Discovery of Three Novel Cytospora Species in Thailand and Their Antagonistic Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Isolation of Fungi
2.2. Morphological Observation
2.3. DNA Extraction, PCR Amplification and Sequencing
2.4. Phylogenetic Analyses
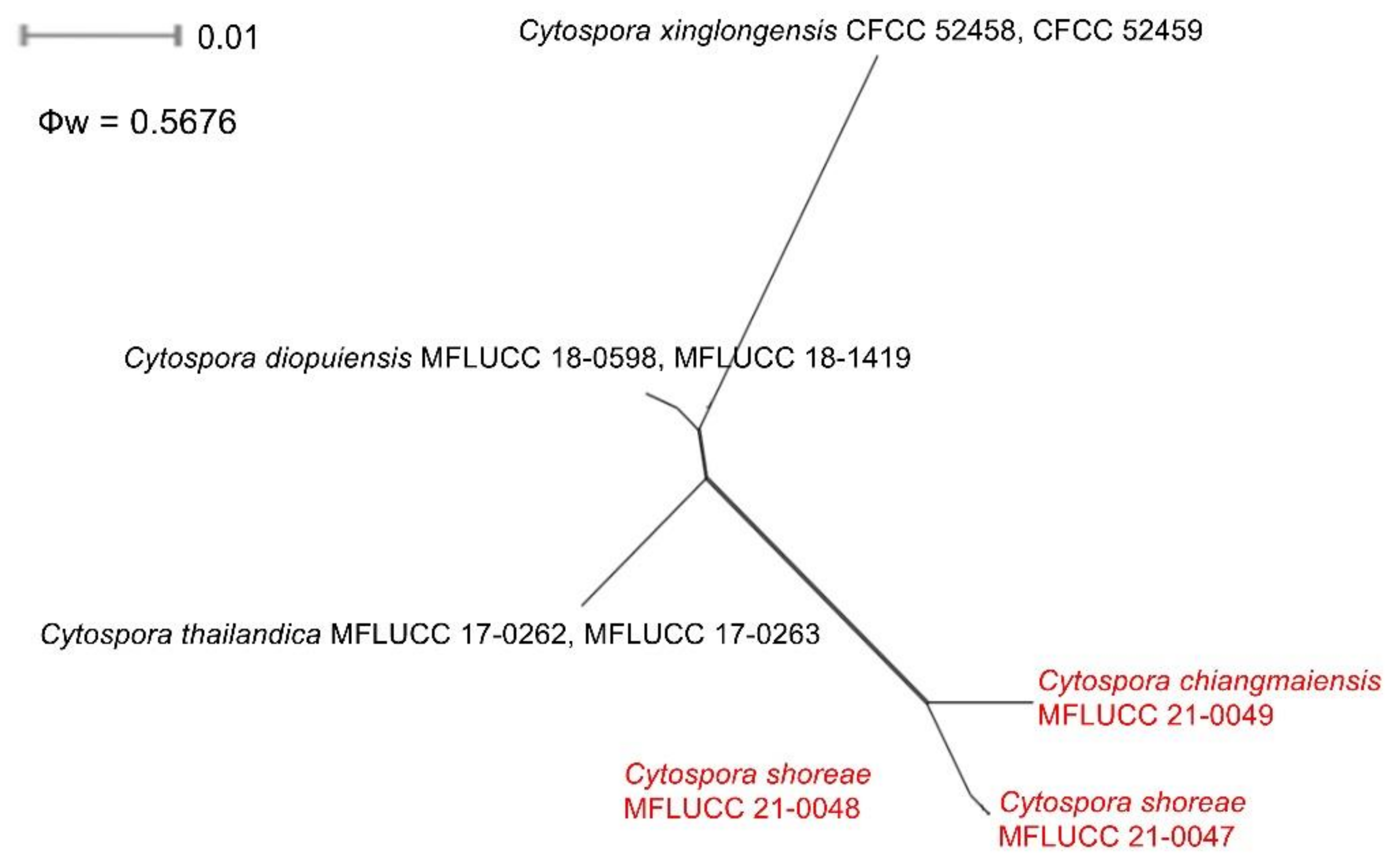
2.5. Genealogical Concordance Phylogenetic Species Recognition Analysis
2.6. Preliminary Screening of Antagonistic Activity against Fungal Pathogens
3. Results
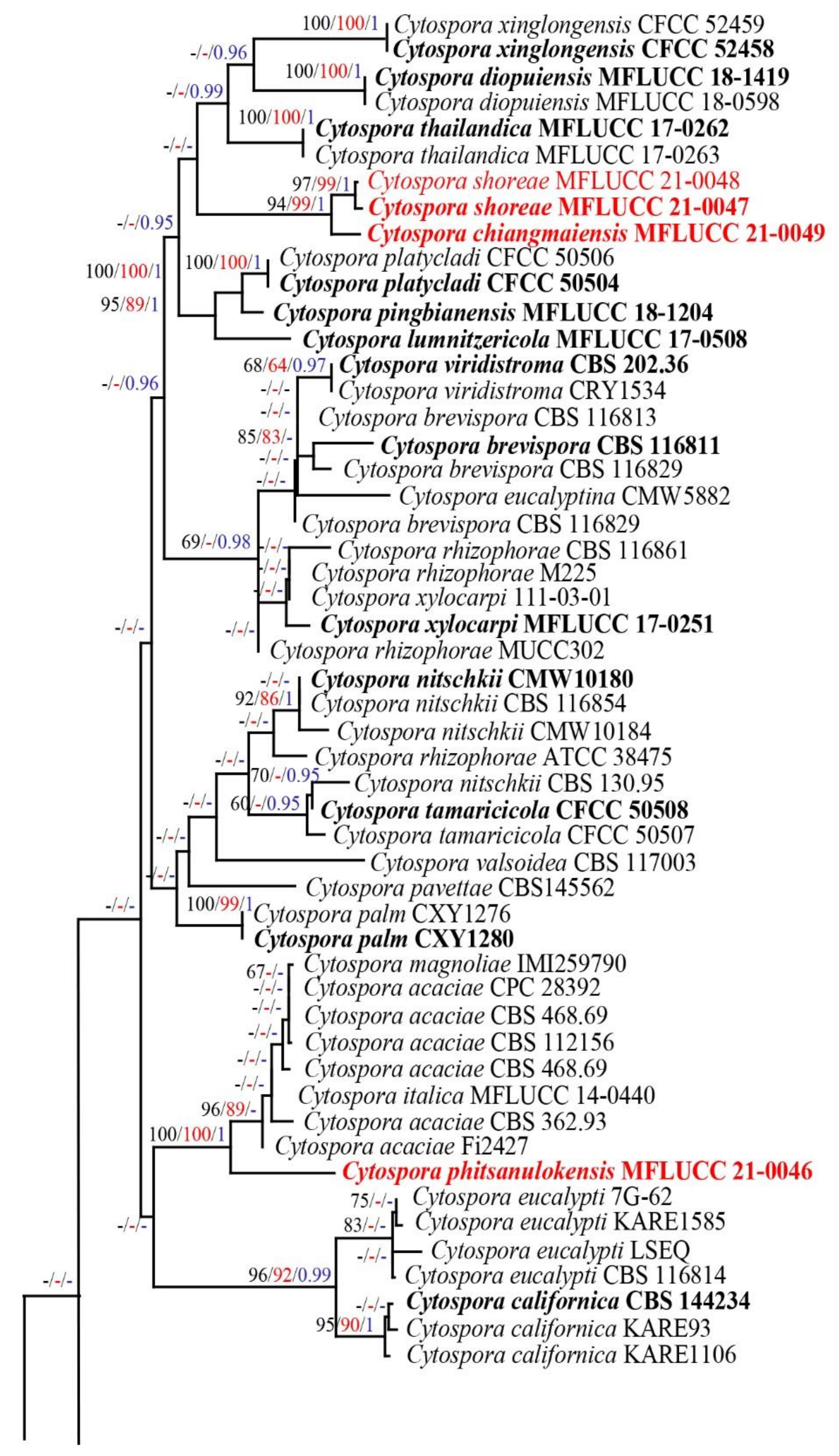
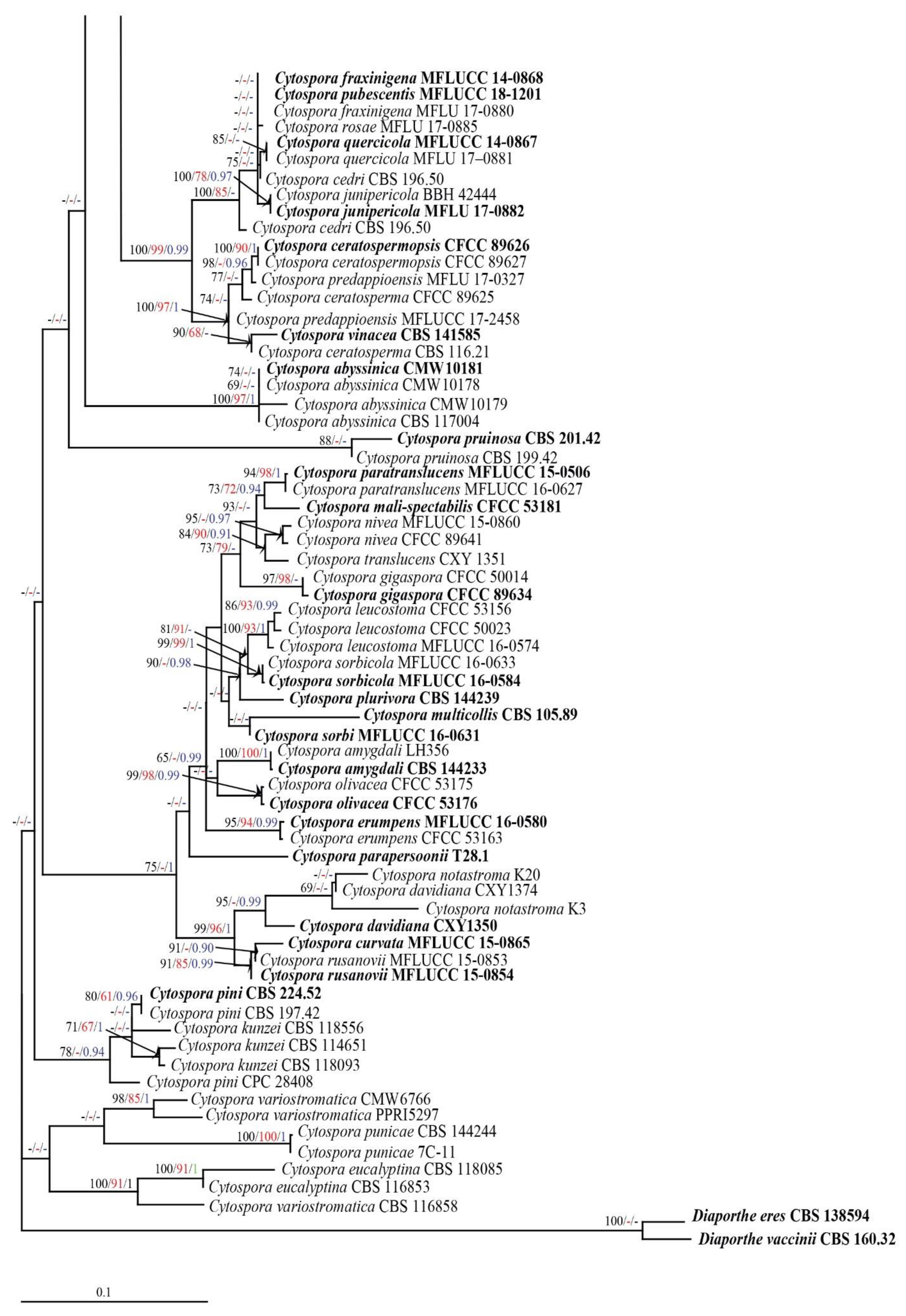
3.1. Phylogenetic Analyses
3.2. Taxonomy
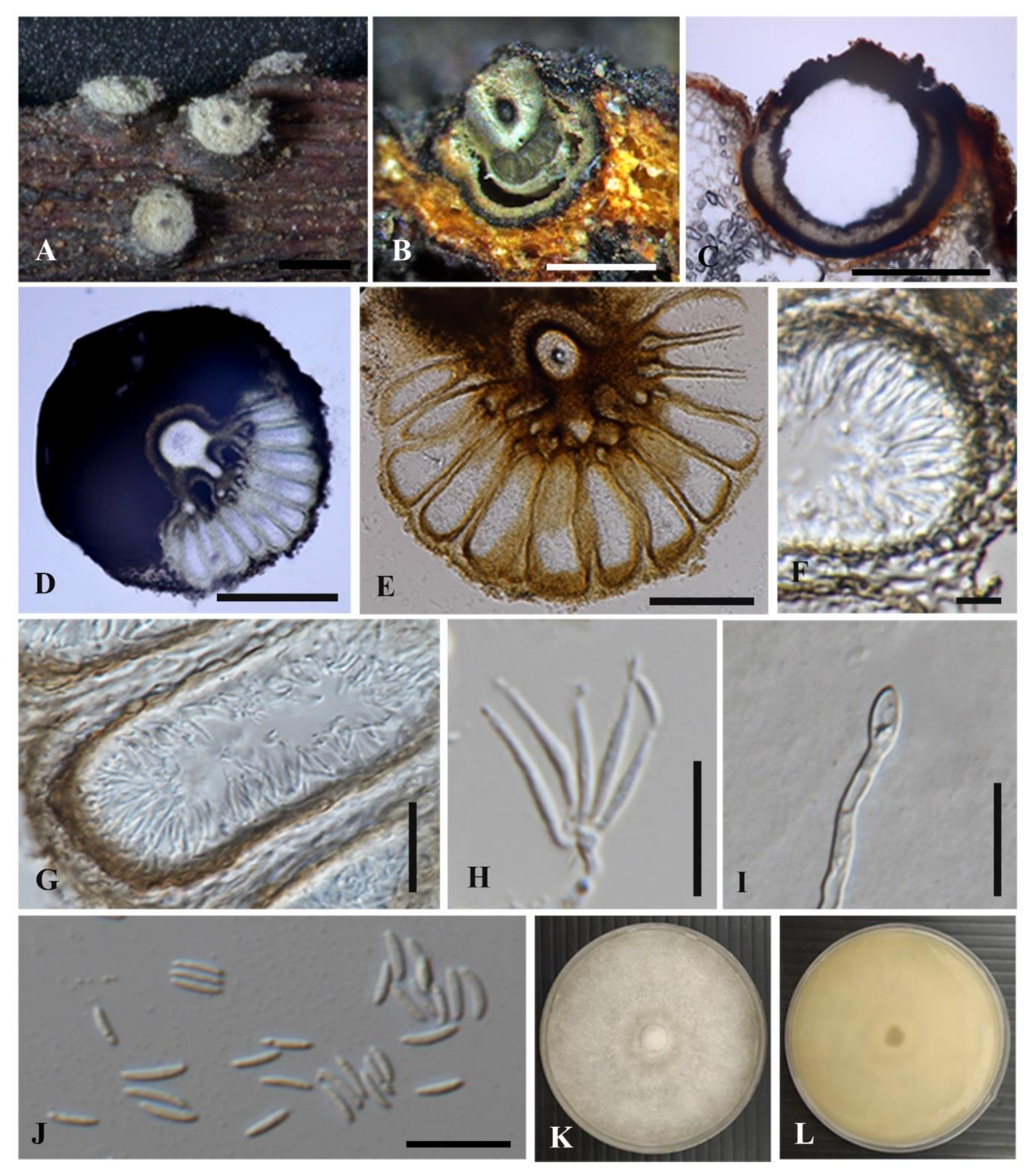
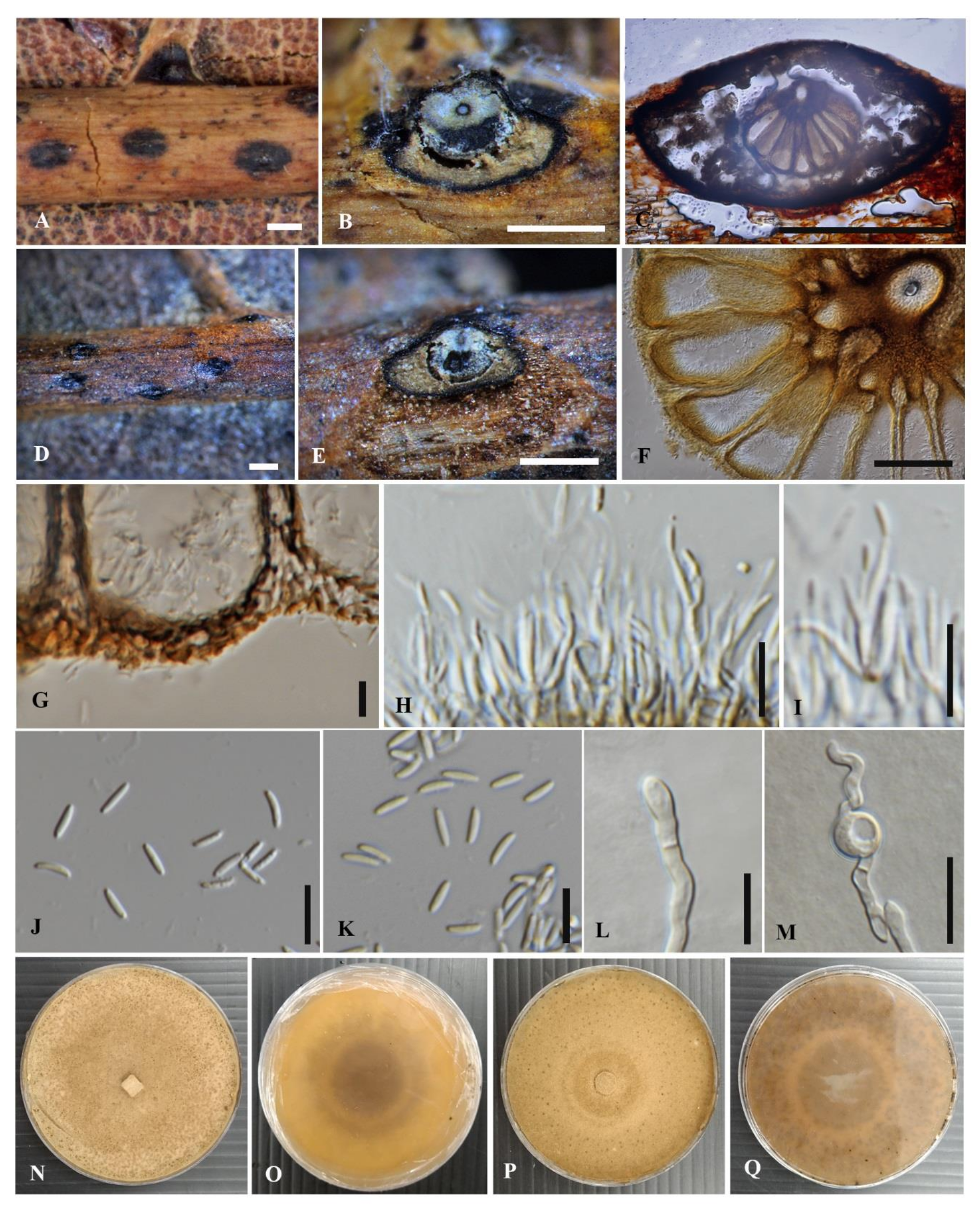
3.2.1. Cytospora chiangmaiensis Monkai and K.D. Hyde, sp. nov.
3.2.2. Cytospora shoreae Monkai and K.D. Hyde, sp. nov.
3.2.3. Cytospora phitsanulokensis Monkai and K.D. Hyde, sp. nov.
3.3. Preliminary Screening of Antagonistic Activity against Fungal Pathogens
4. Discussion
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, X.; Bezerra, J.D.P.; Tian, C.; Crous, P. Cytospora (Diaporthales) in China. Persoonia-Mol. Phylogeny Evol. Fungi 2020, 45, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.J.; Hyde, K.D.; Camporesi, E.; Maharachchikumbura, S.S.N.; Norphanphoun, C.; Brooks, S.; Liu, J.K. Additions to the genus Cytospora with sexual morph in Cytosporaceae. Mycosphere 2020, 11, 189–224. [Google Scholar] [CrossRef]
- Zhu, H.; Pan, M.; Bezerra, J.D.P.; Tian, C.; Fan, X. Discovery of Cytospora species associated with canker disease of tree hosts from Mount Dongling of China. MycoKeys 2020, 62, 97–121. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Bhat, J.D.; Dayarathne, M.C.; Huang, S.-K.; Norphanphoun, C.; Senanayake, I.C.; Perera, R.H.; et al. Families of Sordariomycetes. Fungal Divers. 2016, 79, 1–317. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Raspé, O.; Jeewon, R.; Wen, T.-C.; Hyde, K.D. Morphological and phylogenetic characterisation of novel Cytospora species associated with mangroves. MycoKeys 2018, 38, 93–120. [Google Scholar] [CrossRef]
- Ehrenberg, C.G. Sylvae Mycologicae Berolinenses; Formis Teophili Bruschcke: Berlin, Germany, 1818; pp. 1–32. [Google Scholar]
- Donk, M.A. Nomina conservanda proposita I. Propos. Fungi. Deuteromycetes Regnum Veg. 1964, 34, 7–15. [Google Scholar]
- Rossman, A.Y.; Crous, P.W.; Hyde, K.D.; Hawksworth, D.L.; Aptroot, A.; Bezerra, J.L.; Bhat, J.D.; Boehm, E.; Braun, U.; Boonmee, S.; et al. Recommended names for pleomorphic genera in Dothideomycetes. IMA Fungus 2015, 6, 507–523. [Google Scholar] [CrossRef]
- Norphanphoun, C.; Doilom, M.; Daranagama, D.A.; Phookamsak, R.; Wen, T.C.; Bulgakov, T.S.; Hyde, K.D. Revisiting the genus Cytospora and allied species. Mycosphere 2017, 8, 51–97. [Google Scholar] [CrossRef]
- McNeill, J.; Barrie, F.R.; Buck, W.R.; Demoulin, V.; Greuter, W.; Hawksworths, D.L.; Herendeen, P.S.; Knapp, S.; Marhold, K.; Prado, J.; et al. International code of nomenclature for algae, fungi and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. Regnum Veg. 2012, 154, 1–140. [Google Scholar]
- Adams, G.C.; Wingfield, M.J.; Common, R.; Roux, J. Phylogenetic relationships and morphology of Cytospora species and related teleomorphs (Ascomycota, Diaporthales, Valsaceae) from Eucalyptus. Stud. Mycol. 2005, 52, 1–144. [Google Scholar]
- Wang, X.; Wei, J.; Huang, L.; Kang, Z. Re-evaluation of pathogens causing Valsa canker on apple in China. Mycologia 2011, 103, 317–324. [Google Scholar] [CrossRef]
- Fan, X.; Hyde, K.D.; Liu, M.; Liang, Y.; Tian, C. Cytospora species associated with walnut canker disease in China, with description of a new species C. gigalocus. Fungal Biol. 2015, 119, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Hongsanan, S.; Jeewon, R.; Bhat, D.J.; McKenzie, E.H.C.; Jones, E.B.G.; Phookamsak, R.; Ariyawansa, H.; Boonmee, S.; Zhao, Q.; et al. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016, 80, 1–270. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Holland, L.A.; Nouri, M.T.; Travadon, R.; Abramians, A.; Michailides, T.J.; Trouillas, F.P. Molecular phylogeny of Cytospora species associated with canker diseases of fruit and nut crops in California, with the descriptions of ten new species and one new combination. IMA Fungus 2018, 9, 333–369. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Yang, Q.; Fan, X.-L.; Tian, C.-M. Identification of six Cytospora species on Chinese chestnut in China. MycoKeys 2020, 62, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Pan, M.; Zhu, H.; Bonthond, G.; Tian, C.; Fan, X. High diversity of Cytospora associated with canker and dieback of Rosaceae in China, with 10 new species described. Front. Plant. Sci. 2020, 11, 690. [Google Scholar] [CrossRef]
- Pan, M.; Zhu, H.; Tian, C.; Huang, M.; Fan, X. Assessment of Cytospora isolates from conifer cankers in china, with the descriptions of four new Cytospora species. Front. Plant. Sci. 2021, 12. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org (accessed on 1 August 2021).
- Hyde, K.D.; Norphanphoun, C.; Maharachchikumbura, S.S.N.; Bhat, D.J.; Jones, E.B.G.; Bundhun, D.; Chen, Y.J.; Bao, D.F.; Boonmee, S.; Calabon, M.S.; et al. Refined families of Sordariomycetes. Mycosphere 2020, 11, 305–1059. [Google Scholar] [CrossRef]
- Senanayake, I.; Crous, P.; Groenewald, J.; Maharachchikumbura, S.; Jeewon, R.; Phillips, A.; Bhat, J.; Perera, R.; Li, Q.; Li, W.; et al. Families of Diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 2017, 86, 217–296. [Google Scholar] [CrossRef]
- Adams, G.C.; Roux, J.; Wingfield, M.J. Cytospora species (Ascomycota, Diaporthales, Valsaceae): Introduced and native pathogens of trees in South Africa. Australas. Plant. Pathol. 2006, 35, 521–548. [Google Scholar] [CrossRef]
- Wang, Y.-L.; Lu, Q.; Decock, C.; Li, Y.-X.; Zhang, X.-Y. Cytospora species from Populus and Salix in China with C. davidiana sp. nov. Fungal Biol. 2015, 119, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.F.; Wagenaar, M.M.; Singh, M.P.; Janso, J.E.; Clardy, J. The Cytosporones, new octaketide antibiotics isolated from an endophytic fungus. Org. Lett. 2000, 2, 4043–4046. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.P.; Janso, J.E.; Brady, S.F. Cytoskyrins and cytosporones produced by Cytospora sp. CR200: Taxonomy, fermentation and biological activities. Mar. Drugs 2007, 5, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Sadorn, K.; Saepua, S.; Boonyuen, N.; Boonruangprapa, T.; Rachtawee, P.; Pittayakhajonwut, P. Antimicrobial activity and cytotoxicity of xanthoquinodin analogs from the fungus Cytospora eugeniae BCC42696. Phytochemistry 2018, 151, 99–109. [Google Scholar] [CrossRef]
- Deng, Q.; Li, G.; Sun, M.; Yang, X.; Xu, J. A new antimicrobial sesquiterpene isolated from endophytic fungus Cytospora sp. from the Chinese mangrove plant Ceriops tagal. Nat. Prod. Res. 2018, 34, 1404–1408. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.-C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The faces of fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, S.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guid. to Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef] [Green Version]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; IEEE: New Orleans, LA, USA, 2010; pp. 1–8. [Google Scholar]
- Swofford, D.L. PAUP* Phylogenetic Analysis Using Parsimony * (and Other methods); Version 4.0; Sinauer Associates: Sunderland, UK, 2002. [Google Scholar]
- Nylander, J.A. MrModeltest 2. Program Distributed by the Author. Department of Systematic Zoology; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Zhaxybayeva, O.; Gogarten, J.P. Bootstrap, Bayesian probability and maximum likelihood mapping: Exploring new tools for comparative genome analyses. BMC Genomic. 2002, 3, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hoehna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rambaut, A.; Drummond, A. Tracer v1.4. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 3 August 2021).
- Rambaut, A.; Drummond, A.J. FigTree: Tree Figure Drawing Tool; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, Scotland, 2012. [Google Scholar]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef] [Green Version]
- Quaedvlieg, W.; Binder, M.; Groenewald, J.; Summerell, B.; Carnegie, A.; Burgess, T.; Crous, P. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia-Mol. Phylogeny Evol. Fungi 2014, 33, 1–40. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H. SplitsTree: Analyzing and visualizing evolutionary data. Bioinformatics 1998, 14, 68–73. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2005, 23, 254–267. [Google Scholar] [CrossRef]
- Hamzah, T.N.T.; Lee, S.Y.; Hidayat, A.; Terhem, R.; Faridah-Hanum, I.; Mohamed, R. Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Front. Microbiol. 2018, 9, 1707. [Google Scholar] [CrossRef] [PubMed]
- Jeewon, R.; Hyde, K.D. Establishing species boundaries and new taxa among fungi: Recommendations to resolve taxonomic ambiguities. Mycosphere 2016, 7, 1669–1677. [Google Scholar] [CrossRef]
- Thambugala, K.M.; Daranagama, D.A.; Phillips, A.; Bulgakov, T.; Bhat, D.J.; Camporesi, E.; Bahkali, A.H.; Eungwanichayapant, P.D.; Liu, Z.-Y.; Hyde, K.D. Microfungi on Tamarix. Fungal Divers 2016, 82, 239–306. [Google Scholar] [CrossRef]
- Kepley, J.B.; Reeves, F.B.; Jacobi, W.R.; Adams, G.C. Species associated with Cytospora canker on Populus tremuloides. Mycotaxon 2015, 130, 783–805. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. Available online: https://nt.ars-grin.gov/fungaldatabases/ (accessed on 10 August 2021).
- Wei, C.; Deng, Q.; Sun, M.; Xu, J. Cytospyrone and cytospomarin: Two new polyketides isolated from mangrove endophytic fungus, Cytospora sp. Molecules 2020, 25, 4224. [Google Scholar] [CrossRef] [PubMed]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Doilom, M.; Guo, J.-W.; Phookamsak, R.; Mortimer, P.E.; Karunarathna, S.C.; Dong, W.; Liao, C.-F.; Yan, K.; Pem, D.; Suwannarach, N.; et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China: Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front. Microbiol. 2020, 11, 585215. [Google Scholar] [CrossRef]



| Genes (Primer Pair) | References | PCR Condition |
|---|---|---|
| ITS (ITS5/ITS4) | [30] | An initial denaturation step of 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 50 s at 55 °C and 90 s at 72 °C, and a final extension step of 10 min at 72 °C |
| LSU (LROR/LR5) | [31] | |
| TEF1-α (728F/986R) | [33] | |
| TUB2 (Bt2a/Bt2b) | [34] | |
| ACT (512F/783R) | [33] | An initial denaturation step of 5 min at 96 °C, followed by 35 cycles of 40 s at 95 °C, 30 s at 58 °C and 1 min at 72 °C, and a final extension step of 5 min at 72 °C |
| RPB2 (fRPB2-5F/fRPB2-7cR) | [32] | An initial denaturation step of 5 min at 95 °C, followed by 40 cycles of 1 min at 95 °C, 1 min at 52 °C and 90 s at 72 °C, and a final extension step of 10 min at 72 °C |
| Species Name | Culture Accession No. | Genbank Accession No. | |||||
|---|---|---|---|---|---|---|---|
| ITS | LSU | ACT | RPB2 | TEF1-α | TUB2 | ||
| Cytospora abyssinica | CMW 10181 | AY347353 | N/A | N/A | N/A | N/A | N/A |
| C. abyssinica | CMW 10178 | AY347354 | N/A | N/A | N/A | N/A | N/A |
| C. abyssinica | CMW 10179 | AY347352 | N/A | N/A | N/A | N/A | N/A |
| C. abyssinica | CBS 117004 | KY051833 | KX965299 | KX964737 | KX965495 | KX965098 | N/A |
| C. acaciae | CBS 468.69 | DQ243804 | N/A | N/A | N/A | N/A | N/A |
| C. acaciae | Fi2427 | MG253918 | N/A | N/A | N/A | N/A | N/A |
| C. acaciae | CPC 28392 | KY051980 | KX965438 | KX964858 | KX965579 | KX965220 | KX965027 |
| C. acaciae | CBS 468.69 | KY051937 | KX965394 | KX964817 | N/A | KX965181 | KX964990 |
| C. acaciae | CBS 362.93 | KY051929 | KX965386 | KX964811 | KX965547 | KX965173 | KX964983 |
| C. acaciae | CBS 112156 | KY051776 | KX965241 | KX964688 | N/A | N/A | KX964888 |
| C. amygdali | CBS 144233 | MG971853 | N/A | MG972002 | N/A | MG971659 | N/A |
| C. amygdali | LH356 | MG971852 | N/A | MG972001 | N/A | MG971658 | N/A |
| C. brevispora | CBS 116829 | AF192321 | N/A | N/A | N/A | N/A | N/A |
| C. brevispora | CBS 116811 | AF192315 | N/A | N/A | N/A | N/A | N/A |
| C. brevispora | CBS 116829 | KY051803 | KX965267 | KX964709 | KX965477 | KX965073 | KX964909 |
| C. brevispora | CBS 116813 | KY051788 | KX965252 | KX964698 | KX965470 | KX965061 | KX964898 |
| C. californica | CBS 144234 | MG971935 | N/A | MG972083 | N/A | MG971645 | N/A |
| C. californica | KARE1106 | MG971948 | N/A | MG972094 | N/A | MG971647 | N/A |
| C. californica | KARE93 | MG971930 | N/A | MG972079 | N/A | MG971640 | N/A |
| C. cedri | CBS 196.50 | AF192311 | N/A | N/A | N/A | N/A | N/A |
| C. cedri | CBS 196.50 | KY051905 | KX965364 | KX964790 | KX965534 | KX965153 | N/A |
| C. ceratosperma | CBS 116.21 | AY347335 | N/A | N/A | N/A | N/A | N/A |
| C. ceratosperma | CFCC 89625 | KR045646 | KR045725 | N/A | KU710977 | KP310861 | KR045687 |
| C. ceratospermopsis | CFCC 89626 | KR045647 | KR045726 | KU711011 | KU710978 | KU710934 | KR045688 |
| C. ceratospermopsis | CFCC 89627 | KR045648 | KR045727 | KU711012 | KU710979 | KU710935 | KR045689 |
| C. chiangmaiensis | MFLUCC 21-0049 | MZ356514 | MZ356518 | MZ451157 | MZ451165 | MZ451161 | MZ451169 |
| C. curvata | MFLUCC 15-0865 | KY417728 | N/A | KY417694 | N/A | N/A | N/A |
| C. davidiana | CXY 1350 | KM034870 | N/A | N/A | N/A | N/A | KM034902 |
| C. davidiana | CXY 1374 | KM034869 | N/A | N/A | N/A | N/A | KM034901 |
| C. diopuiensis | MFLUCC 18-1419 | MK912137 | MK571765 | MN685819 | N/A | N/A | N/A |
| C. diopuiensis | MFLUCC 18-0598 | MT215491 | MT215540 | N/A | MT212203 | N/A | N/A |
| C. erumpens | MFLUCC 16-0580 | KY417733 | KY417767 | KY417699 | KY417801 | N/A | N/A |
| C. erumpens | CFCC 53163 | MK673059 | MK673089 | MK673029 | MK673000 | MK672948 | MK672975 |
| C. eucalypti | LSEQ | AY347340 | N/A | N/A | N/A | N/A | N/A |
| C. eucalypti | CBS 144241 | MG971907 | N/A | MG972056 | N/A | MG971617 | N/A |
| C. eucalypti | 7G-62 | MG971910 | N/A | MG972060 | N/A | MG971620 | N/A |
| C. eucalypti | CBS 116814 | KY051789 | KX965253 | KX964699 | N/A | KX965062 | KX964899 |
| C. eucalyptina | CMW 5882 | AY347375 | N/A | N/A | N/A | N/A | N/A |
| C. eucalyptina | CBS 118085 | KY051848 | KX965315 | N/A | N/A | N/A | KX964935 |
| C. eucalyptina | CBS 116853 | KY051822 | KX965288 | KX964727 | N/A | N/A | KX964918 |
| C. fraxinigena | MFLU 17–0880 | MF190134 | MF190079 | N/A | N/A | N/A | N/A |
| C. fraxinigena | MFLUCC 14-0868 | MF190133 | MF190078 | N/A | N/A | N/A | N/A |
| C. gigaspora | CFCC 50014 | KR045630 | KR045710 | KU710999 | KU710959 | KU710922 | KR045671 |
| C. gigaspora | CFCC 89634 | KF765671 | KF765687 | KU711000 | KU710960 | KU710923 | KR045672 |
| C. italica | MFLUCC 14–0440 | KU900329 | KU900301 | N/A | N/A | N/A | N/A |
| C. junipericola | BBH 42444 | MF190126 | MF190071 | N/A | N/A | MF377579 | N/A |
| C. junipericola | MFLU 17-0882 | MF190125 | MF190072 | N/A | N/A | MF377580 | N/A |
| C. kunzei | CBS 118556 | DQ243791 | N/A | N/A | N/A | N/A | N/A |
| C. kunzei | CBS 118093 | KY051855 | KX965322 | KX964754 | N/A | KX965116 | KX964941 |
| C. kunzei | CBS 114651 | KY051780 | KX965243 | KX964691 | N/A | KX965055 | KX964890 |
| C. leucostoma | CFCC 50023 | KR045635 | KR045715 | KU711003 | KU710964 | KU710926 | KR045676 |
| C. leucostoma | CFCC 53156 | MN854447 | MN854658 | MN850762 | MN850748 | MN850755 | MN861117 |
| C. leucostoma | MFLUCC 16-0574 | KY417731 | KY417764 | KY417696 | KY417798 | N/A | N/A |
| C. lumnitzericola | MFLUCC 17-0508 | MG975778 | MH253453 | MH253457 | MH253461 | N/A | N/A |
| C. magnoliae | IMI259790 | JX438623 | N/A | N/A | N/A | JX438565 | N/A |
| C. mali-spectabilis | CFCC 53181 | MK673066 | MK673096 | MK673036 | MK673006 | MK672953 | MK672982 |
| C. multicollis | CBS 105.89 | DQ243803 | N/A | N/A | N/A | N/A | N/A |
| C. nitschkii | CMW 10180 | AY347356 | N/A | N/A | N/A | N/A | N/A |
| C. nitschkii | CMW 10184 | AY347355 | N/A | N/A | N/A | N/A | N/A |
| C. nitschkii | CBS 130.95 | KY051879 | KX965344 | N/A | N/A | N/A | N/A |
| C. nitschkii | CBS 116854 | MH863003 | MH874559 | KX964728 | KX965487 | KX965090 | N/A |
| C. nivea | MFLUCC 15-0860 | KY417737 | KY417771 | KY417703 | KY417805 | N/A | N/A |
| C. nivea | CFCC 89641 | KF765683 | KF765699 | KU711006 | KU710967 | KU710929 | KR045679 |
| C. notastroma | K3 | JX438631 | N/A | N/A | N/A | JX438539 | N/A |
| C. notastroma | K20 | JX438629 | N/A | N/A | N/A | JX438541 | N/A |
| C. olivacea | CFCC 53175 | MK673062 | MK673092 | MK673032 | MK673003 | N/A | MK672978 |
| C. olivacea | CFCC 53176 | MK673068 | MK673098 | MK673038 | MK673008 | MK672955 | MK672984 |
| C. palm | CXY 1276 | JN402990 | N/A | N/A | N/A | KJ781296 | N/A |
| C. palm | CXY 1280 | JN411939 | N/A | N/A | N/A | KJ781297 | N/A |
| C. parapersoonii | T28.1 | AF191181 | N/A | N/A | N/A | JX438545 | N/A |
| C. paratranslucens | MFLUCC 15-0506 | KY417741 | KY417775 | KY417707 | KY417809 | N/A | N/A |
| C. paratranslucens | MFLUCC 16-0627 | KY417742 | KY417776 | KY417708 | KY417810 | N/A | N/A |
| C. pavettae | CBS 145562 | MK876386 | MK876427 | MK876457 | MK876483 | MK876497 | MK876503 |
| C. phitsanulokensis | MFLUCC 21-0046 | MZ356517 | MZ356521 | MZ451160 | MZ451168 | MZ451164 | MZ451172 |
| C. pingbianensis | MFLUCC 18-1204 | MK912135 | MK571763 | MN685817 | MN685826 | N/A | N/A |
| C. pini | CBS 197.42 | AY347332 | N/A | N/A | N/A | N/A | N/A |
| C. pini | CBS 224.52 | AY347316 | N/A | N/A | N/A | N/A | N/A |
| C. pini | CPC 28408 | KY051994 | KX965452 | KX964872 | KX965591 | N/A | KX965041 |
| C. platycladi | CFCC 50504 | MH933645 | MH933679 | MH933552 | MH933610 | MH933516 | MH933581 |
| C. platycladi | CFCC 50506 | MH933647 | MH933681 | MH933554 | MH933612 | MH933518 | MH933583 |
| C. plurivora | CBS 144239 | MG971861 | N/A | MG972010 | N/A | MG971572 | N/A |
| C. predappioensis | MFLU 17-0327 | MH253451 | MH253452 | MH253449 | MH253450 | N/A | N/A |
| C. predappioensis | MFLUCC 17-2458 | MG873484 | MG873480 | N/A | N/A | N/A | N/A |
| C. pruinosa | CBS 201.42 | DQ243801 | N/A | N/A | N/A | N/A | N/A |
| C. pruinosa | CBS 199.42 | KY051911 | KX965369 | KX964795 | N/A | KX965159 | KX964968 |
| C. pubescentis | MFLUCC 18-1201 | MK912130 | MK571758 | MN685812 | MN685821 | N/A | N/A |
| C. punicae | CBS 144244 | MG971943 | N/A | MG972091 | N/A | MG971654 | N/A |
| C. punicae | 7C-11 | MG971942 | N/A | N/A | N/A | MG971653 | N/A |
| C. quercicola | MFLU 17–0881 | MF190128 | MF190074 | N/A | N/A | N/A | N/A |
| C. quercicola | MFLUCC 14-0867 | MF190129 | MF190073 | N/A | N/A | N/A | N/A |
| C. rhizophorae | MUCC302 | EU301057 | N/A | N/A | N/A | N/A | N/A |
| C. rhizophorae | M225 | KR056292 | N/A | N/A | N/A | N/A | N/A |
| C. rhizophorae | ATCC 38475 | DQ996040 | N/A | N/A | N/A | N/A | N/A |
| C. rhizophorae | CBS 116861 | N/A | KX965296 | KX964735 | N/A | N/A | N/A |
| C. rosae | MFLU 17-0885 | MF190131 | MF190076 | N/A | N/A | N/A | N/A |
| C. rusanovii | MFLUCC 15-0853 | KY417743 | KY417777 | KY417709 | KY417811 | N/A | N/A |
| C. rusanovii | MFLUCC 15-0854 | KY417744 | KY417778 | KY417710 | KY417812 | N/A | N/A |
| C. shoreae | MFLUCC 21-0047 | MZ356515 | MZ356519 | MZ451158 | MZ451166 | MZ451162 | MZ451170 |
| C. shoreae | MFLUCC 21-0048 | MZ356516 | MZ356520 | MZ451159 | MZ451167 | MZ451163 | MZ451171 |
| C. sorbi | MFLUCC 16-0631 | KY417752 | KY417786 | KY417718 | KY417820 | N/A | N/A |
| C. sorbicola | MFLUCC 16-0584 | KY417755 | KY417789 | KY417721 | KY417823 | N/A | N/A |
| C. sorbicola | MFLUCC 16-0633 | KY417758 | KY417792 | KY417724 | KY417826 | N/A | N/A |
| C. tamaricicola | CFCC 50507 | MH933651 | MH933686 | MH933559 | MH933616 | MH933525 | MH933587 |
| C. tamaricicola | CFCC 50508 | MH933652 | MH933687 | MH933560 | MH933617 | MH933523 | MH933588 |
| C. thailandica | MFLUCC 17-0262 | MG975776 | MH253455 | MH253459 | MH253463 | N/A | N/A |
| C. thailandica | MFLUCC 17-0263 | MG975777 | MH253456 | MH253460 | MH253464 | N/A | N/A |
| C. translucens | CXY 1351 | KM034874 | N/A | N/A | N/A | N/A | KM034895 |
| C. valsoidea | CBS 117003 | KY051832 | KX965298 | N/A | KX965494 | KX965097 | N/A |
| C. variostromatica | CMW 6766 | AY347366 | N/A | N/A | N/A | N/A | N/A |
| C. variostromatica | PPRI5297 | AF260264 | N/A | N/A | N/A | N/A | N/A |
| C. variostromatica | CBS 116858 | KY051828 | KX965293 | KX964732 | N/A | N/A | KX964921 |
| C. vinacea | CBS 141585 | KX256256 | N/A | N/A | N/A | KX256277 | KX256235 |
| C. viridistroma | CBS 202.36 | MN172408 | MN172388 | N/A | N/A | MN271853 | N/A |
| C. viridistroma | CRY 1534 | AF452120 | N/A | N/A | N/A | N/A | N/A |
| C. xinglongensis | CFCC 52458 | MK432622 | MK429892 | MK442946 | MK578082 | N/A | N/A |
| C. xinglongensis | CFCC 52459 | MK432623 | MK429893 | MK442947 | MK578083 | N/A | N/A |
| C. xylocarpi | MFLUCC 17-0251 | MG975775 | MH253454 | MH253458 | MH253462 | N/A | N/A |
| C. xylocarpi | 111_03_01 | MT507847 | N/A | N/A | N/A | N/A | N/A |
| Diaporthe eres | CBS 138594 | KJ210529 | N/A | KJ420760 | N/A | KJ210550 | KJ420799 |
| D. vaccinii | CBS 160.32 | KC343228 | N/A | JQ807297 | N/A | KC343954 | KC344196 |
| Species | Conidiomata (µm) | Conidiogenous Cell (µm) | Conidia (µm) | References |
|---|---|---|---|---|
| Cytospora chiangmaiensis | 650–800 | 8.5–12.2 × 1–1.7 | 5–7 × 0.9–1.7 | This study |
| C. italica | 580–730 | 10–32 × 0.7–2.4 | 3.7–5.2 × 1–1.3 | [51] |
| C. lumnitzericola | Undetermined | 8–14 × 0.6–1.6 | 3.7–4.5 × 1–1.5 | [5] |
| C. platycladi | 210–330 | 5–12 × 1–1.5 | 4–5.5 × 1–1.5 | [1] |
| C. shoreae | 400–1000 | 6.7–11.8 × 1–1.9 | 5–7 × 1–1.6 | This study |
| C. thailandica | 400–1200 | 3.3–9.1 × 1–1.7 | 3.3–4 × 1–1.5 | [5] |
| C. xinglongensis | Undetermined | 4.5–12 × 1–1.5 | 7.5–10.5 × 1–1.5 | [16] |
| New Taxa/Strains | New Taxa/Strains Compared with | Number of Different Nucleotides/Number of All Nucleotides (% Base Pairs Difference) | |||||
|---|---|---|---|---|---|---|---|
| ITS | LSU | ACT | RPB2 | TEF1-α | TUB2 | ||
| Cytospora shoreae (MFLUCC 21-0047) | C. shoreae (MFLUCC 21-0048) | 0/494 (0%) | 0/510 (0%) | 0/180 (0%) | 4/726 (0.6%) | 0/291 (0%) | 3/375 (0.8%) |
| C. chiangmaiensis (MFLUCC 21-0049) | C.shoreae (MFLUCC 21-0047) | 0/494 (0%) | 0/510 (0%) | 5/180 (2.8%) | 13/726 (1.8%) | 16/291 (5.5%) | 13/375 (3.5%) |
| C. chiangmaiensis (MFLUCC 21-0049) | C. shoreae (MFLUCC 21-0048) | 0/494 (0%) | 0/510 (0%) | 5/180 (2.8%) | 11/726 (1.5%) | 16/291 (5.5%) | 12/375 (3.2%) |
| Fungal Name | % Growth Inhibition | |||
|---|---|---|---|---|
| Cytospora chiangmaiensis (MFLUCC 21-0049) | C. shoreae (MFLUCC 21-0047) | C. shoreaeb (MFLUCC 21-0048) | C. phitsanulokensis (MFLUCC 21-0046) | |
| Colletotrichum fructicola (MFLUCC 18-1160) | 44.2 ± 2.5 a | 49.6 ± 0.0 a | 50.5 ± 5.7 a | 37.4 ± 1.5 a |
| Co. siamense (MFLUCC 18-1162) | 18.1 ± 4.4 a | 31.9 ± 0.4 b | 53.5 ± 1.8 c | 39.5 ± 2.2 b |
| Co. artocarpicola (MFLUCC 18-1167) | 11.7 ± 3.9 a | 14.9 ± 5.5 a | 32.3 ± 3.6 b | 58.0 ± 1.0 c |
| Co. viniferum (MFLUCC 18-1179) | 30.0 ± 3.9 a | 64.0 ± 1.5 b | 61.8 ± 1.4b | 75.1 ± 4.0 b |
| Fusarium sambucinum (MFLUCC 17-1056) | 17.4 ± 3.2 a | 39.1 ± 4.2 b | 33.1 ± 4.4 b | 67.5 ± 1.3c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monkai, J.; Tibpromma, S.; Manowong, A.; Mapook, A.; Norphanphoun, C.; Hyde, K.D.; Promputtha, I. Discovery of Three Novel Cytospora Species in Thailand and Their Antagonistic Potential. Diversity 2021, 13, 488. https://doi.org/10.3390/d13100488
Monkai J, Tibpromma S, Manowong A, Mapook A, Norphanphoun C, Hyde KD, Promputtha I. Discovery of Three Novel Cytospora Species in Thailand and Their Antagonistic Potential. Diversity. 2021; 13(10):488. https://doi.org/10.3390/d13100488
Chicago/Turabian StyleMonkai, Jutamart, Saowaluck Tibpromma, Areerat Manowong, Ausana Mapook, Chada Norphanphoun, Kevin D. Hyde, and Itthayakorn Promputtha. 2021. "Discovery of Three Novel Cytospora Species in Thailand and Their Antagonistic Potential" Diversity 13, no. 10: 488. https://doi.org/10.3390/d13100488
APA StyleMonkai, J., Tibpromma, S., Manowong, A., Mapook, A., Norphanphoun, C., Hyde, K. D., & Promputtha, I. (2021). Discovery of Three Novel Cytospora Species in Thailand and Their Antagonistic Potential. Diversity, 13(10), 488. https://doi.org/10.3390/d13100488








