Recent Trends in Food Quality and Authentication: The Role of Omics Technologies in Dairy and Meat Production
Abstract
1. Introduction
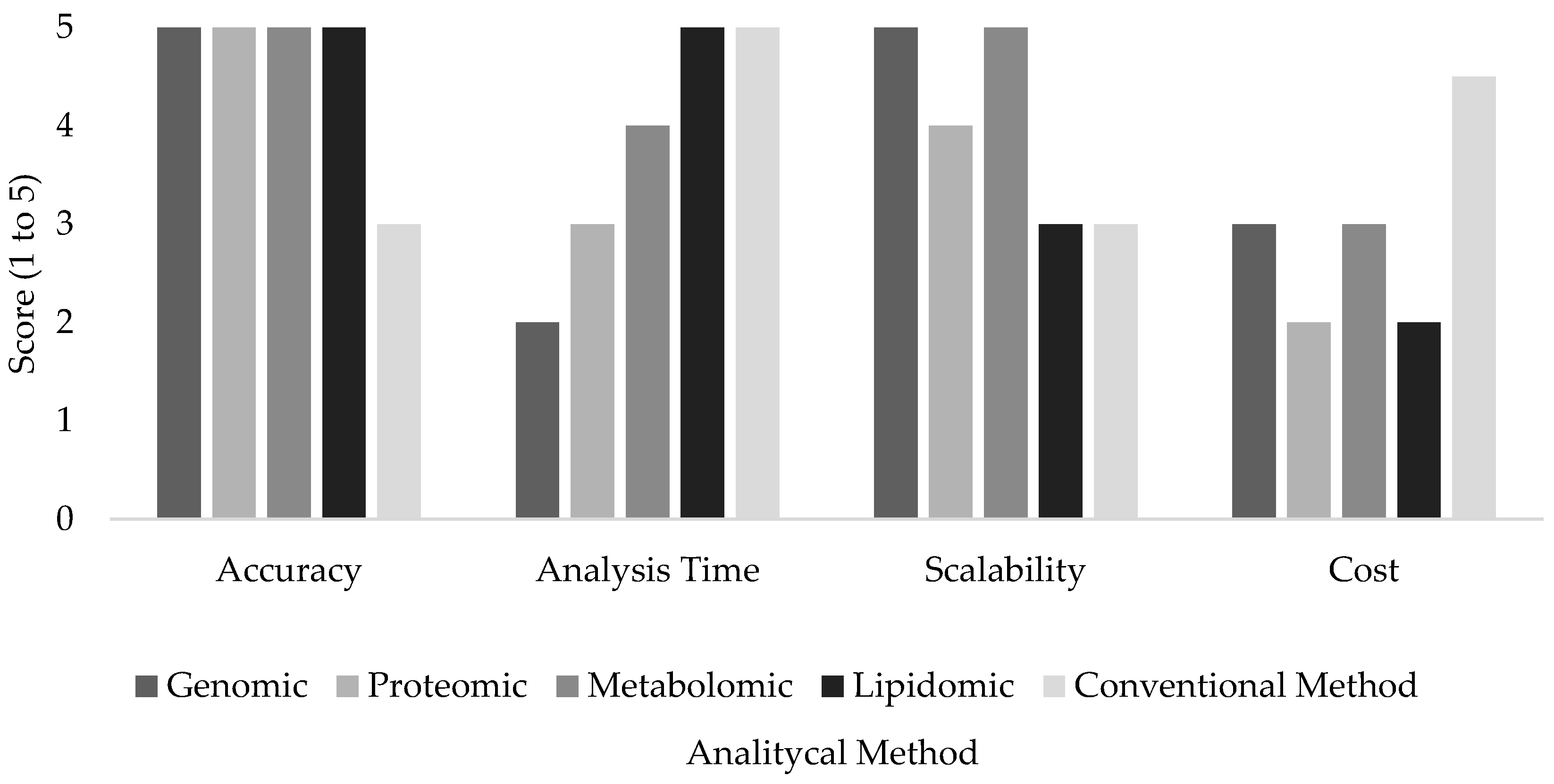
2. Overview of Omics Technologies
2.1. Current Development of Omics Technologies
2.2. Key Omics Disciplines and Analytical Approaches
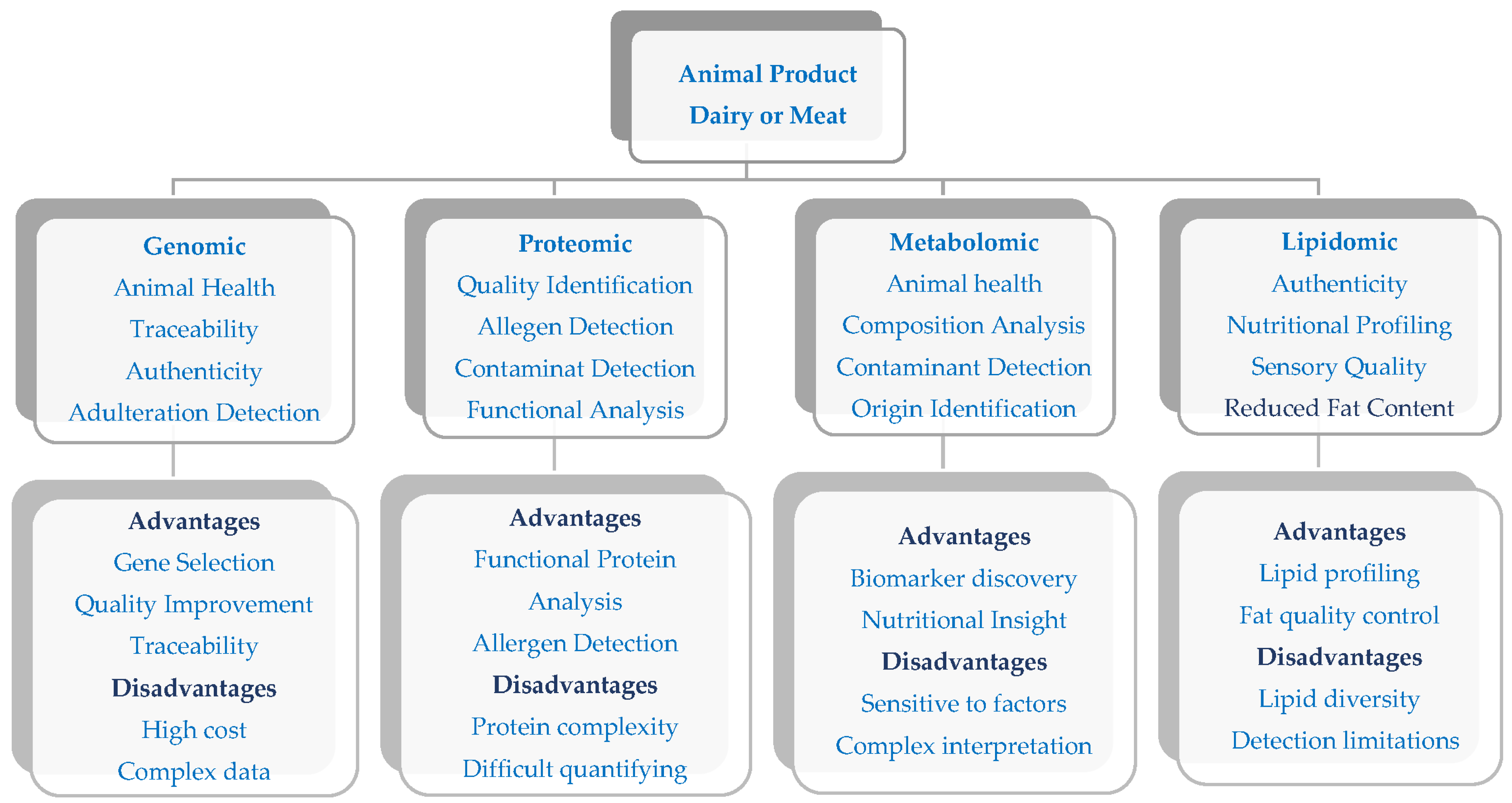
3. Application of Omics Technologies in Dairy and Meat Production
3.1. Integrative Role of Omics in Food Quality, Nutritional Profiling, and Product Authenticity
3.1.1. Genomics
| Food Matriz | Techniques/Methods | Results | Application | References |
|---|---|---|---|---|
| Milk and Dairy Products | Real-time PCR targeting mitochondrial 12S rRNA and cytB genes | High sensitivity and specificity for detecting species such as Bos taurus, Ovis aries, Bubalus bubalis, and Capra hircus. Detection limit <1% | Origin Identification | [85] |
| Specific TaqMan probes for the detection of cow and mare DNA | Accurate and reproducible detection in koumiss and yogurt | Origin Identification | [86] | |
| Weighted single-step GWAS using genomic breeding values | Identification of 141 novel genes related to milk production and 5 associated with Somatic Cell Score | Quality Identification | [87] | |
| PCR-RFLP targeting κ-casein gene (CSN3) | Differentiation between cow, goat, and sheep milk | Authenticity Identification | [88] | |
| Conventional and real-time PCR for milk powder using 12S rRNA gene | Verification of mitochondrial DNA integrity in powdered milk | Adulteration Detection | [62] | |
| Meat | CNV and CNVR detection using Bovine HD SNP array | Identification of 112,198 CNVs and 10,102 CNVRs, including regions related to backfat color and thickness | Composition Analysis | [89] |
| GWAS and pathway-based analysis using GeneSeek Genomic Profiler Bovine LD array | 37 significant SNPs associated with 12 traits in Piedmontese cattle | Quality Identification | [90] | |
| Genome-wide association studies (GWAS) on carcass traits | Identification of SNPs and genes linked to growth, muscle development, and meat quality | Quality Identification | [81] | |
| Multiplex PCR for meat authentication (7 species) | High reproducibility even in heat-processed meat; low detection limits | Authenticity Identification | [91] | |
| Two-tube hexaplex PCR for 12 meat species | Molecular identification of up to 12 species in adulterated meat mixtures | Adulteration Detection | [92] |
3.1.2. Proteomics
| Food Matriz | Techniques/Methods | Results | Application | References |
|---|---|---|---|---|
| Milk | nanoRP-UPLC-ESI-MS/MS; trypsin digestion; DIA and DDA acquisition | Identification of 132 modified peptides in 62 proteins (14 Age types). Increase in AGEs with processing severity, stable during storage. Formyl lysine was predominant | Quality Identification | [118] |
| MALDI-TOF MS with reference spectra from >150 samples | Identification of animal species in feta and mozzarella cheeses. Proteomic modulation observed during mastitis. | Quality Identification | [119] | |
| Meat | MALDI-TOF MS on Longissimus thoracis from heifers and steers | Validation of MALDI-TOF MS to differentiate cow, sheep, goat, and buffalo milk in cheeses | Quality Identification | [120] |
| 2D-PAGE, mass spectrometry, bioinformatics | Identification of Pediococcus and Lactobacillus strains capable of reducing β-lactoglobulin sensitization and hydrolyzing allergenic fragments | Composition Analysis | [121] | |
| OFFGEL electrophoresis (pH 4–7) | Four protein bands (Desmin, Pyruvate kinase, Myosin light/heavy chains) differentiated high vs. normal pH meats | Composition Analysis | [122] | |
| Trypsin/Lys-C digestion, LC-MS/MS (Q Exactive™ HF Orbitrap™) | Identification of 36 peaks in Uniprot database from meat exudates | Allergen Detection | [96] | |
| Shotgun proteomics of Longissimus thoracis in Arouquesa cattle | Proteins like HSP70 and laminin correlated with oxidative muscle stability | Authenticity Identification | [123] | |
| LC-MS for protein identification | Biomarkers such as VIM, FSCN1, SERPINH1, ALDH1A1, MYH4 identified as meat quality indicators | Quality Identification | [124] | |
| SDS-PAGE with image-based protein band quantification | Integration with OFFGEL electrophoresis and MS enabled high-resolution profiling of myofibrillar proteins | Quality Identification | [125] |
3.1.3. Metabolomics
| Food Matriz | Techniques/Methods | Results | Application | References |
|---|---|---|---|---|
| Milk | NMR-based metabolomics; different lactation stages in Friesian and native cows | Identification of 2355 chemical compounds, providing detailed chemical characterization | Composition Analysis | [145] |
| LC-MS, ICP-MS, and NMR | Discriminate metabolites include lipids (fatty acids, phospholipids), amino acids, and plant-derived compounds. | Composition Analysis | [146] | |
| LC-MS/MS targeted metabolomics | 296 metabolites identified in commercial bovine milk, with 1447 unique structures | Contaminant Detection | [147] | |
| Direct injection MS and LC-MS/MS for Aflatoxin M1 (AFM1) detection | AFM1 levels increased in milk from cows fed AFB1-contaminated diets | Contaminant Detection | [148] | |
| GC-FID and LC-MS for raw milk from healthy and subclinical ketosis cows | Increased tyrosine, leucine, carnitine, acetone in subclinical ketosis; reduced galactose-1-phosphate | Animal Health | [149] | |
| LC-MS/MS untargeted metabolomics | Decrease in creatinine, taurine, α-ketoglutarate in cows with subclinical ketosis | Composition Analysis | [150] | |
| Meat | GC-MS and UHPLC-MS for beef and pork adulteration | Biomarkers such as leucine and creatine used for aging assessment | Adulteration Detection | [36] |
| UPLC–MS/MS for beef muscle lipid profile | Correlation between degree of unsaturation in lipids and meat quality (unsaturated fatty acids, melting point) | Quality Identification | [151] | |
| NMR-based metabolomic profiling of beef (Nellore vs. Angus × Nellore) | Identification of 31 metabolites, including carnosine, betaine, and glycerol, correlating with sensory traits like flavor and tenderness | Composition Analysis | [152] | |
| UPLC-Orbitrap-MS and GC-MS for beef origin differentiation | 24 metabolites identified as markers to differentiate beef origin (Australia, Japan, USA) | Origin Identification | [153] |
3.1.4. Lipidomics
| Food Matriz | Techniques/Methods | Results | Application | References |
|---|---|---|---|---|
| Milk | LC-MS/MS analysis of phospholipids, sphingolipids, glycolipids, and ceramides | Identification of 514 lipid species across 15 classes | Composition analysis | [158] |
| Infusion-electrospray mass spectrometry for triacylglycerides | Detection and quantification of over 100 TAG species in milk | Quality identification | [159] | |
| ¹H-NMR and 1D TOCSY | Higher levels of α-linolenic acid, linoleic acid, and unsaturated fatty acids in organic milk; CLA isomers (9-cis, 11-trans) more abundant | Authenticity identification | [160] | |
| UPLC-Q-Exactive Orbitrap-MS | Soy milk: rich in phospholipids (PC, PE, PS, PG); Goat milk: high in MCTs, ω-3 and ω-6; Cow milk: intermediate lipid profile; 14 lipids identified as biomarkers | Adulteration detection | [161] | |
| UHT Milk Reconstituted whole milk | UPLC-Q-Exactive Orbitrap-MS | Major lipid classes: PC (120 µmol/L), PE (150 µmol/L), SM (90 µmol/L) | Composition analysis | [162] |
| Meat | Intelligent surgical knife (iKnife) with REIMS | High precision lipid profiling (CV < 15% for most TAGs) | Quality Identification | [163] |
| DART-QTOF (+) and LC-ESI-QTOF (+/−) | DART-QTOF: 852 peaks, 62 differential; LC-ESI-QTOF: 879 peaks, 165 differential; Clear clustering by country of origin (e.g., Brazil vs Canada) | Origin identification | [164] | |
| UHPLC-HRMS in positive and negative ion modes | Negative mode: detection of fatty acids, phospholipids, sphingolipids; Positive mode: phospholipids and glycerolipids | Authenticity identification | [165] |
3.2. Challenges and Opportunities
3.2.1. Challenges
3.2.2. Opportunities
3.3. Future Perspectives and Research Directions
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lorenzo, J.M.; Munekata, P.E.S.; Gómez, B.; Barba, F.J.; Mora, L.; Pérez, C.; Toldrá, F. Bioactive peptides as natural antioxidants in food products-A review. Trends Food Sci. Technol. 2018, 79, 136–147. [Google Scholar] [CrossRef]
- Khan, I.T.; Nadeem, M.; Imran, M.; Ayaz, M.; Ullah, R.; Ajmal, M.; Jaspal, M.H. Antioxidant properties of Milk and dairy products: A comprehensive review of the current knowledge. Lipids Health Dis. 2019, 18, 41. [Google Scholar] [CrossRef]
- Statista. Cattle Population Worldwide 2012–2022. Available online: https://www.statista.com/statistics/263979/global-cattle-population-since-1990/ (accessed on 6 November 2024).
- Qian, J.; Dai, B.; Wang, B.; Zha, Y.; Song, Q. Traceability in food processing: Problems, methods, and performance evaluations-A review. Crit. Rev. Food Sci. Nutr. 2020, 62, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ozturk-Kerimoglu, B.; He, L.; Zhang, M.; Pan, J.; Liu, Y.; Zhang, Y.; Huang, S.; Wu, Y.; Jin, G. Advanced lipidomics in the modern meat industry: Quality traceability, processing requirement, and health concerns. Front. Nutr. 2022, 9, 925846. [Google Scholar] [CrossRef]
- Espinales, C.; Baldeón, M.; Bravo, C.; Toledo, H.; Carballo, J.; Romero-Peña, M.; Cáceres, P.J. Strategies for Healthier Meat Foods: An Overview. Prev. Nutr. Food Sci. 2024, 29, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Darshna, D.; Tiwari, S.S. Industrial Microbiology and Biotechnology: Emerging concepts in Microbial Technology, 1st ed.; Springer Nature: Singapore, 2023; pp. 1–22. [Google Scholar] [CrossRef]
- Sidira, M.; Smaoui, S.; Varzakas, T. Recent Proteomics, Metabolomics and Lipidomics Approaches in Meat Safety, Processing and Quality Analysis. Appl. Sci. 2024, 14, 5147. [Google Scholar] [CrossRef]
- Farré, M. Mass Spectrometry in Food and Environmental Chemistry. In The Handbook of Environmental Chemistry, 1st ed.; Springer: Cham, Switzerland, 2022; pp. 187–224. [Google Scholar] [CrossRef]
- Suravajhala, P.; Goltsov, A. Three Grand Challenges in High Throughput Omics Technologies. Biomolecules 2022, 12, 1238. [Google Scholar] [CrossRef]
- Gonesh, C.; Saha, M.B.; Khan, M.J.; Saha, H.; Avinash, D.K. Integrative Analysis of Multi-Omics Data with Deep Learning: Challenges and Opportunities in Bioinformatics. J. Propuls. Technol. 2023, 44, 1384–1389. [Google Scholar]
- Molla, G.; Bitew, M. Revolutionizing Personalized Medicine: Synergy with Multi-Omics Data Generation, Main Hurdles, and Future Perspectives. Biomedicines 2024, 12, 2750. [Google Scholar] [CrossRef]
- Kaur, P.; Singh, A.; Chana, I. Computational Techniques and Tools for Omics Data Analysis: State-of-the-Art, Challenges, and Future Directions. Arch. Computat. Methods Eng. 2021, 28, 4595–4631. [Google Scholar] [CrossRef]
- Brito, L.F.; Bedere, N.; Douhard, F.; Oliveira, H.R.; Arnal, M.; Peñagaricano, F.; Schinckel, A.P.; Baes, C.F.; Miglior, F. Review: Genetic selection of high-yielding dairy cattle toward sustainable farming systems in a rapidly changing world. Animal 2021, 15 (Suppl. S1), 100292. [Google Scholar] [CrossRef] [PubMed]
- Zavadilová, L.; Kašná, E.; Krupová, Z.; Klímová, A. Health traits in current dairy cattle breeding: A review. Czech J. Anim. Sci. 2021, 66, 235–250. [Google Scholar] [CrossRef]
- Tarbeeva, S.; Kozlova, A.; Sarygina, E.; Kiseleva, O.; Ponomarenko, E.; Ilgisonis, E. Food for Thought: Proteomics for Meat Safety. Life 2023, 13, 255. [Google Scholar] [CrossRef] [PubMed]
- Mahrous, E.; Chen, R.; Zhao, C.; Farag, M.A. Lipidomics in food quality and authentication: A comprehensive review of novel trends and applications using chromatographic and spectroscopic techniques. Crit. Rev. Food Sci. Nutr. 2023, 64, 9058–9081. [Google Scholar] [CrossRef]
- Su, G.; Yu, C.; Liang, S.; Wang, W.; Wang, H. Multi-omics in food safety and authenticity in terms of food components. Food Chem. 2024, 437, 137943. [Google Scholar] [CrossRef]
- Mao, X.; Bassey, A.P.; Sun, D.; Yang, K.; Shan, K.; Li, C. Overview of omics applications in elucidating the underlying mechanisms of biochemical and biological factors associated with meat safety and nutrition. J Proteom. 2023, 276, 104840. [Google Scholar] [CrossRef]
- Gagaoua, M. Recent Advances in OMICs Technologies and Application for Ensuring Meat Quality, Safety and Authenticity. Foods 2022, 11, 2532. [Google Scholar] [CrossRef]
- Muguruma, Y.; Nunome, M.; Inoue, K. A review on the foodomics based on liquid chromatography mass spectrometry. Chem. Pharm. Bull. 2022, 70, 12–18. [Google Scholar] [CrossRef]
- Maru, D.; Kumar, A. Applications of Omics Technologies in Livestock Production, Improvement and Sustainability. In Sustainable Agriculture Reviews; Kumar Yata, V., Mohanty, A.K., Lichtfouse, E., Eds.; Springer: Cham, Switzerland, 2024; Volume 62, pp. 1–54. [Google Scholar] [CrossRef]
- Munekata, P.E.; Pateiro, M.; López-Pedrouso, M.; Gagauz, M.; Lorenzo, J.M. Foodomics in Meat Quality. Curr. Opin. Food Sci. 2021, 38, 79–85. [Google Scholar] [CrossRef]
- Singh, N.; Barthwal, R.; Negi, A.; Aggarwal, S.; Kathuria, D.; Kumar, V.; Paul, M. Foodomics: Futuristic Omic Strategies to Assess the Impact of Food Nutrients on Human Health and Gut Microbiome. Int. J. Food Sci. Technol. 2024, 59, 4194–4212. [Google Scholar] [CrossRef]
- Hwang, Y.H.; Lee, E.Y.; Lim, H.T.; Joo, S.T. Multi-Omics Approaches to Improve Meat Quality and Taste Characteristics. Food Sci. Anim. Resour. 2023, 43, 1067–1086. [Google Scholar] [CrossRef] [PubMed]
- Valdés, A.; Álvarez-Rivera, G.; Socas-Rodríguez, B.; Herrero, M.; Ibáñez, E.; Cifuentes, A. Foodomics: Analytical Opportunities and Challenges. Anal. Chem. 2022, 94, 366–381. [Google Scholar] [CrossRef] [PubMed]
- Gauly, M.; Ammer, S. Review: Challenges for Dairy Cow Production Systems Arising from Climate Changes. Animal 2020, 14, S196–S203. [Google Scholar] [CrossRef]
- Neto, J.B.S.; Mota, L.F.; Londoño-Gil, M.; Schmidt, P.I.; Rodrigues, G.R.; Ligori, V.A.; Baldi, F. Genotype-by-environment interactions in beef and dairy cattle populations: A review of methodologies and perspectives on research and applications. Anim. Genet. 2024, 55, 871–892. [Google Scholar] [CrossRef]
- Agregan, R.; Echegaray, N.; López-Pedrouso, M.; Kharabsheh, R.; Franco, D.; Lorenzo, J.M. Proteomic Advances in Milk and Dairy Products. Molecules 2021, 26, 3832. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes, A. Comprehensive Foodomics; Elsevier: Amsterdam, Netherlands, 2020; Available online: http://hdl.handle.net/10261/253309 (accessed on 14 January 2025).
- Kovac, J.; Rolon, M.L.; Naum, M.; Lampel, K.A. DNA-Based Assays. In Encyclopedia of Dairy Sciences, 3rd ed.; McSweeney, P.L.H., McNamara, J.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 356–362. [Google Scholar]
- Rao, M.S.; Van Vleet, T.R.; Ciurlionis, R.; Buck, W.R.; Mittelstadt, S.W.; Blomme, E.A.; Liguori, M.J. Comparison of RNA-Seq and Microarray Gene Expression Platforms for the Toxicogenomic Evaluation of Liver from Short-Term Rat Toxicity Studies. Front. Genet. 2019, 9, 636. [Google Scholar] [CrossRef]
- Valdés, A.; Cifuentes, A.; León, C. Foodomics Evaluation of Bioactive Compounds in Foods. Trends Anal. Chem. 2017, 96, 2–13. [Google Scholar] [CrossRef]
- Yu, Q.; Paulo, J.A.; Naverrete-Perea, J.; McAlister, G.C.; Canterbury, J.D.; Bailey, D.J.; Robitaille, A.M.; Huguet, R.; Zabrouskov, V.; Gygi, S.P.; et al. Benchmarking the Orbitrap Tribrid Eclipse for Next Generation Multiplexed Proteomics. Anal. Chem. 2020, 92, 6478–6485. [Google Scholar] [CrossRef]
- Hernández-Mesa, M.; D’Atri, V.; Barknowitz, G.; Fanuel, M.; Pezzatti, J.; Dreolin, N.; Ropartz, D.; Monteau, F.; Vigneau, E.; Rudaz, S.; et al. Interlaboratory and Interplatform Study of Steroids Collision Cross Section by Traveling Wave Ion Mobility Spectrometry. Anal. Chem. 2020, 92, 5013–5022. [Google Scholar] [CrossRef]
- Meier, F.; Park, M.A.; Mann, M. Trapped Ion Mobility Spectrometry and Parallel Accumulation-Serial Fragmentation in Proteomics. Mol. Cell Proteom. 2021, 20, 100138. [Google Scholar] [CrossRef]
- Yan, S.; Bhawal, R.; Yin, Z.; Thannhauser, T.W.; Zhang, S. Recent Advances in Proteomics and Metabolomics in Plants. Mol. Hortic. 2022, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Gross, R.W. Global Analyses of Cellular Lipidomes Directly from Crude Extracts of Biological Samples by ESI Mass Spectrometry: A Bridge to Lipidomics. J. Lipid Res. 2003, 44, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Balkir, P.; Kemahlioglu, K.; Yucel, U. Foodomics: A New Approach in Food Quality and Safety. Trends Food Sci. Technol. 2021, 108, 49–57. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, J.L. Mass-Spectrometry-Based Lipidomics. J. Sep. Sci. 2018, 41, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Han, X. Analytical Challenges of Shotgun Lipidomics at Different Resolution of Measurements. TrAC Trends Anal. Chem. 2019, 121, 115697. [Google Scholar] [CrossRef]
- Luque de Castro, M.D.; Quiles-Zafra, R. Lipidomics: An Omics Discipline with a Key Role in Nutrition. Talanta 2020, 219, 121197. [Google Scholar] [CrossRef]
- Wang, R.; Li, B.; Lam, S.M.; Shui, G. Integration of Lipidomics and Metabolomics for In-Depth Understanding of Cellular Mechanism and Disease Progression. J. Genet. Genom. 2020, 47, 69–83. [Google Scholar] [CrossRef]
- Wu, Z.; Bagarolo, G.I.; Thoröe-Boveleth, S.; Jankowski, J. “Lipidomics”: Mass Spectrometric and Chemometric Analyses of Lipids. Adv. Drug Deliv. Rev. 2020, 159, 294–307. [Google Scholar] [CrossRef]
- Wu, B.; Wei, F.; Xu, S.; Xie, Y.; Lv, X.; Chen, H.; Huang, F. Mass Spectrometry-Based Lipidomics as a Powerful Platform in Foodomics Research. Trends Food Sci. Technol. 2021, 107, 358–376. [Google Scholar] [CrossRef]
- Rohman, A.; Fadzillah, N.A. Application of Spectroscopic and Chromatographic Methods for the Analysis of Non-halal Meats in Food Products. In Multifaceted Protocols in Biotechnology; Amid, A., Ed.; Springer: Cham, Switzerland, 2021; Volume 2, pp. 75–92. [Google Scholar] [CrossRef]
- Trivedi, D.K.; Hollywood, K.A.; Rattray, N.J.; Ward, H.; Trivedi, D.K.; Greenwood, J.; Ellis, D.I.; Goodacre, R. Meat, the Metabolites: An Integrated Metabolite Profiling and Lipidomics Approach for the Detection of the Adulteration of Beef with Pork. Analyst 2016, 141, 2155–2164. [Google Scholar] [CrossRef]
- Sun, T.; Wang, X.; Cong, P.; Xu, J.; Xue, C. Mass Spectrometry-Based Lipidomics in Food Science and Nutritional Health: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2530–2558. [Google Scholar] [CrossRef] [PubMed]
- Brajnik, Z.; Ogorevc, J. Candidate Genes for Mastitis Resistance in Dairy Cattle: A Data Integration Approach. J. Anim. Sci. Biotechnol. 2023, 14, 10. [Google Scholar] [CrossRef]
- Dehelean, A.; Cristea, G.; Puscas, R.; Hategan, A.R.; Magdas, D.A. Assigning the Geographical Origin of Meat and Animal Rearing System Using Isotopic and Elemental Fingerprints. Appl. Sci. 2022, 12, 12391. [Google Scholar] [CrossRef]
- Cardin, M.; Cardazzo, B.; Mounier, J.; Novelli, E.; Coton, M.; Coton, E. Authenticity and Typicity of Traditional Cheeses: A Review on Geographical Origin Authentication Methods. Foods 2022, 11, 3379. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xing, R.; Sun, R.; Ge, Y.; Chen, Y. An Accurate and Rapid Way for Identifying Food Geographical Origin and Authenticity: Editable DNA-Traceable Barcode. Foods 2023, 12, 17. [Google Scholar] [CrossRef]
- Stanojević, D.; Đedović, R.Ć.; Gligović, N. Genomics as a Tool for Improving Dairy Cattle Populations. Ann. Univ. Craiova-Agric. Mont. Cadastre Ser. 2023, 53, 291–297. [Google Scholar] [CrossRef]
- Cardin, M.; Mounier, J.; Coton, E.; Cardazzo, B.; Perini, M.; Bertoldi, D.; Novelli, E. Discriminative Power of DNA-Based, Volatilome, Near Infrared Spectroscopy, Elements and Stable Isotopes Methods for the Origin Authentication of Typical Italian Mountain Cheese Using sPLS-DA Modeling. Food Res. Int. 2024, 178, 113975. [Google Scholar] [CrossRef] [PubMed]
- Safdar, M.; Kaleem, M.; Duarte, P.M.; Tazerji, S.S.; Ozaslan, M.; Hassanpour, S.; Rizwan, M.A. Review on Optimizing Dairy Sector Efficiency: Integrating of Genetic Markers with Managemental Techniques. Ecol. Genet. Genom. 2024, 32, 100259. [Google Scholar] [CrossRef]
- Lee, H.S.; Kim, Y.; Lee, D.H.; Seo, D.; Lee, D.J.; Do, C.H.; Dinh, P.T.N.; Ekanayake, W.; Lee, K.H.; Yoon, D.; et al. Comparison of Accuracy of Breeding Value for Cow from Three Methods in Hanwoo (Korean Cattle) Population. J. Anim. Sci. Technol. 2023, 65, 720–734. [Google Scholar] [CrossRef]
- Won, K.H.; Kim, D.; Hwang, I.; Lee, H.K.; Oh, J.D. Genome-Wide Association Studies on Collagen Contents Trait for Meat Quality in Hanwoo. J. Anim. Sci. Technol. 2023, 65, 311–323. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, J.; Xiao, B.; Sun, X.; Chen, J.; Huang, F.; Chen, A. Amplification-Free Quantitative Detection of Genomic DNA Using Lateral Flow Strips for Milk Authentication. Biosens. Bioelectron. 2024, 252, 116140. [Google Scholar] [CrossRef]
- Cortes, O.; Cañon, J.; Gama, L.T. Applications of Microsatellites and Single Nucleotide Polymorphisms for the Genetic Characterization of Cattle and Small Ruminants: An Overview. Ruminants 2022, 2, 456–470. [Google Scholar] [CrossRef]
- Hossain, K.; Mazumder, B.; Rahman, S.; Hamid, M. Genetic Diversity Analysis of Lactic Acid Bacteria Isolated from Regional Yogurt Samples. Bangladesh J. Livest. Res. 2021, 27, 55–63. [Google Scholar] [CrossRef]
- Nagraik, R.; Sharma, A.; Kumar, D.; Chawla, P.; Kumar, A.P. Milk Adulterant Detection: Conventional and Biosensor-Based Approaches—A Review. Sens. Bio-Sens. Res. 2021, 33, 100433. [Google Scholar] [CrossRef]
- Liao, J.; Liu, Y.F.; Ku, T.; Liu, M.H.; Huang, Y. Qualitative and Quantitative Identification of Adulteration of Milk Powder Using DNA Extracted with a Novel Method. J. Dairy Sci. 2017, 100, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Ansorge, W.J. Next-Generation DNA Sequencing Techniques. New Biotechnol. 2009, 25, 195–203. [Google Scholar] [CrossRef]
- Baptista, M.; Cunha, J.T.; Domingues, L. DNA-based approaches for dairy products authentication: A review and perspectives. Trends Food Sci. Technol. 2021, 109, 386–397. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef]
- Maruki, T.; Lynch, M. Genotype Calling from Population-Genomic Sequencing Data. G3 Genes Genomes Genet. 2017, 7, 1393–1404. [Google Scholar] [CrossRef]
- Leekitcharoenphon, P.; Kaas, R.S.; Thomsen, M.C.F.; Friis, C.; Rasmussen, S.; Aarestrup, F.M. snpTree—A web-server to identify and construct SNP trees from whole genome sequence data. BMC Genom. 2012, 13 (Suppl. S7), S6. [Google Scholar] [CrossRef]
- Mafra, I.; Honrado, M.; Amaral, J.S. Animal species authentication in dairy products. Foods 2022, 11, 1124. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Kushwaha, A.K.; Rai, A.; Chaturvedi, K.K.; Kumar, A.; Pradhan, A.K.; Kumar, S. Genomic selection: A tool for accelerating the efficiency of molecular breeding for development of climate-resilient crops. Front. Genet. 2022, 13, 832153. [Google Scholar] [CrossRef]
- Bovo, S.; Ballan, M.; Schiavo, G.; Gallo, M.; Dall’Olio, S.; Fontanesi, L. Haplotype-based genome-wide association studies reveal new loci for haematological and clinical–biochemical parameters in Large White pigs. Anim. Genet. 2020, 51, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Tian, R.; Mahmoodi, M.; Tian, J.; Esmailizadeh Koshkoiyeh, S.; Zhao, M.; Saminzadeh, M.; Li, H.; Wang, X.; Li, Y.; Esmailizadeh, A. Leveraging Functional Genomics for Understanding Beef Quality Complexities and Breeding Beef Cattle for Improved Meat Quality. Genes 2024, 15, 1104. [Google Scholar] [CrossRef] [PubMed]
- Arikawa, L.M.; Mota, L.F.M.; Schmidt, P.I.; Frezarim, G.B.; Fonseca, L.F.S.; Magalhães, A.F.B.; Silva, D.A.; Carvalheiro, R.; Chardulo, L.A.L.; Albuquerque, L.G. Genome-wide scans identify biological and metabolic pathways regulating carcass and meat quality traits in beef cattle. Meat Sci. 2024, 209, 109402. [Google Scholar] [CrossRef]
- Lu, X.; Arbab, A.A.I.; Abdalla, I.M.; Liu, D.; Zhang, Z.; Xu, T.; Su, G.; Yang, Z. Genetic Parameter Estimation and Genome-Wide Association Study-Based Loci Identification of Milk-Related Traits in Chinese Holstein. Front. Genet. 2022, 12, 799664. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Rehman, M.U.; Ahmad, S.M.; Mehraj, T.; Hussain, I.; Nadeem, A.; Mir, M.U.R.; Ganie, S.A. In Silico Tools for Analysis of Single-Nucleotide Polymorphisms in the Bovine Transferrin Gene. Animals 2022, 12, 693. [Google Scholar] [CrossRef]
- Rahayu, A.P.; Hartatik, T.; Purnomoadi, A.; Kurnianto, E. Association of single nucleotide polymorphisms in the fatty acid synthase, LOC514211, and fat mass and obesity-associated genes with milk traits in Indonesian-Holstein dairy cattle. Vet. World 2019, 12, 1160–1166. [Google Scholar] [CrossRef]
- Erasmus, L.M.; van Marle-Köster, E. Moving towards sustainable breeding objectives and cow welfare in dairy production: A South African perspective. Trop. Anim. Health Prod. 2021, 53, 470. [Google Scholar] [CrossRef]
- Rexroad, C.; Vallet, J.; Matukumalli, L.K.; Reecy, J.; Bickhart, D.; Blackburn, H.; Wells, K. Genome to phenome: Improving animal health, production, and well-being–a new USDA blueprint for animal genome research 2018–2027. Front. Genet. 2019, 10, 327. [Google Scholar] [CrossRef]
- Ruan, D.; Zhuang, Z.; Ding, R.; Qiu, Y.; Zhou, S.; Wu, J.; Xu, C.; Hong, L.; Huang, S.; Zheng, E.; et al. Weighted Single-Step GWAS Identified Candidate Genes Associated with Growth Traits in a Duroc Pig Population. Genes 2021, 12, 117. [Google Scholar] [CrossRef] [PubMed]
- Al-Sharif, M.; Radwan, H.; Hendam, B.; Ateya, A. DNA polymorphisms of FGFBP1, leptin, κ-casein, and αs1-casein genes and their association with reproductive performance in dromedary she-camels. Theriogenology 2022, 178, 18–29. [Google Scholar] [CrossRef]
- Ma, B.; Khan, R.; Raza, S.H.A.; Gao, Z.; Hou, S.; Ullah, F.; Hassan, M.M.; Hassan, M.M.; AlGabbani, Q.; Alotaibi, M.A.; et al. Determination of the Relationship between Class IV Sirtuin Genes and Growth Traits in Chinese Black Tibetan Sheep. Anim. Biotechnol. 2021, 34, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Raza, S.H.A.; Khan, S.; Amjadi, M.; Abdelnour, S.A.; Ohran, H.; Alanazi, K.M.; El-Hack, M.E.A.; Taha, A.E.; Khan, R.; Gong, C.; et al. Genome-wide association studies reveal novel loci associated with carcass and body measures in beef cattle. Arch. Biochem. Biophys. 2020, 694, 108543. [Google Scholar] [CrossRef]
- Uncu, A.O.; Uncu, A.T. A barcode-DNA analysis method for the identification of plant oil adulteration in milk and dairy products. Food Chem. 2020, 326, 126986. [Google Scholar] [CrossRef]
- Cai, Z.; Zhou, S.; Liu, Q.; Ma, H.; Yuan, X.; Gao, J.; Cao, J.; Pan, D. A Simple and Reliable Single Tube Septuple PCR Assay for Simultaneous Identification of Seven Meat Species. Foods 2021, 10, 1083. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolny, S.; Uhlig, S.; Frost, K.; Schlierf, A.; Nichani, K.; Simon, K.; Cichna-Markl, M.; Hochegger, R. Interlaboratory Validation of a DNA Metabarcoding Assay for Mammalian and Poultry Species to Detect Food Adulteration. Foods 2022, 11, 1108. [Google Scholar] [CrossRef]
- Di Domenico, M.; Di Giuseppe, M.; Rodríguez, J.W.; Cammà, C. Validation of a fast real-time PCR method to detect fraud and mislabeling in milk and dairy products. J. Dairy Sci. 2017, 100, 106–112. [Google Scholar] [CrossRef]
- Guo, L.; Qian, J.P.; Guo, Y.S.; Hai, X.; Liu, G.Q.; Luo, J.X.; Ya, M. Simultaneous identification of bovine and equine DNA in milks and dairy products inferred from triplex TaqMan real-time PCR technique. J. Dairy Sci. 2018, 101, 6776–6786. [Google Scholar] [CrossRef]
- Buaban, S.; Lengnudum, K.; Boonkum, W.; Phakdeedindan, P. Genome-wide association study on milk production and somatic cell score for Thai dairy cattle using weighted single-step approach with random regression test-day model. J. Dairy Sci. 2022, 105, 468–494. [Google Scholar] [CrossRef]
- Vafin, R.R.; Galstyan, A.G.; Tyulkin, S.V.; Gilmanov, K.K.; Yurova, E.A.; Semipyatniy, V.K.; Bigaeva, A.V. Species identification of ruminant milk by genotyping of the κ-casein gene. J. Dairy Sci. 2022, 105, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, T.; Xie, X.; Niu, Q.; Zhao, Z.; Zhu, B.; Chen, Y.; Zhang, L.; Gao, X.; Niu, X.; et al. Genetic Association Analysis of Copy Number Variations for Meat Quality in Beef Cattle. Foods 2023, 12, 3986. [Google Scholar] [CrossRef] [PubMed]
- Pegolo, S.; Cecchinato, A.; Savoia, S.; Di Stasio, L.; Pauciullo, A.; Brugiapaglia, A.; Bittante, G.; Albera, A. Genome-wide association and pathway analysis of carcass and meat quality traits in Piemontese young bulls. Animal 2020, 14, 243–252. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, S.; Ju, S.; Zhou, S.; Zeng, X.; Wu, Z.; Pan, D.; Zhong, G.; Cai, Z. Heat-Treated Meat Origin Tracing and Authenticity through a Practical Multiplex Polymerase Chain Reaction Approach. Nutrients 2022, 14, 4727. [Google Scholar] [CrossRef]
- Cai, Z.; Zhong, G.; Liu, Q.; Yang, X.; Zhang, X.; Zhou, S.; Zeng, X.; Wu, Z.; Pan, D. Molecular authentication of twelve meat species through a promising two-tube hexaplex polymerase chain reaction technique. Front. Nutr. 2022, 9, 813962. [Google Scholar] [CrossRef]
- Bouroutzika, E.; Proikakis, S.; Anagnostopoulos, A.K.; Katsafadou, A.I.; Fthenakis, G.C.; Tsangaris, G.T. Proteomics Analysis in Dairy Products: Cheese, a Review. Appl. Sci. 2021, 11, 7622. [Google Scholar] [CrossRef]
- Gagaoua, M.; Franco, D.; Ramanathan, R. Meat Omics: Trends and applications of Omics strategies in meat research. J. Proteomics 2024, 295, 105090. [Google Scholar] [CrossRef]
- López-Pedrouso, M.; Lorenzo, J.M.; Alché, J.d.D.; Moreira, R.; Franco, D. Advanced Proteomic and Bioinformatic Tools for Predictive Analysis of Allergens in Novel Foods. Biology 2023, 12, 714. [Google Scholar] [CrossRef]
- Bianco, M.; Calvano, C.D.; Ventura, G.; Losito, I.; Cataldi, T.R. Determination of hidden milk allergens in meat-based foodstuffs by liquid chromatography coupled to electrospray ionization and high-resolution tandem mass spectrometry. Food Control 2022, 131, 108443. [Google Scholar] [CrossRef]
- Rahman, M.M.; Takashima, S.; Kamatari, Y.O.; Shimizu, K.; Okada, A.; Inoshima, Y. Comprehensive proteomic analysis revealed a large number of newly identified proteins in the small extracellular vesicles of milk from late-stage lactating cows. Animals 2021, 11, 2506. [Google Scholar] [CrossRef]
- Singh, M.K.; Kumar, A.; Nimmanapalli, R.; Pandey, A.K. Proteomics-based milk whey proteome profiling of Indian Jersey crossbreed cows followed by chromosomal mapping. J. Sci. Food Agric. 2023, 103, 5634–5640. [Google Scholar] [CrossRef]
- Zhu, Z.; Bu, S.; Liu, J.; Niu, C.; Wang, L.; Yuan, H.; Zhang, L.; Song, Y. Label-free-based proteomics analysis reveals differential proteins of sheep, goat, and cow milk. J. Dairy Sci. 2024, 107, 8908–8918. [Google Scholar] [CrossRef] [PubMed]
- Verma, M.; Dige, M.S.; Gautam, D.; De, S.; Rout, P.K. Functional milk proteome analysis of genetically diverse goats from different agro-climatic regions. J. Proteom. 2020, 227, 103916. [Google Scholar] [CrossRef] [PubMed]
- Pourjoula, M.; Picariello, G.; Garro, G.; D’Auria, G.; Nitride, C.; Rheza Ghaisari, A.; Ferranti, P. The Protein and Peptide Fractions of Kashk, a Traditional Middle East Fermented Dairy Product. Food Res. Int. 2020, 132, 109060. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.H.; Bovenhuis, H.; Delacroix-Buchet, A.; Miranda, G.; Boichard, D.; Visker, M.H.P.W.; Martin, P. Genetic and Nongenetic Factors Contributing to Differences in AS-Casein Phosphorylation Isoforms and Other Major Milk Proteins. J. Dairy Sci. 2017, 100, 5564–5577. [Google Scholar] [CrossRef]
- Le, T.T.; Deeth, H.C.; Larsen, L.B. Proteomics of Major Bovine Milk Proteins: Novel Insights. Int. Dairy J. 2017, 67, 2–15. [Google Scholar] [CrossRef]
- Picard, B.; Gagaoua, M. Meta-proteomics for the discovery of protein biomarkers of beef tenderness: An overview of integrated studies. Food Res. Int. 2020, 127, 108739. [Google Scholar] [CrossRef]
- Gagaoua, M.; Hughes, J.; Terlouw, E.C.; Warner, R.D.; Purslow, P.P.; Lorenzo, J.M.; Picard, B. Proteomic biomarkers of beef colour. Trends Food Sci. Technol. 2020, 101, 234–252. [Google Scholar] [CrossRef]
- Severino, M.; Gagaoua, M.; Baldassini, W.; Ribeiro, R.; Torrecilhas, J.; Pereira, G.; Curi, R.; Chardulo, L.A.; Padilha, P.; Neto, O.M. Proteomics Unveils Post-Mortem Changes in Beef Muscle Proteins and Provides Insight into Variations in Meat Quality Traits of Crossbred Young Steers and Heifers Raised in Feedlot. Int. J. Mol. Sci. 2022, 23, 12259. [Google Scholar] [CrossRef]
- Zhu, Y.; Gagaoua, M.; Mullen, A.M.; Viala, D.; Rai, D.K.; Kelly, A.L.; Sheehan, D.; Hamill, R.M. Shotgun proteomics for the preliminary identification of biomarkers of beef sensory tenderness, juiciness and chewiness from plasma and muscle of young Limousin-sired bulls. Meat Sci. 2021, 176, 108488. [Google Scholar] [CrossRef]
- Franco, D.; Mato, A.; Salgado, F.J.; López-Pedrouso, M.; Carrera, M.; Bravo, S.; Parrado, M.; Gallardo, J.M.; Zapata, C. Tackling proteome changes in the Longissimus thoracis bovine muscle in response to pre-slaughter stress. J. Proteome 2015, 122, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, P.; Tang, S.; Xiao, C.; Huang, Y.; Wang, L.; Yang, X.; Chen, X.; Shao, B. Detection of chemical contaminants in heat-processed meat products based on UPLC-MS/MS. J. Food Compos. Anal. 2023, 124, 105711. [Google Scholar] [CrossRef]
- Banerjee, R.; Maheswarappa, N.B.; Mohan, K.; Biswas, S.; Batabyal, S. Proteomic technologies and their application for ensuring meat quality, safety, and authenticity. Curr. Proteom. 2022, 19, 128–141. [Google Scholar] [CrossRef]
- Qin, C.; Liu, L.; Wang, Y.; Leng, T.; Zhu, M.; Gan, B.; Chen, Y. Advancement of omics techniques for chemical profile analysis and authentication of milk. Trends Food Sci. Technol. 2022, 127, 114–128. [Google Scholar] [CrossRef]
- Abdelmegid, S.; Kelton, D.; Caswell, J.; Kirby, G. Proteomic 2D-DIGE analysis of milk whey from dairy cows with Staphylococcus aureus mastitis reveals overexpression of host defense proteins. Microorganisms 2020, 8, 1883. [Google Scholar] [CrossRef]
- Nardiello, D.; Natale, A.; Palermo, C.; Quinto, M.; Centonze, D. Milk authenticity by ion-trap proteomics following multi-enzyme digestion. Food Chem. 2018, 244, 317–323. [Google Scholar] [CrossRef]
- Naveena, B.M.; Jagadeesh, D.S.; Jagadeesh Babu, A.; Madhava Rao, T.; Kamuni, V.; Vaithiyanathan, S.; Kulkarni, V.V.; Rapole, S. OFFGEL electrophoresis and tandem mass spectrometry approach compared with DNA-based PCR method for authentication of meat species from raw and cooked ground meat mixtures containing cattle meat, water buffalo meat and sheep meat. Food Chem. 2017, 233, 311–320. [Google Scholar] [CrossRef]
- Pu, K.; Qiu, J.; Tong, Y.; Liu, B.; Cheng, Z.; Chen, S.; Ni, W.-X.; Lin, Y.; Ng, K.-M. Integration of non-targeted proteomics mass spectrometry with machine learning for screening cooked beef adulterated samples. J. Agric. Food Chem. 2022, 71, 2173–2182. [Google Scholar] [CrossRef]
- Villa, C.; Costa, J.; Oliveira, M.B.P.; Mafra, I. Bovine milk allergens: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 137–164. [Google Scholar] [CrossRef]
- Natale, M.; Bisson, C.; Monti, G.; Peltran, A.; Perono Garoffo, L.; Valentini, S.; Fabris, C.; Bertino, E.; Coscia, A.; Conti, A. Cow’s Milk Allergens Identification by Two-Dimensional Immunoblotting and Mass Spectrometry. Mol. Nutr. Food Res. 2004, 48, 363–369. [Google Scholar] [CrossRef]
- Milkovska-Stamenova, S.; Hoffmann, R. Diversity of advanced glycation end products in the bovine milk proteome. Amino Acids 2019, 51, 891–901. [Google Scholar] [CrossRef] [PubMed]
- Malheiros, J.M.; Braga, C.P.; Grove, R.A.; Ribeiro, F.A.; Calkins, C.R.; Adamec, J.; Chardulo, L.A.L. Influence of oxidative damage to proteins on meat tenderness using a proteomics approach. Meat Sci. 2019, 148, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Rau, J.; Korte, N.; Dyk, M.; Wenninger, O.; Schreiter, P.; Hiller, E. Rapid animal species identification of feta and mozzarella cheese using MALDI-TOF mass-spectrometry. Food Control 2020, 117, 107349. [Google Scholar] [CrossRef]
- Nalazek-Rudnicka, K.; Kłosowska-Chomiczewska, I.; Wasik, A.; Macierzanka, A. MRM–MS of marker peptides and their abundance as a tool for authentication of meat species and meat cuts in single-cut meat products. Food Chem. 2019, 283, 367–374. [Google Scholar] [CrossRef]
- Fuente-Garcia, C.; Sentandreu, E.; Aldai, N.; Oliván, M.; Sentandreu, M.Á. Characterization of the myofibrillar proteome as a way to better understand differences in bovine meats having different ultimate pH values. Proteomics 2020, 20, 2000012. [Google Scholar] [CrossRef]
- Pu, K.; Qiu, J.; Li, J.; Huang, W.; Lai, X.; Liu, C.; Lin, Y. MALDI-TOF MS protein profiling combined with multivariate analysis for identification and quantitation of meat adulteration. Res. Sq. 2022, 16, 132–142. [Google Scholar] [CrossRef]
- Setyabrata, D.; Ma, D.; Xie, S.; Thimmapuram, J.; Cooper, B.R.; Aryal, U.K.; Kim, Y.H.B. Proteomics and metabolomics profiling of meat exudate to determine the impact of postmortem aging on oxidative stability of beef muscles. Food Chem. X 2023, 18, 100660. [Google Scholar] [CrossRef]
- Sacarrão-Birrento, L.; Ribeiro, D.M.; Dittmann, A.; Alves, S.P.; Kunz, L.; Silva, S.; Venâncio, C.A.; de Almeida, A.M. The effect of the production system on the proteomics profiles of the Longissimus thoracis muscle in Arouquesa cattle. J. Proteom. 2024, 307, 105265. [Google Scholar] [CrossRef]
- Sen, C.; Ray, P.R.; Bhattacharyya, M. A critical review on metabolomic analysis of milk and milk products. Int. J. Dairy Technol. 2020, 74, 17–31. [Google Scholar] [CrossRef]
- Zhang, J.; Sun, M.; Elmaidomy, A.H.; Youssif, K.A.; Zaki, A.M.; Kamal, H.H.; Abdelmohsen, U.R. Emerging trends and applications of metabolomics in food science and nutrition. Food Funct. 2023, 14, 9050–9082. [Google Scholar] [CrossRef]
- Singh, N.; Joshi, R. Metabolomics applications in food science, technology and nutrition. Int. J. Food Sci. Technol. 2024, 59, 4166–4168. [Google Scholar] [CrossRef]
- Rocchetti, G.; O’Callaghan, T.F. Application of metabolomics to assess milk quality and traceability. Curr. Opin. Food Sci. 2021, 40, 168–178. [Google Scholar] [CrossRef]
- Xu, W.; Vervoort, J.; Saccenti, E.; van Hoeij, R.; Kemp, B.; van Knegsel, A. Milk metabolomics data reveal the energy balance of individual dairy cows in early lactation. Sci. Rep. 2018, 8, 15828. [Google Scholar] [CrossRef]
- Rocchetti, G.; Gallo, A.; Nocetti, M.; Lucini, L.; Masoero, F. Milk metabolomics based on ultra-high-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry to discriminate different cows feeding regimens. Food Res. Int. 2020, 134, 109279. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Zhang, H.; Zhang, Y.; Xiong, B.; Jiang, L. Microbiome and metabolome analyses of milk from dairy cows with subclinical Streptococcus agalactiae mastitis—Potential biomarkers. Front. Microbiol. 2019, 10, 2547. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Zhang, L.; Zheng, X.; Huang, Q.; Farag, M.A.; Zhu, R.; Zhao, C. Emerging applications of metabolomics in food science and future trends. Food Chem. X 2022, 16, 100500. [Google Scholar] [CrossRef]
- Chaji, S.; Olmo-García, L.; Serrano-García, I.; Carrasco-Pancorbo, A.; Bajoub, A. Metabolomic approaches applied to food authentication: From data acquisition to biomarkers discovery. In Food Authentication and Traceability; Academic Press: Cambridge, MA, USA, 2021; pp. 331–378. [Google Scholar] [CrossRef]
- Chen, Y.; MacNaughtan, W.; Jones, P.; Yang, Q.; Williams, H.; Foster, T. Selection of potential molecular markers for cheese ripening and quality prediction by NMR spectroscopy. LWT 2021, 136, 110306. [Google Scholar] [CrossRef]
- Liu, S.J.; Wu, Y.N.; Chan, L. Application of Metabonomics Approach in Food Safety Research—A Review. Food Rev. Int. 2019, 36, 547–558. [Google Scholar] [CrossRef]
- Braconi, D.; Millucci, L.; Parisi, M.L.; Spiga, O.; Santucci, A. Omics-based Technologies for Food Authentication and Traceability. In Food Authentication and Traceability; Academic Press: Cambridge, MA, USA, 2021; pp. 215–245. [Google Scholar] [CrossRef]
- Liu, J.; Zhao, H.; Yin, Z.; Dong, H.; Chu, X.; Meng, X.; Li, Y.; Ding, X. Application and Prospect of Metabolomics-Related Technologies in Food Inspection. Food Res. Int. 2023, 171, 113071. [Google Scholar] [CrossRef]
- Afifah, E.N.; Putri, S.P. Food Metabolomics for Improvement of Nutrition and Well-Being. BIO Web Conf. 2024, 127, 07001. [Google Scholar] [CrossRef]
- Rocchetti, G.; Ghilardelli, F.; Bonini, P.; Lucini, L.; Masoero, F.; Gallo, A. Changes of Milk Metabolomic Profiles Resulting from a Mycotoxins-Contaminated Corn Silage Intake by Dairy Cows. Metabolites 2021, 11, 475. [Google Scholar] [CrossRef]
- Yanibada, B.; Hohenester, U.; Pétéra, M.; Canlet, C.; Durand, S.; Jourdan, F.; Ferlay, A.; Morgavi, D.P.; Boudra, H. Milk metabolome reveals variations on enteric methane emissions from dairy cows fed a specific inhibitor of the methanogenesis pathway. J. Dairy Sci. 2021, 104, 12553–12566. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Kim, M.; Ji, S.Y.; Baek, Y.C.; Lee, S.; Oh, Y.K.; Reddy, K.E.; Seo, H.W.; Cho, S.; Lee, H.J. Metabolomics analysis of the beef samples with different meat qualities and tastes. Food Sci. Anim. Resour. 2020, 40, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Adebo, J.A.; Chinma, C.E.; Omoyajowo, A.O.; Njobeh, P.B. Metabolomics and its application in fermented foods. In Indigenous Fermented Foods for the Tropics; Academic Press: Cambridge, MA, USA, 2023; pp. 361–376. [Google Scholar] [CrossRef]
- Spears, M.; Cooper, G.; Sather, B.; Bailey, M.; Boles, J.A.; Bothner, B.; Miles, M.P. Comparative Impact of Organic Grass-Fed and Conventional Cattle-Feeding Systems on Beef and Human Postprandial Metabolomics—A Randomized Clinical Trial. Metabolites 2024, 14, 533. [Google Scholar] [CrossRef]
- Tomassini, A.; Curone, G.; Solè, M.; Capuani, G.; Sciubba, F.; Conta, G.; Miccheli, G.; Vigo, D. NMR-based metabolomics to evaluate the milk composition from Friesian and autochthonous cows of Northern Italy at different lactation times. Nat. Prod. Res. 2019, 33, 1085–1091. [Google Scholar] [CrossRef]
- Foroutan, A.; Guo, A.C.; Vazquez-Fresno, R.; Lipfert, M.; Zhang, L.; Zheng, J.; Badran, H.; Budinski, Z.; Mandal, R.; Ametaj, B.N.; et al. Chemical composition of commercial cow’s milk. J. Agric. Food Chem. 2019, 67, 4897–4914. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, Y.; Zheng, N.; Guo, L.; Song, X.; Zhao, S.; Wang, J. Biological System Responses of Dairy Cows to Aflatoxin B1 Exposure Revealed with Metabolomic Changes in Multiple Biofluids. Toxins 2019, 11, 77. [Google Scholar] [CrossRef]
- Magan, J.B.; O’Callaghan, T.F.; Zheng, J.; Zhang, L.; Mandal, R.; Hennessy, D.; Fenelon, M.A.; Wishart, D.S.; Kelly, A.L.; McCarthy, N.A. Impact of Bovine Diet on Metabolomic Profile of Skim Milk and Whey Protein Ingredients. Metabolites 2019, 9, 305. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, H.; Li, L.; Tan, J.; Wang, Y.; Liu, M.; Jiang, L. Multi-omics analysis reveals that the metabolite profile of raw milk is associated with dairy cows’ health status. Food Chem. 2023, 428, 136813. [Google Scholar] [CrossRef]
- Gauglitz, J.M.; Aceves, C.M.; Aksenov, A.A.; Aleti, G.; Almaliti, J.; Bouslimani, A.; Brown, E.A.; Campeau, A.; Caraballo-Rodríguez, A.M.; Chaar, R.; et al. Untargeted mass spectrometry-based metabolomics approach unveils molecular changes in raw and processed foods and beverages. Food Chem. 2020, 302, 125290. [Google Scholar] [CrossRef]
- Yu, Q.; Tian, X.; Shao, L.; Li, X.; Dai, R. Targeted metabolomics to reveal muscle-specific energy metabolism between bovine longissimus lumborum and psoas major during early postmortem periods. Meat Sci. 2019, 156, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Antonelo, D.S.; Cônsolo, N.R.B.; Gómez, J.F.M.; Beline, M.; Goulart, R.S.; Corte, R.R.P.S.; Colnago, L.A.; Schilling, M.W.; Gerrard, D.E.; Silva, S.L. Metabolite profile and consumer sensory acceptability of meat from lean Nellore and Angus × Nellore crossbreed cattle fed soybean oil. Food Res. Int. 2020, 132, 109056. [Google Scholar] [CrossRef] [PubMed]
- Man, K.Y.; Chan, C.O.; Tang, H.H.; Dong, N.P.; Capozzi, F.; Wong, K.H.; Kwok, K.W.H.; Chan, H.M.; Mok, D.K. Mass spectrometry-based untargeted metabolomics approach for differentiation of beef of different geographic origins. Food Chem. 2021, 338, 127847. [Google Scholar] [CrossRef]
- Li, M.; Li, Q.; Kang, S.; Cao, X.; Zheng, Y.; Wu, J.; Wu, R.; Shao, J.; Yang, M.; Yue, X. Characterization and comparison of lipids in bovine colostrum and mature milk based on UHPLC-QTOF-MS lipidomics. Food Res. Int. 2020, 136, 109490. [Google Scholar] [CrossRef]
- England, P.; Tang, W.; Kostrzewa, M.; Shahrezaei, V.; Larrouy-Maumus, G. Discrimination of bovine milk from non-dairy milk by lipids fingerprinting using routine matrix-assisted laser desorption ionization mass spectrometry. Sci. Rep. 2020, 10, 5160. [Google Scholar] [CrossRef] [PubMed]
- Ceciliani, F.; Audano, M.; Addis, M.F.; Lecchi, C.; Ghaffari, M.H.; Albertini, M.; Tangorra, F.; Piccinini, R.; Caruso, D.; Mitro, N.; et al. The untargeted lipidomic profile of quarter milk from dairy cows with subclinical intramammary infection by non-aureus staphylococci. J. Dairy Sci. 2021, 104, 10268–10281. [Google Scholar] [CrossRef]
- Song, Y.; Cai, C.; Song, Y.; Sun, X.; Liu, B.; Xue, P.; Zhu, M.; Chai, W.; Wang, Y.; Wang, C.; et al. A comprehensive review of lipidomics and its application to assess food obtained from farm animals. Food Sci. Anim. Resour. 2022, 42, 1–17. [Google Scholar] [CrossRef]
- Liu, Z.; Li, C.; Pryce, J.; Rochfort, S. Comprehensive characterization of bovine milk lipids: Phospholipids, sphingolipids, glycolipids, and ceramides. J. Agric. Food Chem. 2020, 68, 6726–6738. [Google Scholar] [CrossRef]
- Bukowski, M.R.; Picklo, M.J. Simple, rapid lipidomic analysis of triacylglycerols in bovine milk by infusion-electrospray mass spectrometry. Lipids 2021, 56, 243–255. [Google Scholar] [CrossRef]
- Tsiafoulis, C.G.; Papaemmanouil, C.; Alivertis, D.; Tzamaloukas, O.; Miltiadou, D.; Balayssac, S.; Malet-Martino, M.; Gerothanassis, I.P. NMR-based metabolomics of the lipid fraction of organic and conventional bovine milk. Molecules 2019, 24, 1067. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.; Zhu, D.; Pang, X.; Liu, Y.; Frew, R.; Chen, G. Lipidomics profiling of goat milk, soymilk and bovine milk by UPLC-Q-Exactive Orbitrap Mass Spectrometry. Food Chem. 2017, 224, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhu, D.; Su, M.; Tan, D.; Zhang, X.; Jia, M.; Wu, H.; Chen, G. Lipidomics strategy for the identification of Ultra-High temperature and reconstituted milk by UPLC-Q-Exactive orbitrap mass spectrometry. Food Anal. Methods 2021, 14, 1064–1073. [Google Scholar] [CrossRef]
- He, Q.; Yang, M.; Chen, X.; Yan, X.; Li, Y.; He, M.; Liu, T.; Chen, F.; Zhang, F. Differentiation between Fresh and Frozen-Thawed Meat using Rapid Evaporative Ionization Mass Spectrometry: The Case of Beef Muscle. J. Agric. Food Chem. 2021, 69, 5709–5724. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xu, L.; Wang, X.; Chen, A.; Xu, Z. Discrimination of Beef from Different Origins Based on Lipidomics: A Comparison Study of DART-QTOF and LC-ESI-QTOF. LWT 2021, 149, 111838. [Google Scholar] [CrossRef]
- Maritha, V.; Harlina, P.W.; Musfiroh, I.; Muchtaridi, M.; Rafi, M.; Geng, F.; Khan, M.R.; Nawaz, A. Lipidomics Analysis for Halal Authentication of Triceps brachii, Longissimus dorsi, and Biceps femoris Meats: Profiling the Lipid Composition. LWT 2023, 185, 115187. [Google Scholar] [CrossRef]
- Misra, B.B.; Langefeld, C.D.; Olivier, M.; Cox, L.A. Integrated Omics: Tools, Advances, and Future Approaches. J. Mol. Endocrinol. 2018, 13, JME-18-0055. [Google Scholar] [CrossRef]
- ISO/TS 20428:2024; Genomics Informatics—Data Elements and Their Metadata for Describing Structured Clinical Genomic Sequence Information in Electronic Health Records. 2nd ed. ISO: Geneva, Switzerland, 2024; p. 28.
- ISO 14001:2015; Environmental Management Systems—Requirements with Guidance for Use. 3rd ed. ISO: Geneva, Switzerland, 2018; p. 35.
- ISO 22000:2018; Food Safety Management Systems—Requirements for Any Organization in the Food Chain. 2nd ed. ISO: Geneva, Switzerland, 2018; p. 37.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez, A.; Abanto, M.; Días, N.B.; Olate, P.; Pérez Nuñez, I.; Díaz, R.; Sepúlveda, N.; Paz, E.A.; Quiñones, J. Recent Trends in Food Quality and Authentication: The Role of Omics Technologies in Dairy and Meat Production. Int. J. Mol. Sci. 2025, 26, 4405. https://doi.org/10.3390/ijms26094405
Martínez A, Abanto M, Días NB, Olate P, Pérez Nuñez I, Díaz R, Sepúlveda N, Paz EA, Quiñones J. Recent Trends in Food Quality and Authentication: The Role of Omics Technologies in Dairy and Meat Production. International Journal of Molecular Sciences. 2025; 26(9):4405. https://doi.org/10.3390/ijms26094405
Chicago/Turabian StyleMartínez, Ailín, Michel Abanto, Nathalia Baptista Días, Paula Olate, Isabela Pérez Nuñez, Rommy Díaz, Néstor Sepúlveda, Erwin A. Paz, and John Quiñones. 2025. "Recent Trends in Food Quality and Authentication: The Role of Omics Technologies in Dairy and Meat Production" International Journal of Molecular Sciences 26, no. 9: 4405. https://doi.org/10.3390/ijms26094405
APA StyleMartínez, A., Abanto, M., Días, N. B., Olate, P., Pérez Nuñez, I., Díaz, R., Sepúlveda, N., Paz, E. A., & Quiñones, J. (2025). Recent Trends in Food Quality and Authentication: The Role of Omics Technologies in Dairy and Meat Production. International Journal of Molecular Sciences, 26(9), 4405. https://doi.org/10.3390/ijms26094405








