Abstract
The insect superfamily Psylloidea (Hemiptera) includes economically important biocontrol agents, pests and plant pathogen vectors, for which a rapid and accurate identification is fundamental for international biosecurity. Australasia is a hot spot for psyllid diversity, but previous species assessments in the region were largely based on morphology and host plant association. Morphological identification of psyllids remains challenging for a wide number of species and for juvenile insects, while a robust molecular framework for identification is not available. Consequently, knowledge of psyllid biology is compromised. Here, incorporating morphological evidence and host plant associations, insects collected from almost 600 primarily New Zealand locations were linked to 67 previously described species. By applying species delimitation methods including GYMC (General Mixed Yule–Coalescent method), PTP (Poisson Tree Processes), mPTP (multi–rate Poisson Tree Processes) and ABGD (Automatic Barcode Gap Discovery) to a dataset composed of 425 cytochrome oxidase I (COI) DNA barcode sequences, further cryptic diversity was revealed among the psyllid collection; more than 20 undescribed taxa are reported here for the first time, resulting in a total of 90 taxa across 21 genera and six families included in this study. Our improved understanding of psyllid diversity in New Zealand revealed new plant host-psyllid associations and geographical variation. The DNA barcode resource will enable future studies of psyllid ecology and more accurate, rapid identifications of psyllids that pose biosecurity threats to Australasia.
1. Introduction
The superfamily of phloem–feeding insects Psylloidea (Hemiptera: Sternorrhyncha) contains almost 4000 described species worldwide, belonging to more than 270 genera and eight families [1,2]. Psyllids have a rich cultural and scientific history, from the sugary lerps on Eucalyptus leaves used as a food source by Australian aboriginals [3] to the report of Psyllopsis fraxinicola in the 1758 edition of the Systema naturae [4]. More recently, the discovery that psyllids vector economically damaging bacterial crop pathogens [5] has raised the biosecurity threat posed by psyllids and refocused research on important biological, ecological and evolutionary questions that might better inform the threat associated with these insects. Perhaps the most high profile example of an important psyllid pest is the Asian citrus psyllid, Diaphorina citri Kuwayama, vector of Candidatus Liberibacter asiaticus, agent of the Huanglongbing disease (citrus greening) [6,7]. The potential arrivals of such species are closely monitored, especially by those countries with higher risk of invasion. Recent modelling has highlighted the high risk for countries such as Australia of invasion by the South American Potato Psyllid, Russelliana solanicola Tuthill [8]. The arrival of a similar pest species could have economically damaging results, similar to those perpetrated in New Zealand by Bactericera cockerelli Šulc, the tomato/potato psyllid [9,10,11]. To understand the biosecurity risk linked to the arrival of adventive psyllid species, a strong understanding of the psyllid fauna present in a country is required.
Australasia is a “hot spot” for psyllid diversity, with 402 described species belonging to 48 genera identified in Australia [2,12]. In New Zealand, Ferris and Klyver [13] and Tuthill [14] were among the earlier taxonomist revising the New Zealand psyllids, listing 25 and 51 species, respectively. Subsequently, the work of Dale [15] provided a fundamental contribution through a very detailed field survey and taxonomic study of the New Zealand psyllids. Dale listed 81 species, of which 24 were reported as new. More recently, the New Zealand Inventory of Biodiversity [16] reported a total of 95 species while the most recent record indicates a total of 99 species in 26 genera, with representatives across six of the eight Psylloidea families [17]. The majority of species in New Zealand belong to three genera: Trioza (Triozidae), Acizzia (Psyllidae) and Ctenarytaina (Aphalaridae). Trioza is especially noteworthy, as the recorded diversity in this genus spans 52 species on almost 20 host plant genera, with all but one plant species native to New Zealand [17]. Adventive psyllids that were not present in New Zealand prior to European colonization are also present in large numbers. Morphological studies have indicated that the majority of these adventive psyllids are the same as species in Australia or are very close relatives [17]. For instance, 35 species of exotic psyllids are recorded in New Zealand on wattles and eucalypts, plants native to Australia [17]. Their presence in New Zealand has been primarily attributed to incidental introduction of the insects together with their ornamental or forestry host plants [14]. Biogeographical drivers are also likely to have driven dispersal of psyllids in Australasia. For example, westward winds have a strong influence on trans-oceanic insect dispersal in the Southern Hemisphere [18,19] Many adventive species have only been in New Zealand for a matter of decades, such as Creiis lituratus, Cardiaspina fiscella, Eucalyptolyma maideni and Cryptoneossa triangula. These species were not recorded by Dale [15], but were listed in the Biodiversity Index published in 2010 [16]. Prevailing winds from Australia raise the possibility that non-indigenous psyllid species may have established in New Zealand from this country via wind dispersal since the surveys conducted some 30 years ago. This could be hypothesized based on the fact that, in recent times, several invertebrates have been recorded in New Zealand, dispersing from Australia on wind currents [20,21] in what has been defined as a “rain of invertebrates” [22].
Other species, such as Ctenarytaina eucalypti (Maskell) (Aphalaridae), and a number of Acizzia psyllids (Psyllidae) were originally described in New Zealand, only to be later recognized as of Australian origin based on the origin of the host plants [23]. These include A. acaciae (Maskell) (described in 1894), A. albizziae (Ferris & Klyver) and A. uncatoides (Ferris & Klyver) (both described in 1932), A. conspicua (Tuthill), A. dodonaeae (Tuthill), A. exquisita (Tuthill) and A. jucunda (Tuthill) (all described in 1952) [14].
Psyllids have also been introduced to New Zealand from further afield, almost certainly as a result of human-assisted spread. For instance, Bactericera cockerelli, the tomato/potato psyllid that established in New Zealand in 2006 and was recently detected in Western Australia (https://www.agric.wa.gov.au/tomato-potato-psyllid-tpp), originates from North America [24]. B. cockerelli vectors the plant pathogen Candidatus Liberibacter solanacearum. The arrival of this bacterium and its host in New Zealand has resulted in significant economic losses to crop production [11].
The continuing changes in the psyllid fauna of New Zealand and the recent discovery that several species are associated with plant pathogens have reignited research to understand psyllid diversity, interactions with host plants, and associated microbiology. DNA barcoding has become valued as a contributor to determining species identification [25,26]. As such, COI sequences were previously included in multi-marker molecular analyses that have been used to enhance the identification of psyllids and to understand their diversity, including small sets of Australian [27] and New Zealand [28] psyllids. The power of molecular methods to redefine psyllid diversity has been demonstrated for a narrow range of psyllid species worldwide [12,29]. A more comprehensive molecular framework (that includes COI sequences) underpinning the evolution of Psylloidea has only recently been presented [30].
The aim of this study was to develop a DNA reference library of psyllids in New Zealand and to contribute to one for Australia. COI barcoding was performed on an up–to–date set of psyllids from New Zealand and Australia, amassed over three years of field collections. COI sequences were then analyzed using phylogenetic and statistical species-delimitation methods in conjunction with assessment of morphology, host plant associations and geographical information to redefine the diversity of psyllids in New Zealand. The resulting data was used to provide new insights into the relationships of the New Zealand psyllids with Australian relatives and to begin to address important evolutionary, ecological and biological questions. In the future, improvements in our understanding of psyllid ecology (e.g., host plant association and distribution) and biology (e.g., their associations with their microbiome) will enable better recognition of the threats posed by new psyllid invasions and the risk associated with the transmission of bacteria such as plant pathogens from an adventive psyllid species to an endemic one and, consequently, to its host plant.
2. Materials and Methods
2.1. Psyllid Collection
Field collection of psyllids was performed from September 2014 to January 2017. Adult specimens and, where present, immature stages were collected from more than 500 different locations in all regions of New Zealand [31]. In addition, psyllids were collected from 90 locations in Southern and Eastern Australia for comparative purposes. Field collection was informed by records of host plant associations and, to a lesser degree, specific collection locations reported in prior works [15,17]. While mindful of previous host records, all plant species at a collection site were examined wherever possible. Plant association for the described species was determined primarily using the keys of Dale [15] and Hollis [3]. Identification of the host plants was performed in collaboration with Landcare Research Herbarium staff (Lincoln, Canterbury, New Zealand).
Collection was iterative, with each search informed by the progressive results of earlier attempts; for example, when little COI DNA sequence variation was recorded in psyllids from Akeake (Dodonaea viscosa) or Pittosporum spp., less emphasis was placed on continued collections from these hosts. In contrast, when DNA variation highlighted the presence of more taxa than expected on a particular host, more locations were surveyed and more specimens collected. Populations of psyllids were initially defined as the psyllids collected from a single plant. This was straightforward if the plant was isolated from other individuals of the same species (by many kilometers), such as in alpine and subalpine habitats. However, it was more complex if psyllids were found on contiguous plants and were difficult to verify in-field as of the same species. Consequently, a population was defined as insects of the same species collected from a single plant or from a group of contiguous plants of the same species. Populations were confirmed by morphological and/or genetic analysis, retrospectively.
Collection of insects involved dislodging them from the leaves of host plants by beating onto a tray followed by capture using an entomological aspirator. In each instance, as many adults and immature stages as possible were collected, as were males and females. Psyllids were preserved in high grade ethanol (99%) for molecular analysis.
2.2. Identification of Insects Based on Morphology and Host Plant Association
Captured insects were examined morphologically using the keys of Dale [15] and Hollis [3]. The process included the preparation of microscope slide mounted [12] and dry mounted specimens; these were deposited in the Lincoln University Entomological Collection (LUNZ).
2.3. Molecular Analysis
DNA extractions from individual specimens were performed using a CTAB (cetyl trimethylammonium bromide) protocol [32]. The DNA barcode region [25] of subunit 1 of the COI gene was amplified by polymerase chain reaction (PCR). The PCR primer C1–J1709 [33] was paired with HCO2198 [34] to generate an amplicon of 403 bp. PCR was performed using the KAPA3G plant PCR Kit (Kapa Biosystems, Massachusetts, USA). Each 20 µL reaction consisted of 10 µL 1x PCR master mix, 1 μL each primer (10 μM), 0.1 μL of polymerase and 1 µL DNA template. Thermal cycling conditions were an initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s and extension at 72 °C for 1 min, followed by a final extension of 7 min. PCR products were Sanger sequenced on an ABI 3130xl Genetic Analyzer by the Sanger sequencing Unit (Bio-Protection Research Center, Lincoln University, Lincoln, New Zealand).
2.4. COI Barcode Data Analysis
Consensus sequences were assembled by combining and trimming sequence reads for both directions of COI (forward and reverse), and then aligned using MEGA version 6 [35]. The Kimura-2-parameter (K2P) model [36] with a bootstrap of 10,000 replicates was used for phylogenetic analyses by Maximum likelihood (ML) algorithm. K2P was chosen for continuity, because of its wide use in barcoding since the first studies [25,26]. While accepting that it has well documented limitations (e.g., [37]), it has been also reported that the species identification success rates is largely unaffected by model choice [38]. Genetic distances between taxa were visualized in a ML tree. A total of 405 COI sequences of the targeted region were deposited in GenBank with the accession numbers MG132221 to MG132630 (Figure S1).
The consensus COI sequences were compared using several species delimitation software packages to assist identification of the insects. Automatic Barcode Gap Discovery (ABGD) analysis [39] was performed using the web interface (http://wwwabi.snv.jussieu.fr/public/abgd/). To perform GYMC [40], PTP [41] and mPTP [42] tree-based analyses, ultrametric trees were generated with 10 million replicates using BEAST v. 2.4.8. [43]. Yule and coalescent priors with constant population growth were selected, as suggested by Michonneau [44]. A 10% burn-in was applied using the software TreeAnnotator. The trees were saved in NEXUS format using the software FigTree v. 1.4.3. [45]. Trees were then tested with GYMC using the software R v. 3.1.1 [46] and the package “splits” [47]. PTP and mPTP analyses were performed using the bPTP web server (available at http://species.h-its.org/ptp/) and mPTP web server (available at http://mptp.h-its.org/#/tree).
3. Results and Discussion
3.1. Species Delimitation
3.1.1. Field Collection, Host Plant Association and Initial Morphological Examination
The distribution of the populations collected on North (169), South (314), Stewart (22), and Waiheke (5) islands of New Zealand and in Australia (South Australia, Queensland, Victoria, and New South Wales) (102) is reported in Table S1. Of the New Zealand populations, 320 were collected from native plant species and 190 from introduced species (Table S1). Morphological features together with reports of host plants were used to link these taxa with 67 previously described New Zealand psyllids taxa [13,14,15,23]. In most cases, this was straightforward; usually a single psyllid taxon was found on a host plant and these insects were morphologically consistent with reports of psyllids from that host species. The high numbers of individuals found on these plants suggest these are hosts rather than “casual plants” or “food plants” as described by Burckhardt et al. [48]. The taxa collected both in New Zealand and Australia were found on the same hosts in both countries. There were also a few cases of psyllids found on plants not previously regarded as hosts [48]. Nymphs were found only infrequently due to field collections performed quite often when psyllids were already adults, so it was difficult to confirm “true” host associations, which are defined by life cycle completion on a host plant species [48]. Earlier researchers established host relationships for many New Zealand psyllids by securing nymphs and rearing them to adulthood [14]. For the most part, the unusual associations appeared to be casual host associations [48], with low psyllid numbers in contrast to the higher numbers on adjacent plants of the expected hosts. Atypical host associations were observed when high numbers of psyllids were present on an isolated plant, however. A conspicuous example was Trioza bifida, which was collected in substantial numbers from unexpected plant hosts of the genera Dracophyllum, Hebe and Pseudowintera (Table 2) in addition to the expected Olearia species [17]. These may be examples of food plants on which adult psyllids can feed but not breed [48], and is consistent with what Tuthill described as “a very active species” [14]. While the findings here contribute to a better understanding of New Zealand psyllid biology, observations for any one psyllid taxon were insufficiently detailed to draw definitive conclusions about their ecology.
Despite the undoubted value of morphological descriptions for New Zealand psyllids [13,14,15], in several cases, the initial taxonomic assignment to species was incorrect. These were revealed by COI sequence divergence (Figure S1) and led to morphological reassessment of other individuals for the same population to confirm the identification. For example, finding T. subacuta on Olearia instead of Brachyglottis initially led to misidentification as T. subvexa. Similarly, collecting psyllids from Schefflera digitata initially led to the assumption they belonged to the species T. schefflericola [15,17]. However, comparing these specimens with samples of T. irregularis revealed scarce morphological variation and a very low COI genetic divergence (<2%), suggesting they belonged to a single taxon.
3.1.2. COI Genetic Analysis
DNA was successfully extracted from 465 samples representing 346 populations of psyllids. Of those, a COI gene fragment was successfully amplified and sequenced from 425; 20 sequences of which have been previously published [28]. No COI fragment could be amplified from Acizzia solanicola (seven specimens) and Atmetocranium myersi (two specimens). However, these specimens were unambiguously identified using morphology and host information.
Based on a 3% uncorrected p-distance threshold for COI divergence, which is widely used today for the identification of psyllids [12,29,49], 89 taxa could be delimited in the COI dataset. This highlighted an unexpectedly high level of cryptic diversity. To confirm this genetic variation, several species delimitation methods were applied. The most conservative methods recorded 81 taxa (mPTP) and 90 taxa (ABGD). Between 94 and 96 taxa could be separated using GMYC, depending on which prior was adopted (Yule or Coalescent), and an average of 107 taxa could be identified using PTP (Table 1).

Table 1.
Species delimitation. Species delimitation from an ultrametric COI sequence tree generated with a Yule or Coalescent model with constant population size was used by the GMYC, PTP and mPTP methods. Species delimitation from ABGD and 3% distance was based on FASTA sequences only. The number of predicted biological entities is shown in the taxa column.
All methods reported high, unrecognized biodiversity within three groups of psyllids: the genus Psylla on native broom (Carmichaelia), the genus Ctenarytaina on mānuka (Leptospermum) and kānuka (Kunzea) and the genus Ctenarytaina on Fuchsia.
A particularly high level of diversity was found in Ctenarytaina spp. from mānuka and kānuka; 38 psyllids were grouped in a similar number of biological entities using ABGD, GMYC and PTP, but a much smaller number by mPTP (Figure 1).

Figure 1.
A COI gene tree (Kimura 2–Parameters model, Maximum Likelihood algorithm, 10,000 replicates, bootstraps <50% not reported) showing relationships between all Ctenarytaina samples collected from mānuka, kānuka and Olearia. The numbers on the right represent the taxa predicted by each of the species delimitation methods, GMYC, mPTP, ABGD, PTP and ≥3% COI divergence. The tree nodes are supported by bootstrap values. The bar represents a genetic distance of 0.02.
3.1.3. Morphological Reexamination of the Cryptic Taxa Detected
While the description of new species was beyond the aim of this work, we are mindful of the requirements that will need to be addressed in future work to delineate the taxa presented here. Between five and 20 individuals per population may be required to the confirm barcode gap [50,51,52]. Additional field collections will also be required to define and describe new New Zealand psyllid species. Nonetheless, the cryptic diversity highlighted by the species delimitation methods was followed up with a preliminary, morphological comparison of the newly recorded taxa. While this work did not intend to provide a morphological description for the newly delimited taxa, these comparisons aimed to highlight variations present even in those taxa showing cryptic genetic variation, namely the genera Ctenarytaina (mānuka and kānuka complex) and Psylla (both kowhai and Carmichaelia complexes). Several characters were found consistent in individuals amongst populations of the same taxa, highlighting the presence of promising characters that can be used for future morphological assessments and descriptions of these species. Among these characters are the shape of male and female terminalia and the shape and veins of wings, both widely used in psyllid taxonomy and species identification (e.g., [13,14,15]).
3.1.4. A Species Delimitation Method Integrating Host Plant, COI and Morphology
The results of different species delimitation methods (GYMC, PTP, ABGD, and mPTP) were compared. In the PTP delimitation, some individuals belonging to the same population and from the same individual plant were proposed as separate taxa. Therefore, the PTP method appeared to overestimate the number of taxa. On the other hand, the mPTP method appeared to underestimate the diversity, as it clustered together species already described such as Ctenarytaina “Short” and C. pollicaris (Figure 1). The remaining two methods (GMYC and ABGD) grouped the taxa in a similar way which was also coincidentally similar to the COI ≥ 3% threshold (Figure 1). Synthesizing these different species delimitation methods together with information on insect morphological variations and host associations, we were able to link psyllid samples and their sequences with 57 formally described taxa and another 10 taxa described in an unpublished work (Table 2) [15]. For two additional psyllid populations with distinct COI sequences, we had low numbers of insect samples, and could not link these to described species; these may be known taxa for which we have insufficient morphological information (e.g., specimens of a single sex) or may be new taxa. A total of 21 taxa could be then delimited combining both genetic and morphological variation (Figure 2, red branches). Therefore, the species delimitation presented in Table 2 is the result of an integrative taxonomic approach. By coincidence, this list corresponds to a >3% between taxa, with the single exception of Ctenarytaina clavata D. For this species, the genetic distance between two populations (ID334 and ID335) was exactly 3% and the method GMYC considered the populations as separate taxa. However, cryptic morphology and close geographic location, together with a low number of samples collected, did not allow a clear separation between the individuals of population 334 and population 335. Furthermore, the results obtained using the method ABGD considered all the populations to belong to the same taxon (Figure 1). Therefore, population 334 was considered to be from the same lineage of the other two.

Table 2.
New Zealand psyllid taxa delimited using their morphology, plant association, and COI sequence divergence of >3%. Newly recorded taxa (*), not formally described but previously known taxa (ˠ) and taxa not identified to the species level (?) are reported. New Zealand locations are based on Crosby’s work [31]; Australian locations (in bold) are New South Wales (NSW), South Australia (SA), and Victoria (VIC). The number of populations from each region is reported in parentheses. The host plants are differentiated between previously known (black), possible new host plants (blue) and uncertain associations (red).
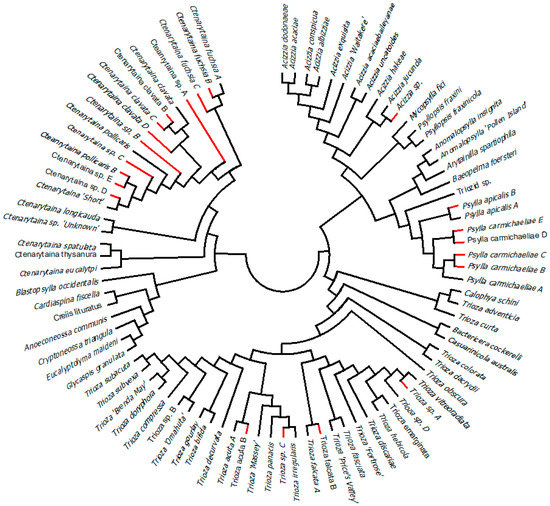
Figure 2.
Graphic representation of the COI gene tree reported in Figure S1. The tree has been collapsed to show the 90 taxa presented in this work and their names. Red branches indicate the 21 taxa reported here for the first time.
Therefore, combining psyllid morphology, distribution, host association and the use of the species delimitation methods GMYC and ABGD, that resulted more accurate for this dataset, a >3% variation was considered supportive of species-level differences for the taxa presented in this work. This resulted in a list that contained 90 New Zealand psyllid taxa, identified from the largest field collection in the last 30 years. These taxa belonged to 21 genera and six families.
3.2. An Updated Biodiversity of the New Zealand Psylloidea Reveals New Insights into Psyllid Biology and Ecology
The list of additional taxa presented in this work enabled the ecological aspects linked to the distribution and host plant association of the New Zealand psyllids to be considered.
In general, the collections performed here confirmed the psyllid/host plant associations reported in the literature ([15,17]; Table 2). Almost all of the newly recorded taxa detected in this study were lineages found on a known host; for example, Trioza acuta A and B were lineages separated by 6–7% from Ozothamnus leptophyllus. In other cases, new lineages were detected on closely related host plants; for example, a Trioza lineage (sp. A) from Pittosporum divaricatum was 4–5% different from Trioza vitreoradiata that was found in multiple populations on other Pittosporum species (see below).
The highest number of newly recorded taxa belonged to the genera Ctenarytaina and Psylla (Table 2), which were mostly closely related to previously described taxa. Five different taxa were detected among Psylla populations collected from different Carmichaelia species growing in close proximity (Table 2; Figure 3c). Geographic separation of taxa was recorded for Ctenarytaina fuchsiae B (Figure 3a) or Ctenarytaina sp. B (Figure 3b). Several closely related taxa were detected among Ctenarytaina clavata and C. pollicaris (Figure 3b).
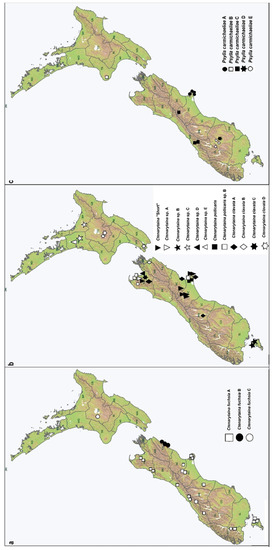
Figure 3.
Distribution of the newly discovered cryptic taxa across New Zealand: Ctenarytaina taxa on Fuchsia (a) and on the mānuka and kānuka tea trees (b); and Psylla spp. on Carmichaelia (c).
3.2.1. The Genus Psylla Showed Examples of Putative Host Plant Specificity and a Syntopic Distribution
Seven Psylla taxa were resolved: two on kowhai and five on native broom (Carmichaelia spp.). Only P. apicalis on kowhai and P. carmichaeliae on Carmichaelia had previously been recognized [15]. Interestingly, once the genetic difference (7–8% COI divergence depending on populations) between the kowhai psyllid taxa became apparent, considerable corroborating morphological differences were observed between the two linages; these included overall dimensions, color and shape of the wings. In at least four Central Otago locations, both taxa were collected from the same individual kowhai plants, highlighting not only a sympatric but also a syntopic distribution (Table 2). Interestingly, when describing P. apicalis for the first time in 1932, Ferris and Klyver recorded the same morphological variations presented here [13]. The presence of two separate taxa was considered but discounted [13].
In contrast, the five Psylla taxa from native broom showed a much broader range of COI variation from 7% to 17%. Each of the five taxa was collected from different Carmichaelia host species; of these hosts, three were identified to the species level using the existing morphological keys [53,54], while the other two, which were morphologically distinct, could not be identified (Table 2). There were no occurrences of more than one of these psyllid taxa on the same Carmichaelia host species. Two of the Carmichaelia species hosting Psylla carmichaeliae D and E grew within 10 km of each other in Cromwell (Central Otago, CO, Figure 3C). This provides preliminary information to suggest that the genetic variation between these psyllid taxa is not due to geographical isolation but to host plant specificity. The sampling of Carmichaelia in this study was insufficiently extensive in terms of either species diversity or geography to confirm this possibility. Eight of the 17 Carmichaelia species recorded in New Zealand [53,54] were examined, from only 11 locations (10 in South Island and one in North Island). In a similar situation to that of P. apicalis, Tuthill (1952) reported morphological variation among P. carmichaelia populations, which he considered indicative of a subspecies, P. carmichaeliae indistincta, and part of what he defined as an “inseparable complex” [14]. An undescribed Psylla aff. carmichaeliae collected from C. torulosa has also been listed amongst the endangered Hemipterans of New Zealand [55]. This study provides a useful start towards resolving this complex of psyllids; a more extensive sampling of psyllids from Carmichaelia spp. is a priority for research and conservation.
3.2.2. The Ctenarytaina fuchsiae Complex and Its Distribution Show a Strict Geographical Isolation
High COI sequence diversity was also found amongst New Zealand Ctenarytaina. A pronounced example was in Ctenarytaina fuchsiae from Fuchsia excorticata. Sampling of Fuchsia in 22 locations across widespread sites in the South and Stewart Islands revealed a single, broadly distributed lineage of C. fuchsia (COI intraspecific divergence at <2%). The exception to this pattern was a lineage showing 12% COI divergence from the other South Island populations, which was restricted to plants growing within a few kilometers of one another on the Kaikoura coastline (C. fuchsia sp. B, Table 2). A single C. fuchsia population collected in the North Island was also very distantly related (22% COI variation) to both South Island taxa (C. fuchsia sp. C, Table 2).
Given that we obtained only a single non-South Island sample, there may be further divergent C. fuchsia lineages in North Island. The Kaikoura population may represent a recent colonization from the north, especially given that the insects are found immediately alongside the main arterial route from North to South. On the other hand, it is puzzling that such a small and apparently mobile insect does not have a more homogenous distribution across the country, especially when considering that some of these insects are known to use wind currents to cover distances of hundreds of kilometers [56]. The basis for the restricted distribution of the Kaikoura C. fuchsia is unclear.
3.2.3. Are Psyllid Cryptic Taxa Associated with Host Plant Cryptic Species?
The greatest number of psyllid cryptic taxa was recorded among Ctenarytaina from the tea trees mānuka and kānuka. Clusters of psyllid taxa were observed to be morphologically consistent with the previous records of C. clavata (four COI lineages) and C. pollicaris (six COI lineages). The C. pollicaris cluster included Ctenarytaina “short” [15], despite the morphology and color of this taxa being distinctive (black with long female terminalia versus yellow with short female terminalia, respectively). These two taxa were found together more than once on the same individual plant, but only in South Island. Identification of the mānuka and kānuka Ctenarytaina taxa was very difficult based on morphology alone. One of the few observed differences was a darker brownish coloration in the body of “Ctenarytaina clavata D” compared with other lineages of C. clavata that tended towards a dark orange. Ctenarytaina clavata D was also collected only from the North Island (in three locations from Wellington to Tongariro). Interestingly, Ctenarytaina clavata D was the only instance where a divergence of 3% between population 334 and 335/402 has been considered intra-specific variation and not inter-specific (Figure 1). Morphological similarity and the immediate proximity of populations 334 and 335 on the same host plant species suggest that these represent a single taxon.
Dale recognized that her accounts of four Ctenarytaina species on mānuka and kānuka was not a complete record of psyllids from these plants species [15] and, further, that observed variation might be due to geographical isolation and/or hybridizations of Ctenarytaina clavata and C. pollicaris [15].
Kānuka was recently reclassified into genus Kunzea while mānuka remains in Leptospermum [57]. Molecular genetic analysis indicated the presence of possible cryptic species within the genus Kunzea [57] while many variants and subspecies have been recorded from mānuka [58]. Certainly, the cryptic genetic divergence of the mānuka and kānuka Ctenarytaina taxa could suggest psyllid speciation at an early stage is underway. This highlights the need for a deeper field collection to determine possible relationships between Ctenarytaina and New Zealand tea tree taxa.
The finding of a new Ctenarytaina taxon, Ctenarytaina sp. “A” from Olearia appears to represent a different evolutionary process within this genus. Olearia is not known to harbor psyllids from Ctenarytaina, although several Trioza and Anomalopsylla possess Olearia hosts. This taxon was clearly genetically distinct (12–17%) from both the “C. clavata complex” and from the “C. pollicaris/short complex” (Figure 1). This Ctenarytaina may well represent a significant host switch within New Zealand. Despite having found only a single population, more than 10 individuals were collected from the plant together with nymphs, which strongly suggests a true host association. Repeat sampling from this location and monitoring the progress of nymphs is warranted.
With the new taxa proposed here, a total of at least 21 Ctenarytaina taxa may be present in New Zealand, making this the second most numerous genus in the country. The much larger psyllid fauna of Australia, comprising more than 400 species, includes only seven Ctenarytaina species [2].
3.2.4. Other Instances of Multiple Taxa on the Same Host Plant
Within Trioza, four lineages were detected which we could not reconcile with prior records from New Zealand. In two of these cases, both morphological and COI data suggested a strong similarity with formerly described species: T. acuta from Ozothamnus leptophyllus and T. falcata from Aristotelia fruticosa. These new taxa are consequently labelled as T. acuta B and T. falcata B (Table 2). It was not possible to label any of the lineages as the previously described species.
The two other new Trioza taxa possibly represent speciation onto closely related host plants. The first Trioza (sp. A) was from Pittosporum divaricartum; the closest relative to this psyllid, based on COI similarity, was T. vitreoradiata, which is widespread and highly abundant on a range of Pittosporum species but has not been reported from P. divaricartum [15]. The second Trioza (sp. C), from Pseudopanax edgerlii, was distinct from other known psyllids on Pseudopanax. Three Trioza taxa have been reported from family Araliaceae: Trioza irregularis, on Pseudopanax arboreus [13], T. panacis on Pseudopanax crassifolius (Maskell 1890) and T. schefflericola on Schefflera digitata (Araliaceae) [14]. In general, delineation of psyllid taxa within the “Pseudopanax-Schefflera host plant group” was poor, from both morphological and molecular assessments. For example, Trioza samples from S. digitata had COI sequences less than 2% different to those from Trioza irregularis, perhaps suggesting within-species variation. The validity of Trioza schefflericola has previously been placed in doubt by Tuthill himself [14,15], with many of the differences between the species described as “slight differences of degree” [14]. The differentiation of Trioza species from Pseudopanax and Schefflera will require more extensive sampling and possibly finer scale molecular markers. Nonetheless, both Trioza taxa were morphologically distinguishable (e.g., variation in the terminalia [15]).
Among psyllids hosted by adventive plant species, a single new psyllid species was identified. Two distinct taxa were detected on Acacia baileyana with COI sequence divergence of 22% and with clear morphological differences (e.g., male parameres longer in the new species compared to A. acaciaebaileyanae). The psyllid Acizzia acaciaebaileyanae has been described from Acacia baileyana (as Psyllia uncata [13]) in both New Zealand and Australia, but a second Acizzia lineage on this host has never been described. This appears to be another example of a native Australian psyllid which is described in New Zealand before being identified among the Australian psyllid fauna, as occurred for A. hakeae [17]. This highlights the incomplete knowledge of both the New Zealand and Australian psyllid faunas.
4. Conclusions
The application of different species delimitation methods produced different results, sometimes in contrast with each other. This suggest that, despite being broadly used for psyllids [12,29,49], a COI 3% uncorrected p-distance between taxa should be combined with at least a species delimitation method, such as ABGD or GMYC, to be considered indicative of species separation. Surely, integration of COI barcoding, different species delimitation methods, and information from host plant association and morphology resulted to be a quick and valuable approach for identification of New Zealand psyllids.
As a result, 21 cryptic taxa are reported here for the first time, confirming COI barcoding as a fundamental tool for understanding New Zealand psyllid biodiversity while providing new material for the study of the evolution of New Zealand terrestrial fauna [59].
This COI dataset will provide an improved ability to recognize new psyllid introductions and host plant associations. Any biosecurity issues may thus be acknowledged sooner. The molecular data from this work, together with additional gene sequences and biological observation provides the foundation for more sophisticated phylogenetic and ecological studies such as those recently conducted on other psyllid fauna [30] and their associated microbiomes [27,60].
Supplementary Materials
Figure S1: COI gene tree (ML, 10,000 replicates, nodes <70% not reported). Table S1: Field collections.
Author Contributions
Conceptualization, F.M., S.B., A.P., K.A. and G.T.; Methodology, F.M.; Formal Analysis, F.M.; Resources, S.B., A.P. and K.A.; Writing–Original Draft Preparation, F.M.; and Writing–Review and Editing, F.M., A.P., S.B. and K.A.
Funding
This research was sponsored by the Australian Plant Biosecurity Cooperative Research Center (PBCRC) and by a Bio-Protection Research Center (BPRC) Writing Scholarship awarded to FM.
Acknowledgments
The authors would like to thank three anonymous reviewers for the useful comments and helpful suggestions. Thanks to Ines Schoenberger and the staff of the Allan Herbarium (Lincoln, New Zealand) for the help in the plant identification. For the help in the psyllid collections, the authors would also thank Samuel and William Brown; Hannah Evans and Simon Fowler (AgResearch, New Zealand); Peter Gillespie (Orange Agricultural Institute, Australia); Sarah Thompson (Plant and Food Research, New Zealand); Mark Blacket, Isabel Valenzuela and Alan Yen (AgriBio, Melbourne, Australia); and Shaun Bennett (MPI, New Zealand). Part of the data published in this article was reported in FM’s PhD thesis [61].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Burckhardt, D.; Ouvrard, D. A revised classification of the jumping plant–lice (Hemiptera: Psylloidea). Zootaxa 2012, 3509, 1–34. [Google Scholar] [CrossRef]
- Ouvrard, D. Psyl’list—The World Psylloidea Database. Available online: Http://www.Hemiptera-databases.Com/psyllist (accessed on 17 April 2018).
- Hollis, D. Australian Psylloidea: Jumping Plant Lice and Lerp Insects; Australian Biological Resources Study: Canberra, Australia, 2004; pp. 1–216. [Google Scholar]
- Linné, C. Systema Naturae per Regna tri Naturae, Secundum Classes, Ordines, Genera, Species, cum Characteribus, Differentiis, Synonymis, Locis, 10th ed.; Impensis Direct: Stockholm, Sweden, 1758; pp. 1–824. [Google Scholar]
- EPPO/CABI. Quarantine Pests for Europe, 2nd ed.; Smith, I.M., McNamara, D.G., Scott, P.R., Holderness, M., Eds.; CABI International: Wallingford, UK, 1997; p. 1425. [Google Scholar]
- Aubert, B. Trioza erytreae Del Guercio and Diaphorina citri Kuwayama (Homoptera: Psylloidea), the two vectors of citrus greening disease: Biological aspects and possible control strategies. Fruits 1987, 42, 149–162. [Google Scholar]
- Capoor, S.P.; Rao, D.G.; Viswanath, S.M. Diaphorina citri Kuwayama, a vector of the greening disease of citrus in India. Indian J. Agric. Sci. 1967, 37, 572–576. [Google Scholar]
- Syfert, M.M.; Serbina, L.; Burckhardt, D.; Knapp, S.; Percy, D.M. Emerging new crop pests: Ecological modelling and analysis of the South American potato psyllid Russelliana solanicola (Hemiptera: Psylloidea) and its wild relatives. PLoS ONE 2017, 12, e0167764. [Google Scholar] [CrossRef] [PubMed]
- Liefting, L.W.; Weir, B.S.; Pennycook, S.R.; Clover, G.R.G. ‘Candidatus Liberibacter solanacearum’, associated with plants in the family solanaceae. Int. J. Syst Evol. Microbiol. 2009, 59, 2274–2276. [Google Scholar] [CrossRef] [PubMed]
- Teulon, D.A.J.; Workman, P.J.; Thomas, K.L.; Krause Nielsen, M.-C. Bactericera cockerelli: Incursion, dispersal and current distribution on vegetable crops in New Zealand. N. Z. Plant. Prot. 2009, 62, 136–144. [Google Scholar]
- Vereijssen, J.; Smith, G.R.; Weintraub, P.G. Bactericera cockerelli (Hemiptera: Triozidae) and Candidatus Liberibacter solanacearum in potatoes in New Zealand: Biology, transmission, and implications for management. J. Integr. Pest. Manag. 2018, 9, 1–21. [Google Scholar] [CrossRef]
- Taylor, G.S.; Fagan-Jeffries, E.P.; Austin, A.D. A new genus and twenty new species of Australian jumping plant-lice (Psylloidea: Triozidae) from Eremophila and Myoporum (Scrophulariaceae: Myoporeae). Zootaxa 2016, 4073, 1–84. [Google Scholar] [CrossRef] [PubMed]
- Ferris, G.F.; Klyver, F.D. Report upon a collection of chermidae (Homoptera) from New Zealand. Trans. N. Z. Inst. 1932, 63, 34–61. [Google Scholar]
- Tuthill, L.D. On the psyllidae of New Zealand (Homoptera). Pac. Sci. 1952, 6, 83–125. [Google Scholar]
- Dale, P.J. A review of the Psylloidea (Insecta: Hemiptera) of the New Zealand Subregion. Unpublished Ph.D. Thesis, University of Auckland, Auckland, New Zealand, 1985. [Google Scholar]
- Macfarlane, R.P.; Andrew, I.G.; Berry, J.A.; Johns, P.M.; Hoare, R.J.B.; Lariviere, M.-C.; Grenslade, P.; Henderson, R.C.; Smithers, C.N.; Palma, R.L.; et al. Phylum Arthropoda, subphylum Hexapoda: Protura, springtails, Diplura, insects. N. Z. Inventory Biodiv. 2010, 2, 233–467. [Google Scholar]
- Martoni, F.; Burckhardt, D.; Armstrong, K. An annotated checklist of the psyllids of New Zealand (Hemiptera: Psylloidea). Zootaxa 2016, 4144, 556–574. [Google Scholar] [CrossRef] [PubMed]
- Munoz, J.; Felicisimo, A.M.; Cabezas, F.; Burgaz, A.R.; Martinez, I. Wind as a long-distance dispersal vehicle in the Southern Hemisphere. Science 2004, 304, 1144–1147. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.D.B. Gone with the wind: Westward dispersal across the Indian Ocean and island speciation in Hemicordulia dragonflies (Odonata: Corduliidae). Zootaxa 2007, 1438, 27–48. [Google Scholar] [CrossRef]
- Close, R.C.; Moar, N.T.; Tomlinson, A.I.; Lowe, A.D. Aerial dispersal of biological material from Australia to New Zealand. Int. J. Biometeorol. 1978, 22, 1–19. [Google Scholar] [CrossRef]
- Tomlinson, A.I. Meteorological aspects of trans-Tasman insect dispersal. N. Z. Entomol. 1973, 5, 253–268. [Google Scholar] [CrossRef]
- Buckley, T.R.; Krosch, M.; Leschen, R.A.B. Evolution of New Zealand insects: Summary and prospectus for future research. Aust. Entomol. 2015, 54, 1–27. [Google Scholar] [CrossRef]
- Maskell, W.M. On some species of Psyllidae in New Zealand. Trans. N. Z. Inst. 1890, 22, 157–170. [Google Scholar]
- Šulc, K. Trioza cockerelli n. Sp, novinka ze severní ameriky, mající i hospodářský význam [Trioza cockerelli n. Sp., a novelty from North America, being also of economic importance]. Acta Soc. Entomol. Bohem. 1909, 6, 102–108. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B-Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Gregory, T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005, 54, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.A.G.; Morrow, J.L.; Fromont, C.; Steinbauer, M.J.; Taylor, G.S.; Johnson, S.N.; Cook, J.M.; Riegler, M. Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts. Environ. Microbiol. 2016, 18, 2591–2603. [Google Scholar] [CrossRef] [PubMed]
- Martoni, F.; Bulman, S.R.; Pitman, A.R.; Armstrong, K. Elongation factor-1α accurately reconstructs relationships amongst psyllid families (Hemiptera: Psylloidea), with possible diagnostic implications. J. Econ. Entomol. 2017, 110, 2618–2622. [Google Scholar] [CrossRef] [PubMed]
- Percy, D.M. Making the mosts of your host: The Metrosideros-feeding psyllids (Hemiptera: Psylloidea) of the Hawaiian Islands. ZooKeys 2017, 649, 1–163. [Google Scholar] [CrossRef] [PubMed]
- Percy, D.M.; Crampton-Platt, A.; Sveinsson, S.; Lemmon, A.R.; Moriarty Lemmon, E.; Ouvrard, D.; Burckhardt, D. Resolving the psyllid tree of life: Phylogenomic analyses of the superfamily Psylloidea (Hemiptera). Syst. Entomol. 2018. [Google Scholar] [CrossRef]
- Crosby, T.K.; Dugdale, J.S.; Watt, J.C. Area codes for recording specimen localities in the New Zealand subregion. N. Z. J. Zool. 1998, 25, 175–183. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Simon, C.; Frati, F.; Beckenbach, A.; Crespi, B.; Liu, H.; Flook, P. Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann. Entomol. Soc. Am. 1994, 87, 651–701. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit i from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–297. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. Mega6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Srivathsan, A.; Meier, R. On the inappropriate use of Kimura-2-parameter (K2P) divergences in the DNA–barcoding literature. Cladistics 2012, 28, 190–194. [Google Scholar] [CrossRef]
- Collins, R.A.; Boykin, L.M.; Cruickshank, R.H.; Armstrong, K. Barcoding’s next top model: An evaluation of nucleotide substitution models for specimen identification. Methods Ecol. Evol. 2012, 3, 457–465. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. Abgd, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.; Barraclough, T.; Gomez-Zurita, J.; Cardoso, A.; Duran, D.; Hazell, S.; Kamoun, S.; Sumlin, W.; Vogler, A. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Kapli, P.; Lutteropp, S.; Zhang, J.; Kobert, K.; Pavlidis, P.; Stamatakis, A.; Flouri, T. Multi-rate poisson tree processes for singlelocus species delimitation under maximum likelihood and markov chain monte carlo. Bioinformatics 2017, 33, 1630–1638. [Google Scholar] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with Beauti and the Beast 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Michonneau, F. Cryptic and not-so-cryptic species in the complex “Holothuria (Thymiosycia) imaptiens” (Forsskål, 1775) (Echinodermata: Holothuroidea: Holothuriidae). bioRxiv 2015, 014225. [Google Scholar]
- Rambaut, A. Figtree v1.4.3. 2016. Available online: http://tree.Bio.Ed.Ac.Uk/software/figtree/ (accessed on 4 October 2016).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Ezard, T.; Fujisawa, T.; Barraclough, T.G. Splits: Species’ Limits by Threshold Statistics. R Package Version 1.0-18/r45. Available online: http://r-forge.R-project.Org/projects/splits/ (accessed on 15 January 2009).
- Burckhardt, D.; Ouvrard, D.; Queiroz, D.; Percy, D. Psyllid host-plants (Hemiptera: Psylloidea): Resolving a semantic problem. Fla. Entomol. 2014, 97, 242–246. [Google Scholar] [CrossRef]
- Wonglersak, R.; Cronk, Q.C.; Percy, D. Salix transect of Europe: Structured genetic variation and isolation-by-distance in the nettle psyllid, Trioza urticae (Psylloidea, Hemiptera), from Greece to Arctic Norway. Biodivers. Data J. 2017, 5, e10824. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; He, L.J.; Zhang, A.B. A simple 2d non-parametric resampling statistical approach to assess confidence in species identification in DNA barcoding-an alternative to likelihood and bayesian approaches. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Luo, A.R.; Lan, H.Q.; Ling, C.; Zhang, A.B.; Shi, L.; Ho, S.Y.W.; Zhu, C.D. A simulation study of sample size for DNA barcoding. Ecol. Evol. 2015, 5, 5869–5879. [Google Scholar] [CrossRef] [PubMed]
- Ross, H.A.; Murugan, S.; Li, W.L.S. Testing the reliability of genetic methods of species identification via simulation. Syst. Biol. 2008, 57, 216–230. [Google Scholar] [CrossRef] [PubMed]
- Heenan, P.B. Taxonomic revision of Carmichaelia (Fabaceae-Galegeae) in New Zealand 1. N. Z. J. Bot. 1995, 33, 455–475. [Google Scholar] [CrossRef]
- Heenan, P.B. A taxonomic revision of Carmichaelia (Fabaceae-Galegeae) in New Zealand 2. N. Z. J. Bot. 1996, 34, 157–177. [Google Scholar] [CrossRef]
- Stringer, I.A.N.; Hitchmough, R.A.; Lariviere, M.C.; Eyles, A.C.; Teulon, D.A.J.; Dale, P.J.; Henderson, R.C. The conservation status of New Zealand Hemiptera. N. Z. Entomol. 2012, 35, 110–115. [Google Scholar] [CrossRef]
- Yen, A.L.; Finlay, K.J.; Weiss, J.; Vereijssen, J. Understanding the Significance of Natural Pathways into Australia and New Zealand; Plant Biosecurity Cooperative Research Centre: Bruce, Australia, 2014; p. 182. [Google Scholar]
- De Lange, P.J. A revision of the New Zealand Kunzea ericoides (Myrtaceae) complex. Phytokeys 2014, 40, 1–185. [Google Scholar] [CrossRef] [PubMed]
- Stephens, J.M.C.; Molan, P.C.; Clarkson, B.D. A review of Leptospermum scoparium (Myrtaceae) in New Zealand. N. Z. J. Bot. 2005, 43, 431–449. [Google Scholar] [CrossRef]
- Goldberg, J.; Trewick, S.A.; Paterson, A.M. Evolution of New Zealand’s terrestrial fauna: A review of molecular evidence. Philos. Trans. R. Soc. B 2008, 363, 3319–3334. [Google Scholar] [CrossRef] [PubMed]
- Morrow, J.L.; Hall, A.A.G.; Riegler, M. Symbionts in waiting: The dynamics of incipient endosymbiont complementation and replacement in minimal bacterial communities of psyllids. Microbiome 2017, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Martoni, F. Biodviersity, Evolution and Microbiome of the New Zealand Psylloidea (Hemiptera: Sternorrhyncha). Ph.D. Thesis, Lincoln University, Lincoln, New Zealand, 2017. [Google Scholar]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).