Abstract
Oregano (Origanum vulgare L.) is a flowering plant that belongs to the mint family (Lamiaceae). It is used as a culinary herb and is often commercialized as a fine powder or a mixture of small fragments of dried leaves, which makes morphological recognition difficult. Like other commercial preparations of drugs and spices, the contamination of oregano mixtures with vegetable matter of lower quality, or the use of generic misleading names, are frequent and stress the need to develop a molecular traceability system to easily, quickly, and cheaply unveil these scams. The DNA-based analytical approach known as cpDNA barcoding is particularly suited for fraud identification in crop plant species (fresh products and food derivatives), and it represents a promising traceability tool as an alternative or complement to traditional detection methods. In the present study, we used a combined approach based on both qualitative and quantitative cpDNA barcoding with end-point and real-time polymerase chain reaction (PCR) analyses to assess the type and degree of contamination in commercial batches of common oregano. In a preliminary qualitative screening, we amplified, cloned, and sequenced a number of universal trnH-psbA- and trnL-barcoded regions, to identify the main contaminants in the samples under investigation. On the basis of these findings, we then developed and validated a species-specific and sequence-targeted method of testing for the quantitative assessment of contaminants, using trnL gene intron assays. Surprisingly, the results obtained in our case study indicated an almost total absence of O. vulgare in the commercial batches analyzed, but a high presence of group I contaminants (Satureja pilosa Velen.), and a moderate presence of group II contaminants (Cistus lanidifer L./Cistus albidus).
1. Introduction
Frequent contaminations of food matrices with vegetable matter of lower quality, combined with the use of generic names used for labeling commercial preparations, stress the need to develop and implement molecular traceability systems for foods of vegetal origin. In fact, besides representing an economic loss, all of these adulterations can be considered to be a potential risk for consumer health. While all major agricultural products that provide approximately 95% of human food energy needs (e.g., rice, wheat, maize, and potato) are widely monitored and well characterized using DNA marker analysis specifically developed for each cultivar, reliable characterization tools for minor crop plants are far from defined [1]. Minor agricultural species include plant varieties that are cultivated for food, pharmaceutical, cosmetic, and ornamental purposes, with a modest production in terms of the cultivated areas and the quantity of the final product. For most of these species, the absence of suitable traceability protocols leads to frequent cases of plant substitution, and accidental or deliberate contamination. There are many documented examples of commercial frauds where minor crops were substituted with related taxa with a higher productivity or biomass, but without the agronomical and nutritional characteristics of the original species/cultivars [2,3,4]. Traceability studies on spices where frauds have been described were conducted on a limited number of species including Tymus spp. [5], Crocus sativus [6], Cinnamomun spp. [7], Salvia spp. [8], and Origanum spp. [9]. For most of these spices, molecular tools used in species identification were based on a DNA barcoding approach, whereas in the specific case of Oregano (Oregano vulgare L.), a RAPD fingerprinting approach was utilized [9].
Oregano is an exemplary case of how food markets and customers could benefit from the implementation of an appropriate molecular traceability approach. In fact, in this specific case, the scenario is complicated by the large heterogeneity of the genus, and by the denomination of different botanical genera under a single generic name, namely Origanum in the Mediterranean countries, and Lippia in the Mexican regions. This fact that has led to a market distinction between Mediterranean oregano and Mexican oregano [10]. From a botanical point of view, oregano is a perennial and shrubby herb belonging to the Lamiaceae family, which typically grows in mountainous rocky soils typical of East Mediterranean areas. Although the genus Origanum is comprised of up to 43 species and 18 hybrids characterized by a high morphological and chemical diversity [11], the most variable and predominant species of the genus is Origanum vulgare L. [12], which is commonly known and commercialized as “oregano” in many European countries. This species is further subdivided into six subspecies: Origanum vulgare L. subsp. glandulosum (Desfontaines) Ietsw., subsp. hirtum (Link) Ietsw., subsp. gracile (Koch) Ietsw., subsp. virens (Hoffmannsegg et Link) Ietsw., subsp. vulgare, and subsp. viride (Boissier) Hayek [12]. One of the considerable morphological characteristics of oregano plants is the presence of numerous glandular hairs on the upper surface of the leaves, which secrete an essential oil that is of main interest from both a nutritional and herbalist point of view. The content of essential oil is more than 2% in most commercial oregano plants, and is mainly characterized by the presence of phenolic monoterpenoids such as carvacrol and thymol [13].
Most Origanum species have been used since ancient times as culinary and medicinal herbs, as ornamental garden plants, and in some cases, in the production of dyes. Currently, oregano is principally used in the food industry for its strong aroma, to flavor meat, vegetables and fish. Amongst all the Origanum species, only O. vulgare, O. onites, O. majorana, and O. dictamnus are considered GRAS (generally recognized as safe). In particular, O. vulgare leaves are GRAS at 320–2.800 ppm [14]. From a nutritional point of view, O. vulgare is a source of vitamins, minerals, and flavonoids, which have been associated with anticarcinogenic properties that could benefit human health [14]. Medicinally, according to the “Dictionary of herbal medicine and medicinal plants” [15] oregano has been used for thousands of years as a stimulant, carminative, expectorant, and tonic, and to cure asthma, coughs, indigestion, rheumatism, toothache, headache, and spider bites.
The quality of an oregano spices is defined using different standards, and based on the European Pharmacopoeia (PhEur), only two species can be commercialized as true oregano: Origanum vulgare L. subsp. hirtum (Link) Ietsw. and Origanum onites L. Within the food market, specifications internationally approved by the American Spice Trade Association (ASTA) and European Spice Association (ESA) define the quality criteria for spices such as oregano, and are limited to the amount and phytochemical profile of the essential oils. Further authentication analyses that are required, such as weight by weight and acid-insoluble ash contents, are not discriminative and are time-consuming when there are large numbers of samples to be analyzed. True oregano contamination is generally perpetrated with species belonging to the same Origanum genera, and with similar essential oil profiles (e.g., Origanum majorana L., Origanum syriacum L., O. vulgare L. subsp. virens (Hoffmanns & Link) Ietsw., Satureja montana L., and Thymus capitatus L.). In other cases, Mediterranean oregano has been described to be sophisticated, with plants having a similar silvery gray color and size of leaves, as is also the case for Rhus coriaria L. and Cistus spp. [16,17]. These plants are added as bulk, cheap material that are concealed to illicitly increase the volume and, subsequently, the producers’ or traders’ income. Whereas essential oil-bearing plants can be detected by routine GC-MS and other chromatographic or spectroscopic techniques [18,19], the detection of nonaromatic contaminants relies almost completely on the manual application of molecular techniques for the authentication and detection of sophistication in commercial plant material [1,20].
In this sense, DNA barcoding can be considered to be a useful molecular tool with regard to its low operating cost, and the ability to reliably distinguish between different botanical species. Moreover, the strong standardization of protocols used worldwide for DNA barcoding makes this technology particularly suitable for the routine analyses that are required by agencies to safeguard food safety and quality [21].
This paper reports on a survey of commercial samples of dried Mediterranean oregano batches collected by a local food company. We implemented a combined approach based on cpDNA barcoding and quantitative polymerase chain reaction (qPCR) analysis for the identification of oregano, and for estimating the level of contamination in oregano stocks. In a preliminary qualitative screening, to identify the main contaminants in our study samples, we amplified, cloned, and sequenced a number of trnH-psbA and trnL amplicons. Based on our preliminary results, we then developed a species-specific targeted assay for real-time PCR quantitative determination of the types and levels of oregano contamination.
2. Materials and Methods
2.1. Plant Material
Representative samples of dried spices labeled as “pure oregano”, properly collected in three large commercial batches (R1, R2, and R3) imported from the Mediterranean area, were provided by an Italian food company to check the authenticity of the traded product. We took advantage of this case study to implement in our laboratory a methodological procedure for assessing the genetic identity of Origanum vulgare L., and for tracing and quantifying non-Origanum herbs, such as Satureja pilosa Velen. and Cistus lanidifer L. (group I and II contaminants, respectively). For each of the commercial batches (including several dozens of kilograms each), a sample was represented by approximately 500 g of small fragments of dried leaves. The operations for collecting these representative samples were carried out by skilled technicians of the company (the Italian importer). Then, for genomic DNA isolation and PCR amplification, in our laboratory, each sample was analyzed using three technical replicates per experiment. So, n = 3 × 3. In particular, for genomic DNA extraction and purification, approximately 50 mg of ground tissue were used for each of the subsamples of the technical replicates. Seeds of O. vulgare, S. pilosa and C. ladanifer that were used as references for the pure oregano and the two main oregano contaminants identified in our study, were kindly provided by the Botanical Garden of the University of Padua, Italy (www.ortobotanicopd.it). Seeds were incubated in Petri dishes on 3 mm wet filter paper for approximately three weeks, and seedlings were subsequently transferred into pots with soil. Fresh leaflets were collected from growing plants and immediately freeze-dried in liquid nitrogen and stored at −80 °C until molecular analysis.
2.2. Nucleic Acid Purification
Total genomic DNA was extracted from plant material using a modified CTAB protocol [22]. Approximately 50 mg of ground tissue for each sample were mixed with 882 µL of extraction buffer (100 mM Tris-HCl pH 8, 20 mM EDTA pH 8, 1.4 M NaCl, 2% w/v CTAB, 2% w/v PVP, H2O) and 18 µL of β-mercaptoethanol, and incubated for 30 min at 60 °C. After the addition of an equal volume of chloroform/isoamyl alcohol (24:1, v/v), the suspension was mixed and centrifuged at 20,000 g for 10 min. The supernatant was carefully removed to a new 1.5 mL tube, and an equal volume of isopropanol was added to the tube. DNA was precipitated by centrifugation (1 h, 20,000 g) and washed twice with 400 µL ethanol (70% v/v). The precipitate was left to dry for 5 min, eluted in 200 µL TE buffer (10 mM Tris-HCl, 1 mM EDTA pH 8.0), and incubated with RNAse at 37 °C for 30 min. An equal volume of chloroform/isoamyl alcohol (24:1, v/v) was added and mixed. A centrifugation step (10 min, 20,000 g) was performed, and the aqueous supernatant was removed to a new 1.5 mL tube. Sodium acetate (3 M, pH 5.2, 1/10 volumes) and ethanol (100%, 2 volumes) were added and carefully mixed. The DNA was precipitated by centrifugation (30 min, 20,000 g), and the supernatant was removed. The precipitated pellet was washed twice with 200 µL ethanol (70% v/v), and after centrifugation (5 min, 20,000 g), the supernatant was removed. The DNA pellet was dried for 5 min and then resuspended in 50 µL of water. The DNA quality was checked by gel electrophoresis (0.8% agarose). Finally, DNA concentration and purity were spectrophotometrically measured at the ratios of 260/280 nm and 260/230 nm using the NanoDrop 2000c (Thermo Fisher ScientificTM, Waltham, MA, USA).
2.3. Barcode Regions Amplification, Cloning and Sequencing
Purified DNA was amplified using universal primers for the intergenic spacer trnH-psbA and trnL genic intron regions (Table S1). Reactions were performed in a final volume (V) of 50 µL, containing 1x Buffer for KOD Hot Start DNA Polymerase (Sigma-Aldrich Corporation, Saint Louis, Missouri, USA), 1.5 mM MgSO4, 0.2 mM dNTPs (Sigma-Aldrich), 100 µg/µL UltraPureTM BSA (Thermo Fisher ScientificTM, Waltham, MA, USA), 0.3 µM forward primer and reverse primer (Invitrogen Corporation, Carlsbad, CA, USA), 0.02 U/µL KOD Hot Star DNA Polymerase (Sigma-Aldrich), 10 ng DNA, and H2O to volume. PCR thermal conditions consisted of the initial denaturation step for 2 min at 95 °C followed by 30 cycles of 20 s at 95 °C, 10 s at 58 °C, and 10 s at 70 °C. A final extension of 7 min at 72 °C terminated the reaction, filling in the protruding ends of the newly synthesized strands. Bands of interest were purified from the agarose gel using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA fragments were cloned into the pCR-Blunt plasmid using the Zero Blunt® PCR Cloning Kit (Invitrogen Corporation, Carlsbad, CA, USA). A total of 36 colonies transformed with the trnL region, and 36 colonies transformed with the trnH-psbA region were selected, grown overnight (O/N) in LB liquid media, plasmid-purified, and sequenced using M13 forward and reverse primers. The obtained sequences were manually verified and edited using Geneious Software [23], and used as queries in a BLASTn search (www.ncbi.nlm.nih.gov/BLAST) [24]. Multiple sequence alignments (MSA) with O. vulgare L. (FR726132), S. pilosa Velen. (KR063642), F. procumbens (Dunal) Gren. & Godr. (FR865097), and A. unedo L. (KU205821) were performed using the MUSCLE alignment tool of the Geneious Suite using default parameters. UPGMA (unweighted pair group method with arithmetic mean) trees with 1000 bootstrap replicates were applied to measure stability of the obtaining branches. This analysis was performed by MEGA7 [25].
2.4. Quantitative PCR (qPCR)
Standard curves for determining the different levels of contamination of oregano with Cistus and Satureja species were prepared as described in Table S2. The Analyses reported in this study were performed using the standard curve quantification method. PCR experiments were performed using StepOnePlus™ PCR (Applied Biosystems, Foster City, CA, USA). The software StepOne Software v.2.3 (Applied Biosystems) was used for defining the standard curves, their dilution factors, and the quantification of the considered target DNA. Each sample was tested in three technical replicates. Negative controls were implemented in each PCR run. The 10 µL PCR reaction volumes included 5 µL KiCqStart® SYBR® Green qPCR ReadyMix™ with ROX™ (Sigma-Aldrich Corporation, Saint Louis, Missouri, USA), 0.6 µL forward primer, 0.6 µL reverse primer, 1 µL DNA, and 2.8 µL H2O. The temperature profile consisted of a holding stage at 95 °C for 20 s, a denaturation step at 95 °C for 3 s, an annealing step at 60 °C for 30 s, an extension step at 95 °C for 15 s, and a melt curve step at 60 °C for 1 min. Species-specific primers for trnL and trnH-psbA regions are reported in Supplementary Table S1.
3. Results and Discussion
3.1. Qualitative Determination of Contaminants in Oregano Samples
Genomic DNA was isolated and purified from sub-portions of the samples provided by the company, with a modified CTAB protocol [22]. For each sample (R1, R2, R3) three extractions were performed from different starting amounts of plant tissues, in order to optimize the main steps of the protocol. A NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific) was used to quantify the total DNA and its contaminations of carbohydrate carryover, residual phenol, residual guanidine, and glycogen (260/280 and 260/230 ratios). Both quantitative and qualitative analyses confirmed that the genomic DNA was of high quality and high molecular weight, i.e., not contaminated and not degraded. Both the trnH-psbA and trnL barcode regions were amplified using genomic DNA purified from dry oregano samples collected from the commercial batches, and they produced amplicons of the expected sizes (450 bp and 550 bp, respectively) (Figure S1). After electrophoresis, PCR products were purified from the agarose gel and subcloned for DNA sequencing. The PCR products obtained by amplification of the trnH-psbA and trnL regions using universal primers and DNA templates from “unknown samples” were potentially composed by a pool of sequences representing the two barcodes in the different species potentially contained in each of the sample (provided that other contaminants rather than oregano were present in the sample). In this sense, cloning of the amplicons into a commercial plasmid vector and sequencing a number of subcloned fragments represents a quick and cost-effective method for a preliminary qualitative screening in order to ascertain the presence of different species in the food matrix under examination.
In our case, we considered 12 colonies for each sample (R1, R2, and R3) for a total of 36 colonies for the trnH-psbA region and 36 colonies for the trnL region. After trimming and quality checking the DNA sequences obtained by Sanger sequencing, a BLAST analysis was performed to check the correspondence between our processed sequences and sequences deposited in GenBank databases. Table 1 reports a list of the best hits obtained by the BLAST search for the trnL barcode. Surprisingly, amongst all sequences analyzed, no sequence showed significant similarity to the oregano trnL and/or trnH-psbA regions. The majority of sequences (28 sequences out of 36) scored a very high sequence identity with the trnL region of S. pilosa Velen./S. montana L., generally known as winter savory or mountain savory (Table 1 and Figure 1).

Table 1.
List of the best hits obtained by BLASTn analysis of trnL gene–intron amplicons.
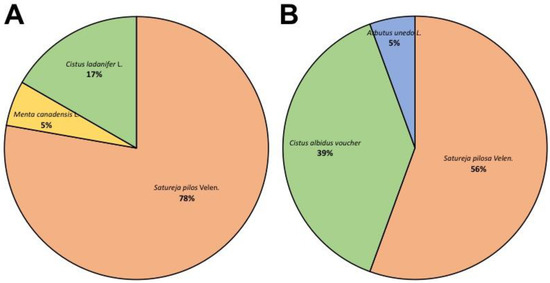
Figure 1.
Pie chart showing the specific proportion of barcode sequences for trnL (A) and trnH-psbA (B) regions in all putative oregano samples considered in this study. Satureja spp. accessions were found to be the most abundant sequences, with a proportion ranging from 56% to 78%.
Winter savory is a perennial, semi-evergreen herb in the family Lamiaceae that is native to the warm temperate regions of southern Europe, the Mediterranean, and Africa. It is one of the oregano Group I contaminants, representing a partially tolerated substitution (in terms of the law) because it belongs to the same family of Origanum and possess a certain commercial value. It is often used in the kitchen and has antiseptic, aromatic, carminative, and digestive properties [9]. However, differences in the composition of the essential oils of this species allow for its discrimination from oregano. Six trnL sequences were found to be related to C. ladanifer L./C. creticus L., with a high percentage of sequence coverage and identity. C. ladanifer is a species of flowering plant belonging to the Cistaceae family. It is native to the western Mediterranean region, and it is commonly known as gum rockrose or laudanum. In contrast to savory, it is considered to be almost free from essential oils, and it therefore cannot be identified by a simple gas chromatographic analysis. Thus, this species belongs to contaminants of Group II and represents a very serious commercial fraud (Table 1 and Figure 1). Finally, two sequences were identified as Mentha canadensis L., which belongs to the Lamiaceae family, like oregano.
A UPGMA tree was constructed as an additional tool to provide graphic representation of the results obtained, using a sequence similarity search (Figure 2, panel A). The phylogenetic tree illustrates the relationships among the sequences obtained from commercial samples and that are related to the trnL reference regions, including O. vulgare L. (AY506614), S. pilosa Velen. (KR063656), C. lanidifer L. (FM179538) and C. creticus L. (EU684550). To confirm that the lack of O. vulgare trnL sequence was due to the inability of trnL universal primers to amplify from this species, we tried to use them with genomic DNA purified from glass-house grown O. vulgare L. plants. Results confirmed that the universal primers worked properly also in this species, and an alignment of sequences obtained gave a 99% identity with the NCBI-deposited O. vulgare trnL sequence (AY506614).
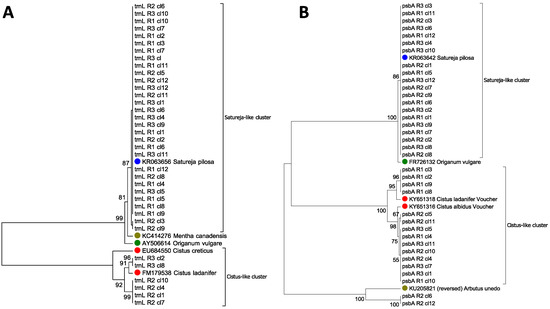
Figure 2.
UPGMA trees constructed using the 36 chloroplast trnL sequences (A) and trnH-psbA sequences (B) obtained from oregano samples. Numbers above nodes represent the bootstrap support after 1000 permutations.
Concerning the trnH-psbA barcode, similarly to what observed for the trnL region, none of the 36 sequences analyzed originated from O. vulgare L. (Table 2). Most of them (20 out of 36) corresponded to the trnH-psbA region of S. pilosa Velen./S. montana L., confirming what was determined by the trnL barcode analysis. A large percentage of sequences (12 out of 36) was mapped to Cistus albidus voucher SEV:286739, while two sequences matched with Cistus ladanifer voucher SEV:286741. It is worth noting that in this case, the query coverage was lower, ranging from 46.3% to 55.98%, because the trnH-psbA sequences deposited were quite short (233 and 230 bp, respectively). Nonetheless, these data clearly confirm and complement the results that were obtained using the trnL region, which demonstrates a partial contamination of a species belonging to the Cistaceae family. Finally, we found two sequences that were related to Arbutus unedo L. (commonly called strawberry tree), which belongs to the Ericaceae family. As previously described for the trnL barcode, the ability of universal primers for trnH-psbA to amplify from genomic DNA purified from O. vulgare L. plants was tested to exclude the possibility that the lack of sequences belonging to this species was due to the inability of primers to work properly in oregano. The presence of this species in low quantities within our samples was not surprising because the strawberry tree is predominant in Mediterranean areas and a small cross-contamination of samples could be explained by chance. The UPGMA tree constructed using the 36 sequences of the trnL-psbA region, together with the references from O. vulgare L. (FR726132), S. pilosa Velen. (KR063642), C. ladanifer (KY651318), C. albidens (KY651316), and A. unedo L. (KU205821) is reported in Figure 2, panel B.

Table 2.
List of the best hits obtained by BLASTn analysis for the intergenic spacer trnH-psbA amplicons.
As a general conclusion, the qualitative analysis of cpDNA barcodes indicated that the samples analyzed were adulterated with species other than O. vulgare L., which was totally absent. In particular, samples revealed a high level of contamination with S. pilosa Velen./S. montana L. (67%), which represents a contaminant belonging to Group I, which is partially tolerated by law under a certain quantity. We also observed a partial level of contamination with Cistus spp., which represents a very serious contaminant belonging to Group II that is not tolerated by law. Moreover, and most importantly, samples analyzed did not showed any presence of the O. vulgare L., as none of the sequences from the cloned amplicons could be associated with any species of the genus Origanum.
3.2. Quantitative Determination of Contaminants on Oregano Samples
The qualitative analysis of samples R1, R2, and R3 with three technical replicates for two distinct barcodes clearly indicated that the commercial oregano batches that were analyzed were adulterated. In particular, we documented a total absence of O. vulgare L. in the analyzed bulks, together with a marked presence of S. pilosa Velen./S. montana L. and a modest presence of C. ladanifer L., which represents a very serious fraud. To obtain a more detailed quantification of the level of contamination, we designed species-specific primers by aligning the trnL sequences obtained by Sanger sequencing with those of O. vulgare L., S. pilosa Velen. and C. ladanifer L. (Figure 3). Species-specific trnL amplification from O. vulgare L, C. ladanifer and S. pilosa Velen. were then performed to evaluate the efficiencies of the primers and the ability to selectively amplify the targeted regions from one species rather than another (Figure S2).
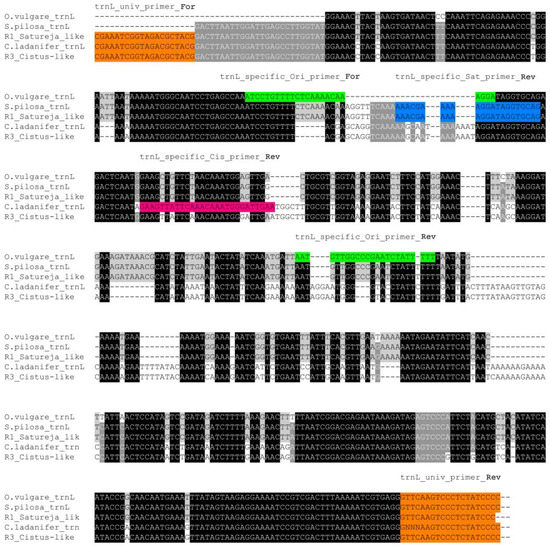
Figure 3.
Alignment of Satureja-like and Cistus-like tnrL consensus sequences obtained by amplicon sequencing with those of O. vulgare L., S. pilosa Velen., and C. ladanifer. Both universal (orange) and species-specific primers for trnL amplification from O. vulgare L. (green), S. pilosa Velen. (blue), and C. ladanifer L. (violet) are indicated.
As a preliminary step, we performed replicated semi-quantitative end-point PCR experiments using genomic DNA aliquots obtained from samples R1, R2, and R3 to get an idea of the specificity of the primers in the “unknown samples”. Specific primers for O. vulgare L., with particular reference to the trnL region, were used for PCR on the samples in our study. The results confirmed that genomic DNA from S. pilosa Velen. and C. ladanifer L. species were actually present in all samples, giving the robust amplicons of the expected size, whereas the primers specifically designed for the Origanum species did not detect any of the specific target regions, producing smeared patterns and non-specific amplicons (Figure 4). These findings further demonstrated an unexpected total lack or low concentration of oregano DNA over all of the samples analyzed.
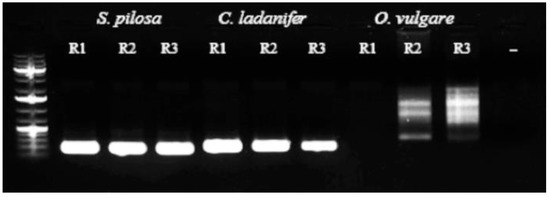
Figure 4.
Results of PCR amplification of the trnL intron target region in the samples (R1, R2, and R3) using species-specific primers: robust amplicons of the expected size were obtained using genomic DNA from the species S. pilosa and C. ladanifer, whereas only faint and non-specific amplicons were obtained for Origanum (- refers to the negative control).
As second step, we performed quantitative real-time PCR on selected samples, in order to quantify the level of contamination. To this aim, we produced preliminary standard curves representing different levels of contamination for each species analyzed. A 1:2 serial dilution of DNA from O. vulgare L., S. pilosa Velen., and C. ladanifer L. was then analyzed as reported in Table S2. The use of standard dilutions representing 100%, 50%, 25%, 12.5%, 6.25%, and 0% of a given species was used to obtain a quantitative idea of the levels of DNA from that particular species in the sample. Then, quantitative PCR runs were performed on both the standard dilutions and unknown samples (R1, R2, R3).
The dissociation curves were analyzed after each experiment to avoid non-specific amplification. The results showed 100% specificity (no false positives) and 100% sensitivity (no false negatives) in the identification of different species from the different batches analyzed. The amplification curves obtained from the test samples and the oregano standard dilutions using oregano trnL specific-primers showed very different Ct values. In particular, the R1, R2, and R3 Ct values were between the Ct values of the standard dilution, with 6.25% and 0% O. vulgare DNA, indicating the presence of a very low quantity of oregano DNA in our samples. Similarly, we performed the same analysis with the trnL specific primers for Cistus and Satureja, together with the relative standard dilutions. The results indicated a modest presence of Cistus in unknown samples, with Ct values in the between those of the standard dilutions with 6.25 and 0% C. ladanifer DNA. We also observed a higher content of S. pilosa, with Ct values close to the STD50 corresponding standard (1:2 dilution of DNA from Satureja). Table 3 reports the relative quantification of each contaminant with respect to the standards utilized.

Table 3.
Results of the quantification of DNA by quantitative PCR (qPCR) for the species Origanum and each of the main contaminants, Cistus and Satureja, compared to the standards.
We found the percentage of contaminants to be lower using this method, compared to the qualitative analysis. This discrepancy could be due to the preferential amplification of universal primers for the genomic DNA of one given species, with respect to the others. In fact, it must be considered that in the mix of DNA templates from different species, universal primers could work in different ways, depending on the quality of the DNA of a given species and on the presence of impurities. Alternatively, the overestimation observed in the qualitative determination could be due to the relatively low number of colonies sequenced. Nevertheless, our quantitative analysis confirmed what was observed in the qualitative analysis, which was an almost total absence of O. vulgare in the samples, a modest contamination of the Group II contaminant C. ladanifer L., and a marked contamination of the Group I contaminant S. pilosa Velen (Table 3).
4. Conclusions
In the present study, we implemented a methodological approach for detecting the degree of contamination of a complex food matrix of plant origin, such as common oregano, commercialized as a mixture of small fragments of dried leaves. We applied a universal cpDNA barcoding assay for a preliminary qualitative analysis aimed at identifying the possible contaminants (i.e., non-oregano species) in commercial batch samples. Then, on the basis of our findings, we developed and validated a species-specific and sequence-targeted qPCR assay for the analytical quantitative measurement of the proportion of contaminants of common oregano. Although, based on a number of preliminary studies, the CBOL Plant Working group initially recommended a core barcode consisting of portions of two plastid-coding regions, rbcL and matK, to be eventually supplemented with additional markers if needed, many lines of evidence have demonstrated that matK is very difficult to amplify using existing universal primer pairs, and that rbcL is characterized by modest discriminatory power, although it is easy to amplify and may provide a useful backbone to the barcode dataset. So, despite their high universality in terms of PCR amplification and DNA amplicon sequencing success, the analysis of these coding regions often fails due to the interspecific sharing of sequences [21]. For these reasons, we focus on the plastid intergenic spacer regions trnH-psbA and the trnL genic intron, since they are known to increase the identification performance of DNA barcoding protocols—especially when dealing with the identification and/or authentication of foodstuffs for genetic traceability purposes and not for genetic diversity studies. These two latter markers are straightforward to amplify among land plants, and they also show high variability across their homologous non-coding regions in plants, even among closely related taxa [26].
It is worth mentioning that we found the universal sequence of the trnL-intron barcode of different Origanum species, including the common oregano (O. vulgare) and four of the main oregano contaminants (O. dictamnus, O. onites, O. syriacum and O. majorana), matched with 100% identity (data not showed). Therefore, while this methodological approach is able to reliably detect and assess the qualitative and quantitative presence of important contaminants, such as Cistus spp. and Satureja spp., it is unable to discriminate impurities belonging to the genus Origanum if the trnL and trnH-psbA barcode regions are to be used.
Although our data revealed several similarities and differences between the qualitative and quantitative assays on the estimates of the contamination rate, our methodological approach relied on robust protocols and revealed sound applications for the traceability of a complex food matrix. We are confident that cpDNA barcoding by combined universal end-point and specific real-time PCR analyses can be profitable and cost-effective for the identification and quantification of the main contaminants of commercially available oregano. Our intent is to apply this approach and to extend this strategy to many other complex foodstuffs of plant origin.
Supplementary Materials
The following are available online at http://www.mdpi.com/1424-2818/10/3/98/s1, Figure S1: Amplification of (A) psbA-trnH locus in R1, R2, R3 samples; (B) trnL locus in R1, R2, R3 samples, Figure S2: Amplification of (A) O. vulgare L. (O), (B) S. pilosa Velen. (S) and (C) C. ladanifer L. (C) trnL region using species-specific primers, Table S1: trnL and trnH-psbA universal and species-specific primers used in this study, Table S2. Serial dilutions used in quantitative PCR (qPCR) for contaminants estimation.
Author Contributions
Conceptualization, G.B. and A.V.; Methodology, A.V.; Software, A.V.; Validation, A.V.; Formal Analysis, A.V.; Investigation, A.V.; Resources, G.B.; Data Curation, A.V.; Writing—Original Draft Preparation, A.V.; Writing—Review & Editing, G.B., M.L.; Visualization, A.V., G.B.; Supervision, G.B.; Project Administration, G.B.; Funding Acquisition, G.B.
Funding
This research was funded by BreedOmics, a service laboratory of genomics for food traceability and variety breeding (https://www.dafnae.unipd.it/servizi/servizi-il-territorio).
Acknowledgments
Authors would like to thank Mery Campalto for her valuable technical support in experimental procedures including DNA isolation, amplification and sequencing.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Galimberti, A.; Labra, M.; Sandionigi, A.; Bruno, A.; Mezzasalma, V.; de Mattia, F. DNA Barcoding for Minor Crops and Food Traceability. Adv. Agric. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Woolfe, M.; Primrose, S. Food forensics: Using DNA technology to combat misdescription and fraud. Trends Biotechnol. 2004, 22, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Dhanya, K.; Sasikumar, B. Molecular marker based adulteration detection in traded food and agricultural commodities of plant origin with special reference to spices. Curr. Trends Biotechnol. Pharm. 2010, 4, 454–489. [Google Scholar]
- Posadzki, P.; Watson, L.; Ernst, E. Contamination and adulteration of herbal medicinal products (HMPs): An overview of systematic reviews. Eur. J. Clin. Pharmacol. 2013, 69, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Federici, S.; Galimberti, A.; Bartolucci, F.; Bruni, I.; de mattia, F.; Cortis, P.; Labra, M. DNA barcoding to analyse taxonomically complex groups in plants: The case of Thymus (Lamiaceae). Bot. J. Linn. Soc. 2013, 171, 687–699. [Google Scholar] [CrossRef]
- Gismondi, A.; Fanali, F.; Martínez Labarga, J.M.; Caiola, M.G.; Canini, A. Crocus sativus L. Genomics and different DNA barcode applications. Plant Syst. Evol. 2013, 299, 1859–1863. [Google Scholar] [CrossRef]
- Kojoma, M.; Kurihara, K.; Yamada, K.; Sekita, S.; Satake, M.; Iida, O. Genetic identification of cinnamon (Cinnamomum spp.) based on the trnL-trnF chloroplast DNA. Planta Med. 2002, 68, 94–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, H.X.; Wang, L.; Wang, T.; Yang, R.W.; Wang, X.L.; Zhou, Y.H.; Ding, C.B.; Zhang, L. Potential use of DNA barcoding for the identification of Salvia based on cpDNA and nrDNA sequences. Gene 2013, 528, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Marieschi, M.; Torelli, A.; Poli, F.; Sacchetti, G.; Bruni, R. RAPD-Based method for the quality control of mediterranean oregano and its contribution to pharmacognostic techniques. J. Agric. Food Chem. 2009, 57, 1835–1840. [Google Scholar] [CrossRef] [PubMed]
- Kintzios, S.E. Medicinal and aromatic plants—Industrial profiles. In Oregano: The Genera Origanum and Lippia; CRC Press: London, UK, 2002; Volume 25, pp. 3–9. ISBN 0-415-36943-6. [Google Scholar]
- Skoula, M.; Harborne, J.B. The taxonomy and chemistry of Origanum. In Oregano: The Genera Origanum and Lippia; Kintzios, S.E., Ed.; Taylor & Francis, CRC Press: London, UK, 2002; pp. 67–108. [Google Scholar]
- Ietswaart, J.H. A Taxonomic Revision of the Genus Origanum (Labiatae); Leiden University Press: Leiden, The Netherlands, 1980; Volume 4. [Google Scholar]
- Baricevic, D.; Bartol, T. The biological/pharmacological activity of the Origanum genus. In Oregano: The Genera Origanum and Lippia; Kintzios, S.E., Ed.; Taylor & Francis: London, UK, 2002; Chapter 8; pp. 177–213. [Google Scholar]
- Tucker, A.O.; DeBaggio, T. The Big Book of Herbs: A Comprehensive Illustrated Reference to Herbs of Flavor and Fragrance; Interweave Press: Loveland, CO, USA, 2000. [Google Scholar]
- Campanini, E. Dizionario di Fitoterapia e Piante Medicinali; Tecniche Nuove: Milano, Italy, 1998. [Google Scholar]
- Kintzios, S.E. Oregano. In Handbook of Herbs and Spices, 1st ed.; Peter, K.V., Ed.; Woodhead Publishing: Cambridge, UK, 2004; Volume 2, pp. 215–229. [Google Scholar]
- Tainter, D.R.; Grenis, A.T. The spices. In Spices and Seasonings: A Food Technology Handbook, 2nd ed.; Tainter, D.R., Grenis, A.T., Eds.; Wiley-VCH: New York, NY, USA, 2001; pp. 116–119. [Google Scholar]
- Huie, C.W. A review of modern sample-preparation techniques for the extraction and analysis of medicinal plants. Anal. Bioanal. Chem. 2002, 373, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Schultz, H.; Quilitzsch, R.; Kruger, H. Rapid evaluation and quantitative analysis of thyme, origano and chamomile essential oils by ATR-IR and NIR spectroscopy. J. Mol. Struct. 2003, 661–662, 299–306. [Google Scholar] [CrossRef]
- Techen, N.; Crockett, S.L.; Khan, I.A.; Scheffler, B.E. Authentication of medicinal plants using molecular biology techniques to compliment conventional methods. Curr. Med. Chem. 2004, 11, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA barcoding as a molecular tool to track down mislabeling and food piracy. Diversity 2016, 8. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of Plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Galimberti, A.; De Mattia, F.; Losa, A.; Bruni, I.; Federici, S.; Casiraghi, M.; Martellos, S.; Labra, M. DNA barcoding as a new tool for food traceability. Food Res. Int. 2013, 50, 55–63. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).