V.; Zervakis, G.I. Basidiomycetes associated with Alnus glutinosa habitats in Andros Island (Cyclades, Greece). Diversity 2020, 12, 232. [Google Scholar] [CrossRef]
- Ogura-Tsujita, Y.; Tetsuka, K.; Tagane, S.; Kubota, M.; Anan, S.; Yamashita, Y.; Tone, K.; Yukawa, T. Differing life-history strategies of two mycoheterotrophic orchid species associated with leaf litter- and wood-decaying fungi. Diversity 2021, 13, 161. [Google Scholar] [CrossRef]
- Dai, Y.C. A revised checklist of corticioid and hydnoid fungi in China for 2010. Mycoscience 2011, 52, 69–79. [Google Scholar] [CrossRef]

| Species Name | Specimen No. | GenBank Accession No. | References | Country | |
|---|---|---|---|---|---|
| ITS | nLSU | ||||
| Byssomerulius corium | FP 102382 | KP135007 | KP135230 | [11] | USA, Wisconsin |
| Hapalopilus eupatorii | Dammrich 10744 | KX752620 | KX752620 | [6] | Germany |
| H. nidulans | JV 0206/2 | KX752623 | KX752623 | [6] | Sweden |
| H. percoctus | Miettinen 2008 | KX752597 | KX752597 | [6] | Botswana |
| Phanerochaete affinis | KHL 11839 | EU118652 | EU118652 | [20] | Sweden |
| P. ericina | HHB 2288 | KP135167 | KP135247 | [11] | USA, North Carolina |
| P. laevis | HHB 15519 | KP135149 | KP135249 | [11] | USA, Alabama |
| P. rhodella | FD-18 | KP135187 | KP135258 | [11] | USA, Massachusetts |
| P. velutina | LE 298547 | KP994360 | KP994385 | [21] | Russia |
| Phaeophlebiopsis caribbeana | HHB-6990 | KP135415 | KP135243 | [11] | USA, Florida |
| P. peniophoroides | FP 150577 | KP135417 | KP135273 | [11] | USA, Hawaii |
| Phlebiopsis crassa | KKN 86 | KP135394 | KP135215 | [11] | USA, Arizona |
| P. crassa | MAFF 420737 | AB809163 | AB809163 | [22] | Japan |
| P. flavidoalba | KHL 13055 | EU118662 | EU118662 | [20] | Costa Rica |
| P. gigantea | FBCC 315 | LN611131 | LN611131 | [23] | Sweden |
| Rhizochaete americana | FP-102188 | KP135409 | KP135277 | [11] | USA, Illinois |
| R. americana | HHB 2004 | AY219391 | AY219391 | [9] | USA, Georgia |
| R. belizensis | FP 150712 | KP135408 | KP135280 | [11] | Belize |
| R. borneensis | WEI 16-426 | MZ637070 | MZ637270 | Unpublished | China |
| R. brunnea | MR 11455 | AY219389 | AY219389 | [9] | Argentina |
| R. filamentosa | FP 105240 | KP135411 | AY219393 | [8] | USA, Indiana |
| R. filamentosa | HHB 3169 | KP135410 | KP135278 | [11] | USA, Maryland |
| R. fissurata | CLZhao 2200 | MZ713640 | MZ713844 | Present study | China |
| R. fissurata | CLZhao 7965 | MZ713641 | MZ713845 | Present study | China |
| R. fissurata | CLZhao 10407 | MZ713642 | MZ713846 | Present study | China |
| R. fissurata | CLZhao 10418 | MZ713643 | MZ713847 | Present study | China |
| R. flava | PR 1141 | KY273030 | KY273033 | [8] | Puerto Rico |
| R. flava | PR 3148 | KY273029 | - | [8] | Puerto Rico |
| R. fouquieriae | KKN 121 | AY219390 | GU187608 | [8] | USA, Arizona |
| R. fouquieriae | KKN-121-sp | KY948786 | KY948858 | [24] | United States |
| R. grandinosa | CLZhao 3117 | MZ713644 | MZ713848 | Present study | China |
| R. radicata | FD 123 | KP135407 | KP135279 | [11] | USA, Massachusetts |
| R. radicata | FD 338 | KP135406 | - | [11] | USA, Massachusetts |
| R. radicata | HHB 1909 | AY219392 | AY219392 | [9] | USA, North Carolina |
| R. rubescens | Wu 0910-45 | LC387335 | MF110294 | [25] | China |
| R. sulphurina | DLL 2014-176 | KY273032 | - | [8] | USA, Idaho |
| R. sulphurina | HHB 5604 | KY273031 | GU187610 | [8] | USA, Montana |
| R. sulphurosa | KHL 16087 | KT003523 | - | [10] | Brazil |
| R. sulphurosa | URM 87190 | KT003522 | KT003519 | [10] | Brazil |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, Z.-R.; Zhao, C.-L. The Hidden Wood-Decaying Fungal Diversity: Rhizochaete from East Asia. Diversity 2021, 13, 503. https://doi.org/10.3390/d13100503
Gu Z-R, Zhao C-L. The Hidden Wood-Decaying Fungal Diversity: Rhizochaete from East Asia. Diversity. 2021; 13(10):503. https://doi.org/10.3390/d13100503
Chicago/Turabian StyleGu, Zi-Rui, and Chang-Lin Zhao. 2021. "The Hidden Wood-Decaying Fungal Diversity: Rhizochaete from East Asia" Diversity 13, no. 10: 503. https://doi.org/10.3390/d13100503
APA StyleGu, Z.-R., & Zhao, C.-L. (2021). The Hidden Wood-Decaying Fungal Diversity: Rhizochaete from East Asia. Diversity, 13(10), 503. https://doi.org/10.3390/d13100503