Transcriptome and Metabolome Analyses Offer New Insights into Bolting Time Regulation in Broccoli
Abstract
1. Introduction
2. Results
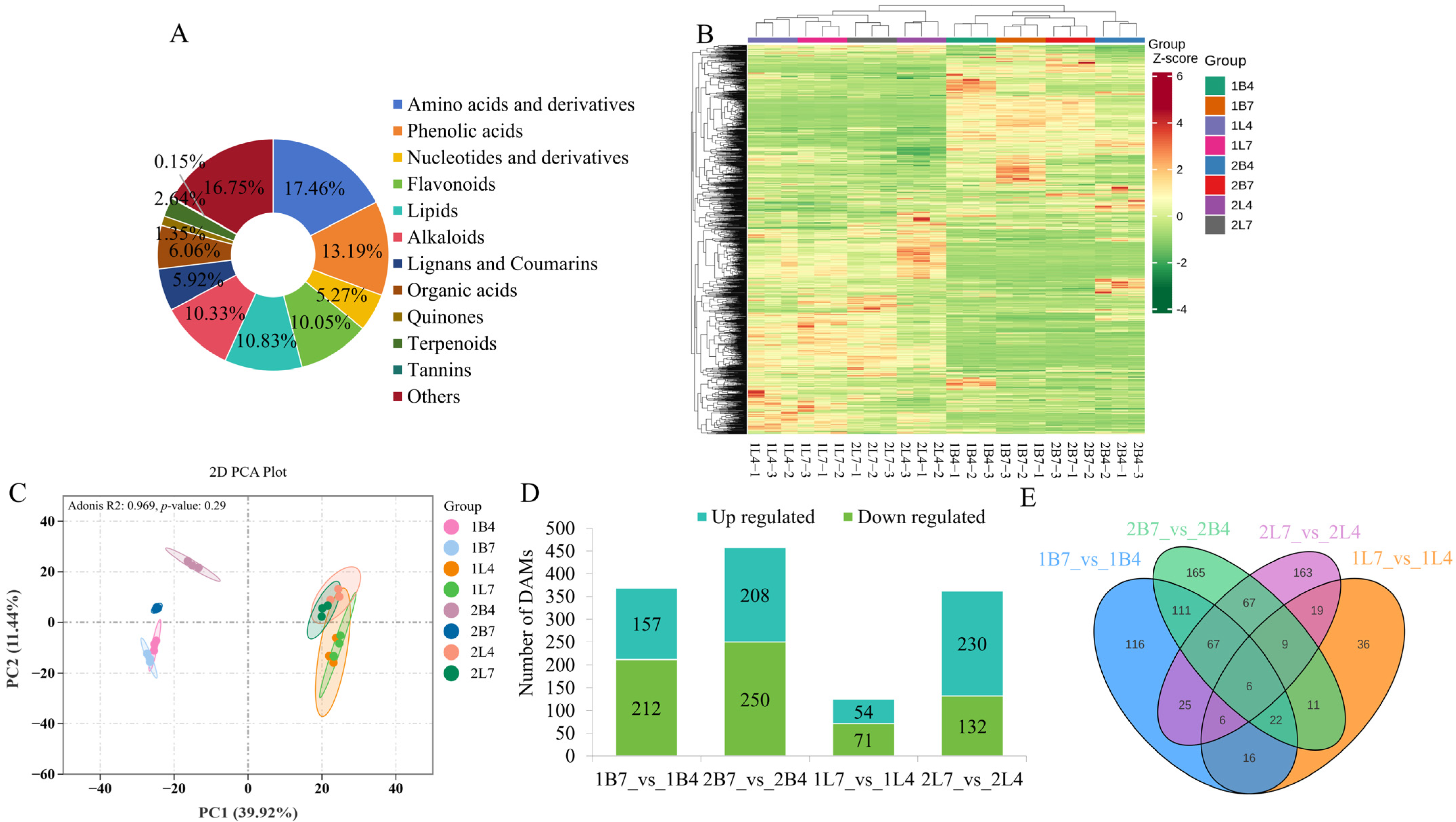
2.1. Overview of the Metabolites Involved in the Development of Early- and Late-Bolting Broccoli
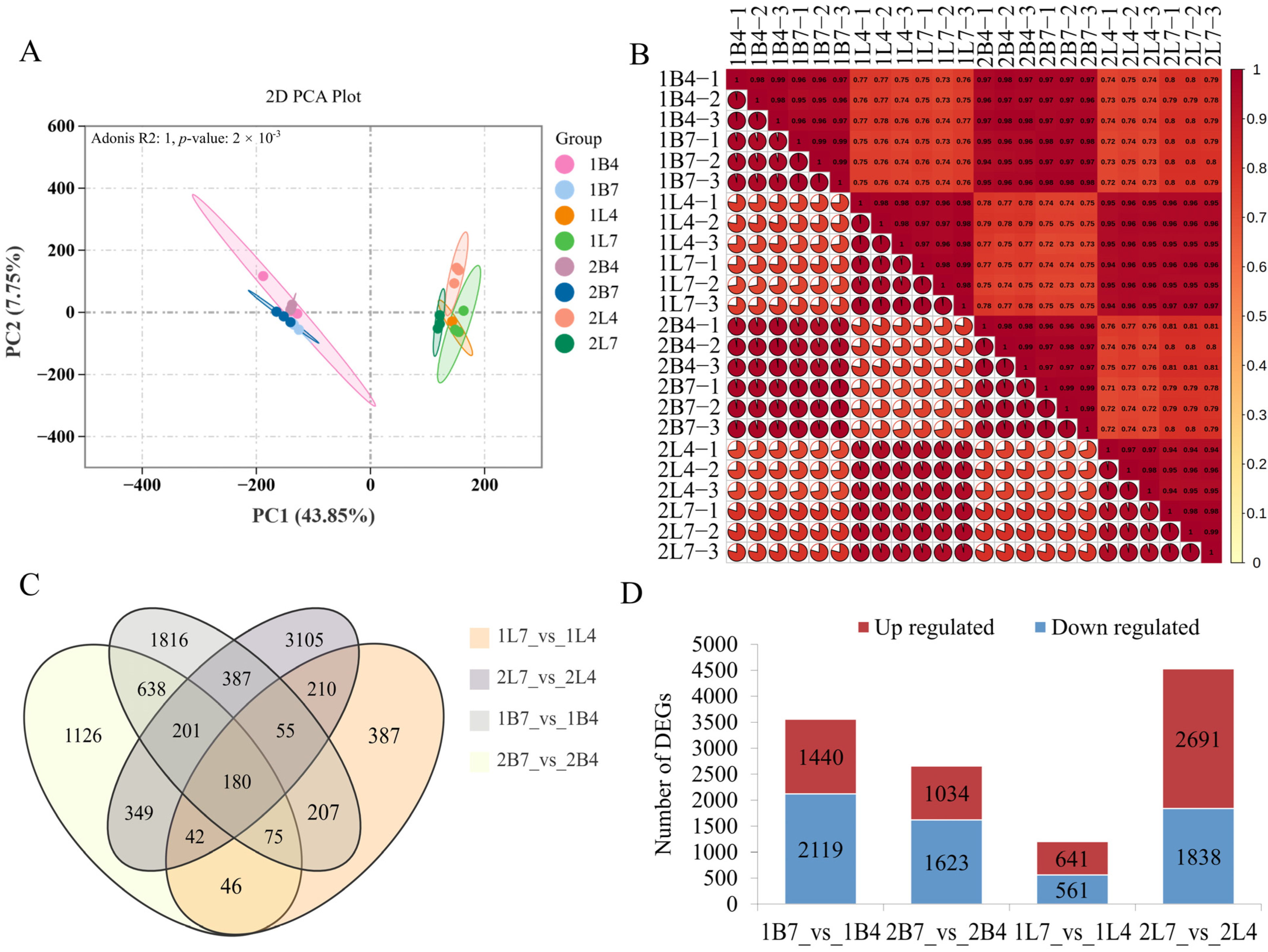
2.2. Transcriptome Analysis of the Development of Early- and Late-Bolting Broccoli
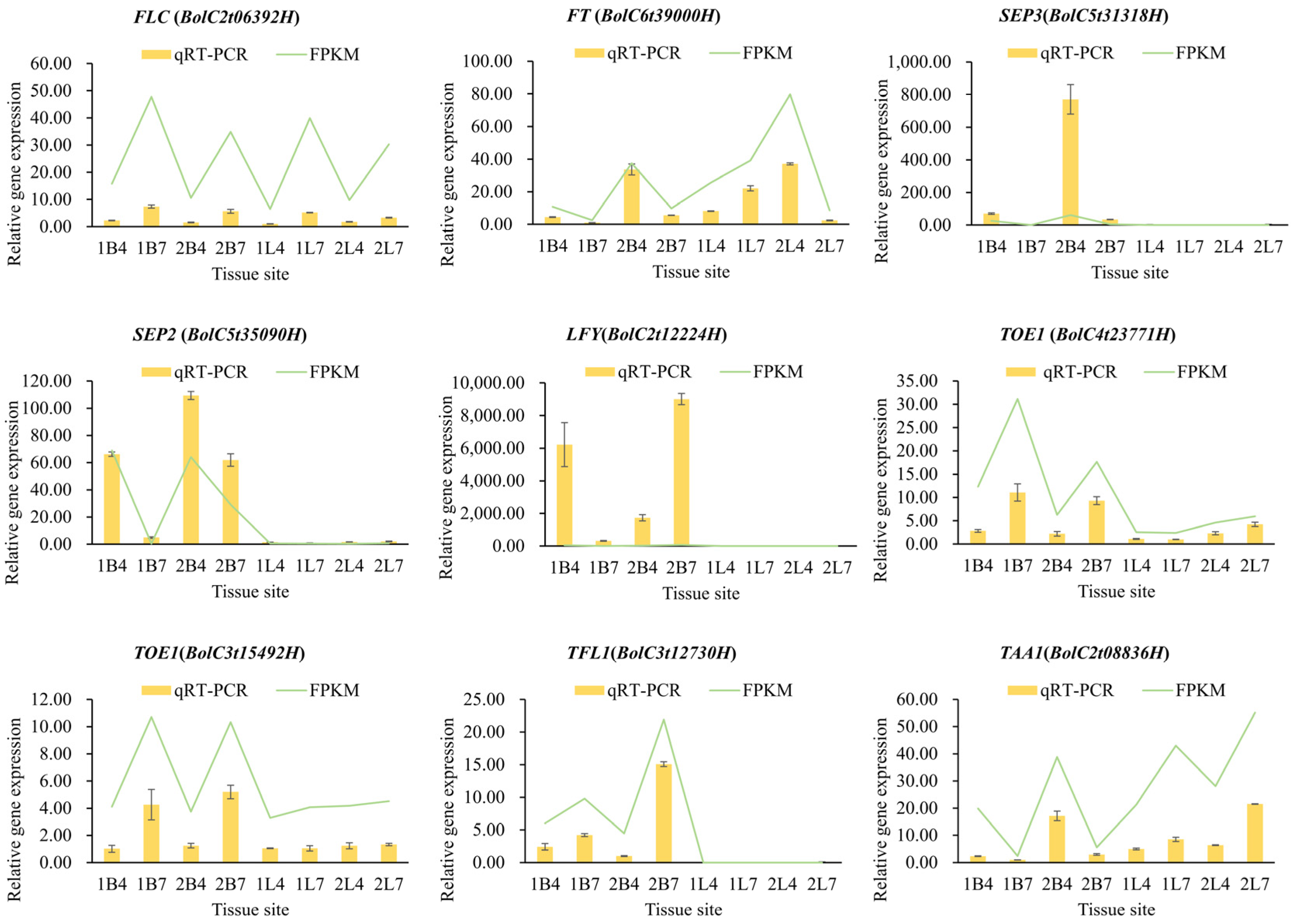
2.3. Validation and Analysis by qRT-PCR
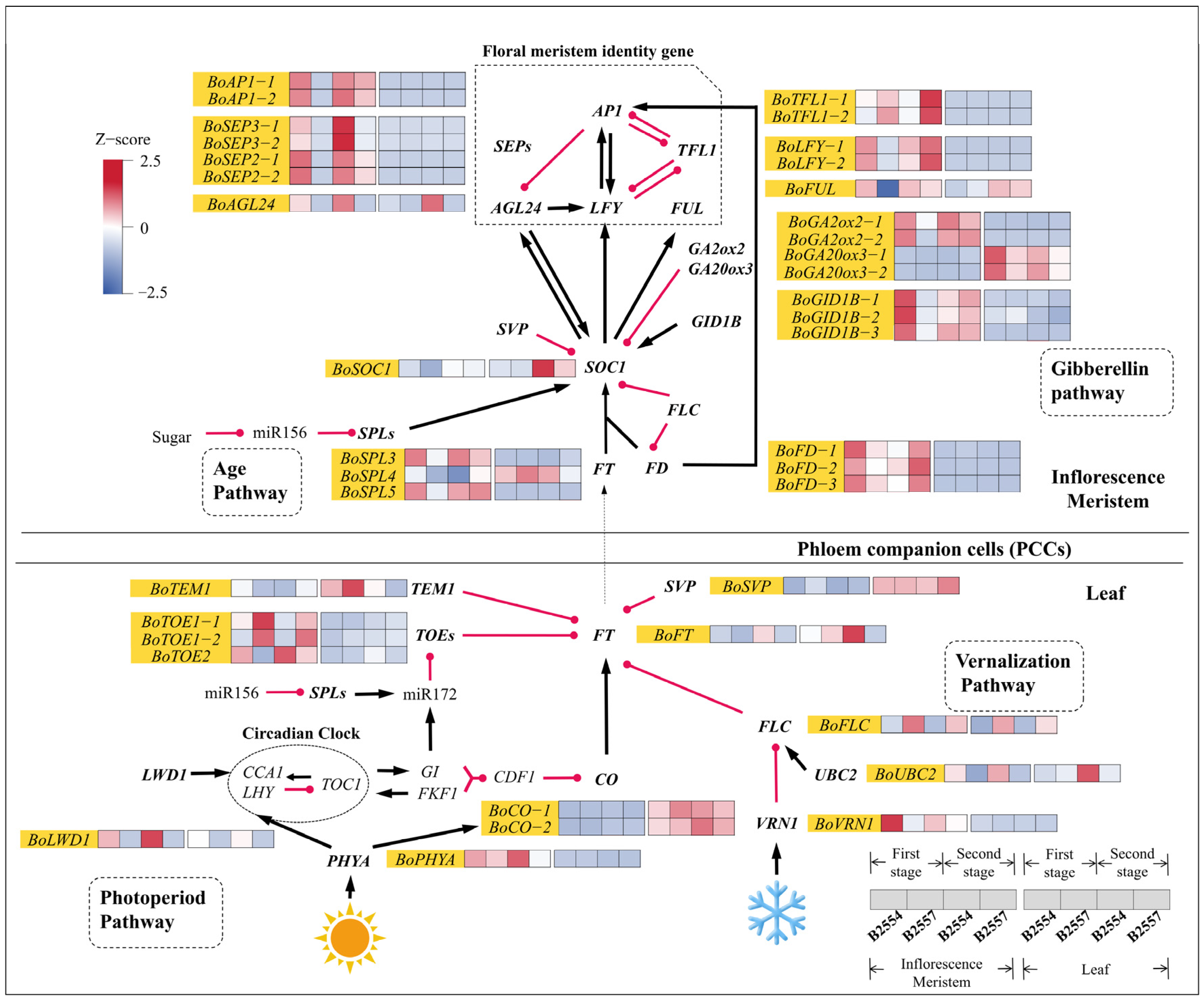
2.4. Differential Expression in the Flowering Pathway
2.5. Differential Expression Analysis in Phytohormone Signal Transduction Pathways
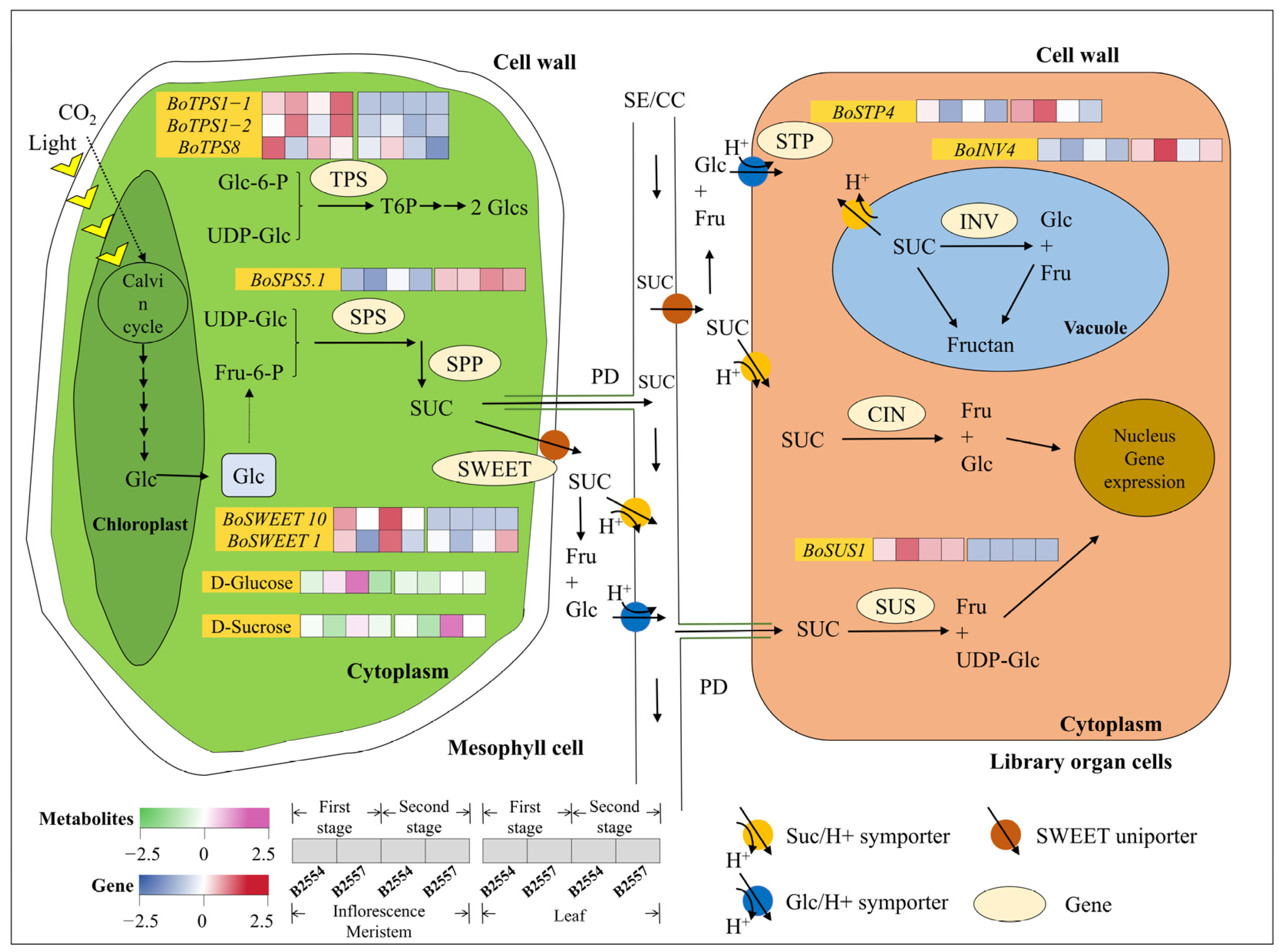
2.6. Differential Regulation in Sucrose Metabolism and Transport Pathways
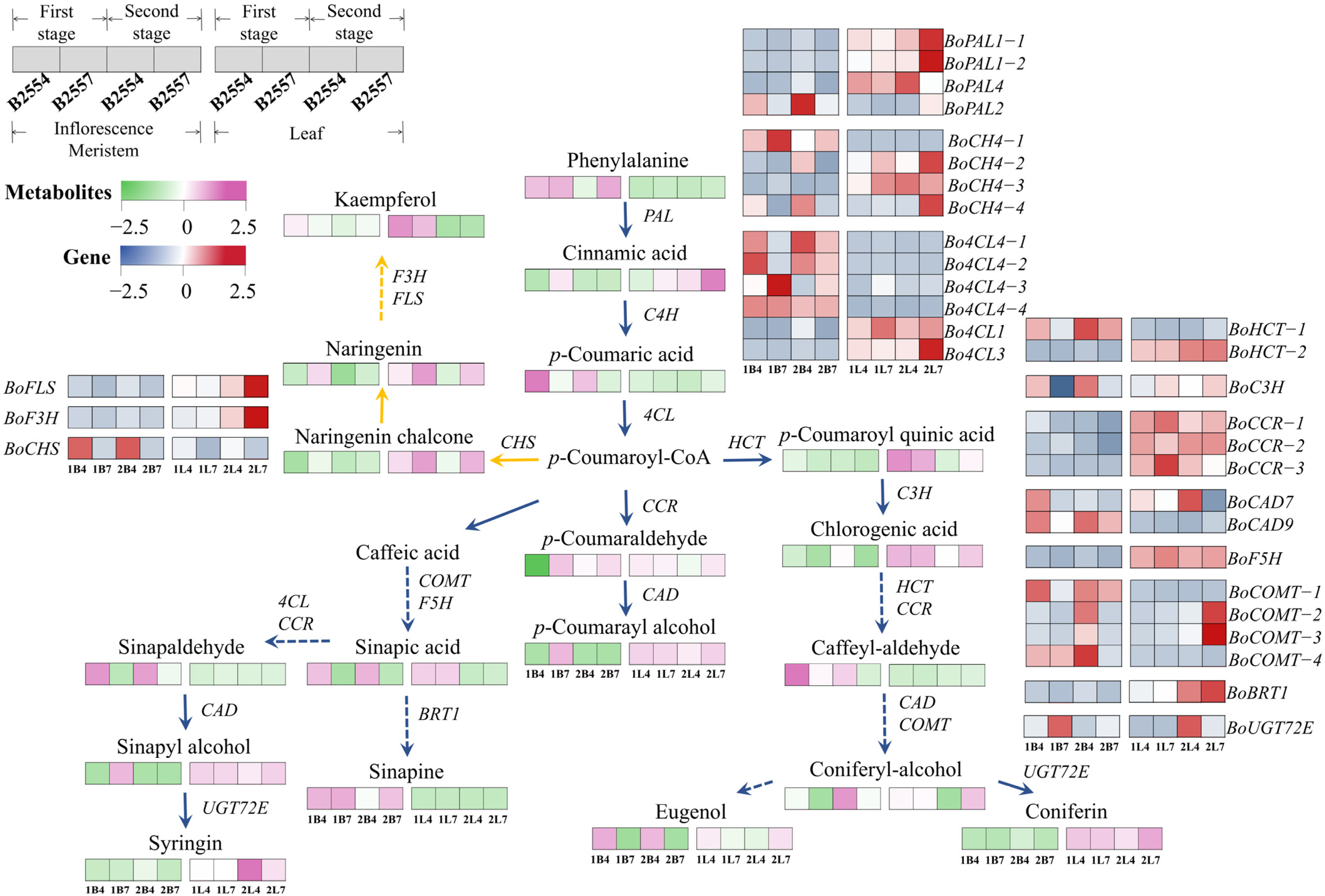
2.7. Differential Regulation in Phenylpropanoid and Flavonoid Biosynthesis Pathways
3. Discussion
3.1. The Interactions Between Flowering Locus T and Its Promoter/Repressors Regulate Broccoli Bolting
3.2. Activated Flower Meristem Genes Regulate the Bolting of the Apical Flower Buds in Broccoli
3.3. The Relationship Between Hormone Levels and the Early- and Late-Bolting Phenotypes
3.4. Sugar-Exporting Transporters Likely Regulate the Early Bolting of the Apical Flower Buds of Broccoli and Play a Crucial Role in the Long-Distance Transportation Process of Sugar
3.5. Phenolic Compounds May Affect the Early Bolting of Broccoli
4. Materials and Methods
4.1. Plant Materials
4.2. Metabolomics Analysis
4.3. Transcriptome Analysis
4.4. qRT-PCR Validation of Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, L.; Ma, P.; Nirasawa, S.; Liu, H. Formation, immunomodulatory activities, and enhancement of glucosinolates and sulforaphane in broccoli sprouts: A review for maximizing the health benefits to human. Crit. Rev. Food Sci. Nutr. 2024, 64, 7118–7148. [Google Scholar] [CrossRef] [PubMed]
- Ruhee, R.T.; Suzuki, K. The Integrative Role of Sulforaphane in Preventing Inflammation, Oxidative Stress and Fatigue: A Review of a Potential Protective Phytochemical. Antioxidants 2020, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.; Wellmer, F. Molecular regulation of flower development. Curr. Top. Dev. Biol. 2019, 131, 185–210. [Google Scholar] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of Flowering in Arabidopsis. Cell 2010, 141, 550–550.e2. [Google Scholar] [CrossRef]
- Ionescu, I.A.; Møller, B.L.; Sánchez-Pérez, R. Chemical control of flowering time. J. Exp. Bot. 2016, 68, 369–382. [Google Scholar] [CrossRef]
- Cai, K.; Zhu, S.; Jiang, Z.; Xu, K.; Sun, X.; Li, X. Biological macromolecules mediated by environmental signals affect flowering regulation in plants: A comprehensive review. Plant Physiol. Biochem. 2024, 214, 108931. [Google Scholar] [CrossRef]
- Cheng, Y.; Zhao, Y. A Role for Auxin in Flower Development. J. Integr. Plant Biol. 2007, 49, 99–104. [Google Scholar] [CrossRef]
- Cheng, H.; Qin, L.; Lee, S.; Fu, X.; Richards, D.E.; Cao, D.; Luo, D.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 2004, 131, 1055–1064. [Google Scholar] [CrossRef]
- Zhang, C.; Jian, M.; Li, W.; Yao, X.; Tan, C.; Qian, Q.; Hu, Y.; Liu, X.; Hou, X. Gibberellin signaling modulates flowering via the DELLA-BRAHMA-NF-YC module in Arabidopsis. Plant Cell 2023, 35, 3470–3484. [Google Scholar] [CrossRef]
- Achard, P.; Baghour, M.; Chapple, A.; Hedden, P.; Van Der Straeten, D.; Genschik, P.; Moritz, T.; Harberd, N.P. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 2007, 104, 6484–6489. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Cheng, H.; De Grauwe, L.; Decat, J.; Schoutteten, H.; Moritz, T.; Van Der Straeten, D.; Peng, J.R.; Harberd, N.P. Integration of plant responses to environmentally activated phytohormonal signals. Science 2006, 311, 91–94. [Google Scholar] [CrossRef]
- Creelman, R.; Mullet, J. Biosynthesis and Action of Jasmonates in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 355–381. [Google Scholar] [CrossRef]
- Hong, G.J.; Xue, X.Y.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Arabidopsis MYC2 Interacts with DELLA Proteins in Regulating Sesquiterpene Synthase Gene Expression. Plant Cell 2012, 24, 2635–2648. [Google Scholar] [CrossRef]
- Guo, W.J.; Pommerrenig, B.; Neuhaus, H.E.; Keller, I. Interaction between sugar transport and plant development. J. Plant Physiol. 2023, 288, 154073. [Google Scholar] [CrossRef]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.; Stitt, M.; Schmid, M. Regulation of Flowering by Trehalose-6-Phosphate Signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.; Onai, K.; Furukawa, Y.; Aoki, E.; Araki, T.; Nakamura, K. Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 2001, 127, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Heyer, A.G.; Raap, M.; Schroeer, B.; Marty, B.; Willmitzer, L. Cell wall invertase expression at the apical meristem alters floral, architectural, and reproductive traits in Arabidopsis thaliana. Plant J. 2004, 39, 161–169. [Google Scholar] [CrossRef]
- Ponnu, J.; Wahl, V.; Schmid, M. Trehalose-6-phosphate: Connecting plant metabolism and development. Front. Plant Sci. 2011, 2, 70. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Reinders, A.; Johnson, M.E.; Krentz, A.D.; Grof, C.P.; Perroux, J.M.; Ward, J.M. Arabidopsis sucrose transporter AtSUC9. High-affinity transport activity, intragenic control of expression, and early flowering mutant phenotype. Plant Physiol. 2007, 143, 188–198. [Google Scholar] [CrossRef]
- Yang, L.; Xu, M.; Koo, Y.; He, J.; Poethig, R.S. Sugar promotes vegetative phase change in Arabidopsis thaliana by repressing the expression of MIR156A and MIR156C. Elife 2013, 2, e00260. [Google Scholar] [CrossRef] [PubMed]
- Cho, L.H.; Pasriga, R.; Yoon, J.; Jeon, J.S.; An, G. Roles of Sugars in Controlling Flowering Time. J. Plant Biol. 2018, 61, 121–130. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, B.; Guo, H.; Chen, L.; Han, F.; Yan, C.; Yang, L.; Zhuang, M.; Lv, H.; Wang, Y.; et al. Transcriptome Analysis Reveals Key Genes and Pathways Associated with the Regulation of Flowering Time in Cabbage (Brassica oleracea L. var. capitata). Plants 2023, 12, 3413. [Google Scholar] [CrossRef]
- Wei, X.; Rahim, M.A.; Zhao, Y.; Yang, S.; Wang, Z.; Su, H.; Li, L.; Niu, L.; Harun-Ur-Rashid, M.; Yuan, Y.; et al. Comparative Transcriptome Analysis of Early- and Late-Bolting Traits in Chinese Cabbage (Brassica rapa). Front. Genet. 2021, 12, 590830. [Google Scholar] [CrossRef]
- Nie, S.; Li, C.; Wang, Y.; Xu, L.; Muleke, E.M.; Tang, M.; Sun, X.; Liu, L. Transcriptomic Analysis Identifies Differentially Expressed Genes (DEGs) Associated with Bolting and Flowering in Radish (Raphanus sativus L.). Front. Plant Sci. 2016, 7, 682. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.Y.; Park, H.J.; Lee, A.; Lee, S.S.; Kim, Y.S.; Cho, H.S. Identification of Flowering-Related Genes Responsible for Differences in Bolting Time between Two Radish Inbred Lines. Front. Plant Sci. 2016, 7, 1844. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Valverde, F.; Mouradov, A.; Soppe, W.; Ravenscroft, D.; Samach, A.; Coupland, G. Photoreceptor Regulation of CONSTANS Protein in Photoperiodic Flowering. Science 2004, 303, 1003–1006. [Google Scholar] [CrossRef]
- Hu, H.; Tian, S.; Xie, G.; Liu, R.; Wang, N.; Li, S.; He, Y.; Du, J. TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2103895118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, L.; Zeng, L.; Zhang, C.; Ma, H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes. Dev. 2015, 29, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Bartrina, I.; Schmülling, T. Cytokinin regulates vegetative phase change in Arabidopsis thaliana through the miR172/TOE1-TOE2 module. Nat. Commun. 2021, 12, 5816. [Google Scholar] [CrossRef]
- Wu, J.F.; Tsai, H.L.; Joanito, I.; Wu, Y.C.; Chang, C.W.; Li, Y.H.; Wang, Y.; Hong, J.C.; Chu, J.W.; Hsu, C.P.; et al. LWD-TCP complex activates the morning gene CCA1 in Arabidopsis. Nat. Commun. 2016, 7, 13181. [Google Scholar] [CrossRef]
- Levy, Y.Y.; Mesnage, S.; Mylne, J.S.; Gendall, A.R.; Dean, C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 2002, 297, 243–246. [Google Scholar] [CrossRef]
- Luo, X.; Chen, T.; Zeng, X.; He, D.; He, Y. Feedback Regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5′ FLC Promoter Region in Arabidopsis. Mol. Plant 2019, 12, 285–288. [Google Scholar] [CrossRef]
- Mateos, J.L.; Madrigal, P.; Tsuda, K.; Rawat, V.; Richter, R.; Romera-Branchat, M.; Fornara, F.; Schneeberger, K.; Krajewski, P.; Coupland, G. Combinatorial activities of SHORT VEGETATIVE PHASE and FLOWERING LOCUS C define distinct modes of flowering regulation in Arabidopsis. Genome Biol. 2015, 16, 31. [Google Scholar] [CrossRef]
- Rieu, I.; Ruiz-Rivero, O.; Fernandez-Garcia, N.; Griffiths, J.; Powers, S.J.; Gong, F.; Linhartova, T.; Eriksson, S.; Nilsson, O.; Thomas, S.G.; et al. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. For. Cell Mol. Biol. 2008, 53, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Porri, A.; Torti, S.; Romera-Branchat, M.; Coupland, G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198–2209. [Google Scholar] [CrossRef]
- Griffiths, J.; Murase, K.; Rieu, I.; Zentella, R.; Zhang, Z.L.; Powers, S.J.; Gong, F.; Phillips, A.L.; Hedden, P.; Sun, T.P.; et al. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 2006, 18, 3399–3414. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Xue, M.; Xu, J.; Liao, R.; Shang, S.; Yang, X.; Zhao, Y.; Li, C.; Liu, Z.; et al. BrKAO2 mutations disrupt leafy head formation in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Theor. Appl. Genet. 2022, 135, 2453–2468. [Google Scholar] [CrossRef] [PubMed]
- Hugouvieux, V.; Silva, C.S.; Jourdain, A.; Stigliani, A.; Charras, Q.; Conn, V.; Conn, S.J.; Carles, C.C.; Parcy, F.; Zubieta, C. Tetramerization of MADS family transcription factors SEPALLATA3 and AGAMOUS is required for floral meristem determinacy in Arabidopsis. Nucleic Acids Res. 2018, 46, 4966–4977. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Wu, M.F.; Winter, C.M.; Berns, M.C.; Nole-Wilson, S.; Yamaguchi, A.; Coupland, G.; Krizek, B.A.; Wagner, D. A molecular framework for auxin-mediated initiation of flower primordia. Dev. Cell 2013, 24, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Strader, L.C. An auxin research odyssey: 1989–2023. Plant Cell 2024, 36, 1410–1428. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Liu, Y.; Wang, Y.; Li, H.; Liu, J.; Tan, J.; He, J.; Bai, J.; Ma, H. Evolution Analysis of the Aux/IAA Gene Family in Plants Shows Dual Origins and Variable Nuclear Localization Signals. Int. J. Mol. Sci. 2017, 18, 2107. [Google Scholar] [CrossRef]
- Wang, X.; Xu, N.; Dong, K.; Li, H.; Shi, S.; Liu, Z.; Cao, X.; Cui, X.; Dai, S. SlNCED1 affects pollen maturation in tomato by regulating the expression of anther-specific genes. Plant Growth Regul. 2021, 95, 191–205. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhang, X.; Wu, F.; Feng, H.; Deng, L.; Xu, L.; Zhang, M.; Wang, Q.; Li, C. Transcriptional Mechanism of Jasmonate Receptor COI1-Mediated Delay of Flowering Time in Arabidopsis. Plant Cell 2015, 27, 2814–2828. [Google Scholar] [CrossRef]
- Torrens-Spence, M.P.; Bobokalonova, A.; Carballo, V.; Glinkerman, C.M.; Pluskal, T.; Shen, A.; Weng, J.K. PBS3 and EPS1 Complete Salicylic Acid Biosynthesis from Isochorismate in Arabidopsis. Mol. Plant 2019, 12, 1577–1586. [Google Scholar] [CrossRef]
- Wada, K.C.; Yamada, M.; Shiraya, T.; Takeno, K. Salicylic acid and the flowering gene FLOWERING LOCUS T homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil. J. Plant Physiol. 2010, 167, 447–452. [Google Scholar] [CrossRef]
- Winter, H.; Huber, S.C. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 253–289. [Google Scholar] [CrossRef]
- Abelenda, J.A.; Bergonzi, S.; Oortwijn, M.; Sonnewald, S.; Du, M.; Visser, R.G.F.; Sonnewald, U.; Bachem, C.W.B. Source-Sink Regulation Is Mediated by Interaction of an FT Homolog with a SWEET Protein in Potato. Curr. Biol. 2019, 29, 1178–1186.e6. [Google Scholar] [CrossRef]
- Andrés, F.; Kinoshita, A.; Kalluri, N.; Fernández, V.; Falavigna, V.S.; Cruz, T.M.D.; Jang, S.; Chiba, Y.; Seo, M.; Mettler-Altmann, T.; et al. The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 53. [Google Scholar] [CrossRef] [PubMed]
- Ylstra, B.; Busscher, J.; Franken, J.; Hollman, P.C.H.; Mol, J.N.M.; Van Tunen, A.J. Flavonols and fertilization in Petunia hybrida: Localization and mode of action during pollen tube growth. Plant J. 2003, 6, 201–212. [Google Scholar] [CrossRef]
- Mo, Y.; Nagel, C.; Taylor, L.P. Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 1992, 89, 7213–7217. [Google Scholar] [CrossRef] [PubMed]
- Malik, N.S.A.; Bradford, J.M. Changes in oleuropein levels during differentiation and development of floral buds in ‘Arbequina’ olives. Sci. Hortic. 2006, 110, 274–278. [Google Scholar] [CrossRef]
- Bongi, O. Oleuropein: An Olea europaea secoiridioid active on growth regulation. Acta Hort. 1986, 179, 245–249. [Google Scholar] [CrossRef]
- Ishimaru, A.; Ishimaru, K.; Ishimaru, M. Correlation of Flowering Induced by Low Temperature and Endogenous Levels of Phenylpropanoids inPharbitis nil: A Study with a Secondary-metabolism Mutant. J. Plant Physiol. 1996, 148, 672–676. [Google Scholar] [CrossRef]
- Pei, L.; Gao, Y.; Feng, L.; Zhang, Z.; Liu, N.; Yang, B.; Zhao, N. Phenolic Acids and Flavonoids Play Important Roles in Flower Bud Differentiation in Mikania micrantha: Transcriptomics and Metabolomics. Int. J. Mol. Sci. 2023, 24, 16550. [Google Scholar] [CrossRef]
- Boudet, A.-M. Evolution and current status of research in phenolic compounds. Phytochemistry 2007, 68, 2722–2735. [Google Scholar] [CrossRef]
- Ehlting, J.; Büttner, D.; Wang, Q.; Douglas, C.J.; Somssich, I.E.; Kombrink, E. Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 2002, 19, 9–20. [Google Scholar] [CrossRef]
- Zou, J.; Wang, P.; Xu, H.; Gan, X.; Zhang, H.; Chen, L.; Chen, H.; Wang, F.; Hu, Y.; Liu, Y. Metabolic profile and bioactivity of the peel of Zhoupigan (Citrus reticulata cv. Manau gan), a special citrus variety in China, based on GC–MS, UPLC-ESI-MS/MS analysis, and in vitro assay. Food Chem. X 2024, 23, 101719. [Google Scholar] [CrossRef] [PubMed]
- Mu, W. RNA Extraction for Plant Samples Using CTAB-pBIOZOL; Springer Nature: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuang, L.; Zhang, Y.; Zhang, N.; Zhang, Y.; Lv, H.; Wang, Y.; Zhuang, M.; Yang, L.; Huang, K.; Li, Z.; et al. Transcriptome and Metabolome Analyses Offer New Insights into Bolting Time Regulation in Broccoli. Int. J. Mol. Sci. 2025, 26, 3726. https://doi.org/10.3390/ijms26083726
Kuang L, Zhang Y, Zhang N, Zhang Y, Lv H, Wang Y, Zhuang M, Yang L, Huang K, Li Z, et al. Transcriptome and Metabolome Analyses Offer New Insights into Bolting Time Regulation in Broccoli. International Journal of Molecular Sciences. 2025; 26(8):3726. https://doi.org/10.3390/ijms26083726
Chicago/Turabian StyleKuang, Linqian, Yue Zhang, Nan Zhang, Yangyong Zhang, Honghao Lv, Yong Wang, Mu Zhuang, Limei Yang, Ke Huang, Zhansheng Li, and et al. 2025. "Transcriptome and Metabolome Analyses Offer New Insights into Bolting Time Regulation in Broccoli" International Journal of Molecular Sciences 26, no. 8: 3726. https://doi.org/10.3390/ijms26083726
APA StyleKuang, L., Zhang, Y., Zhang, N., Zhang, Y., Lv, H., Wang, Y., Zhuang, M., Yang, L., Huang, K., Li, Z., & Ji, J. (2025). Transcriptome and Metabolome Analyses Offer New Insights into Bolting Time Regulation in Broccoli. International Journal of Molecular Sciences, 26(8), 3726. https://doi.org/10.3390/ijms26083726








