Current Perspectives on Functional Involvement of Micropeptides in Virus–Host Interactions
Abstract
1. Introduction
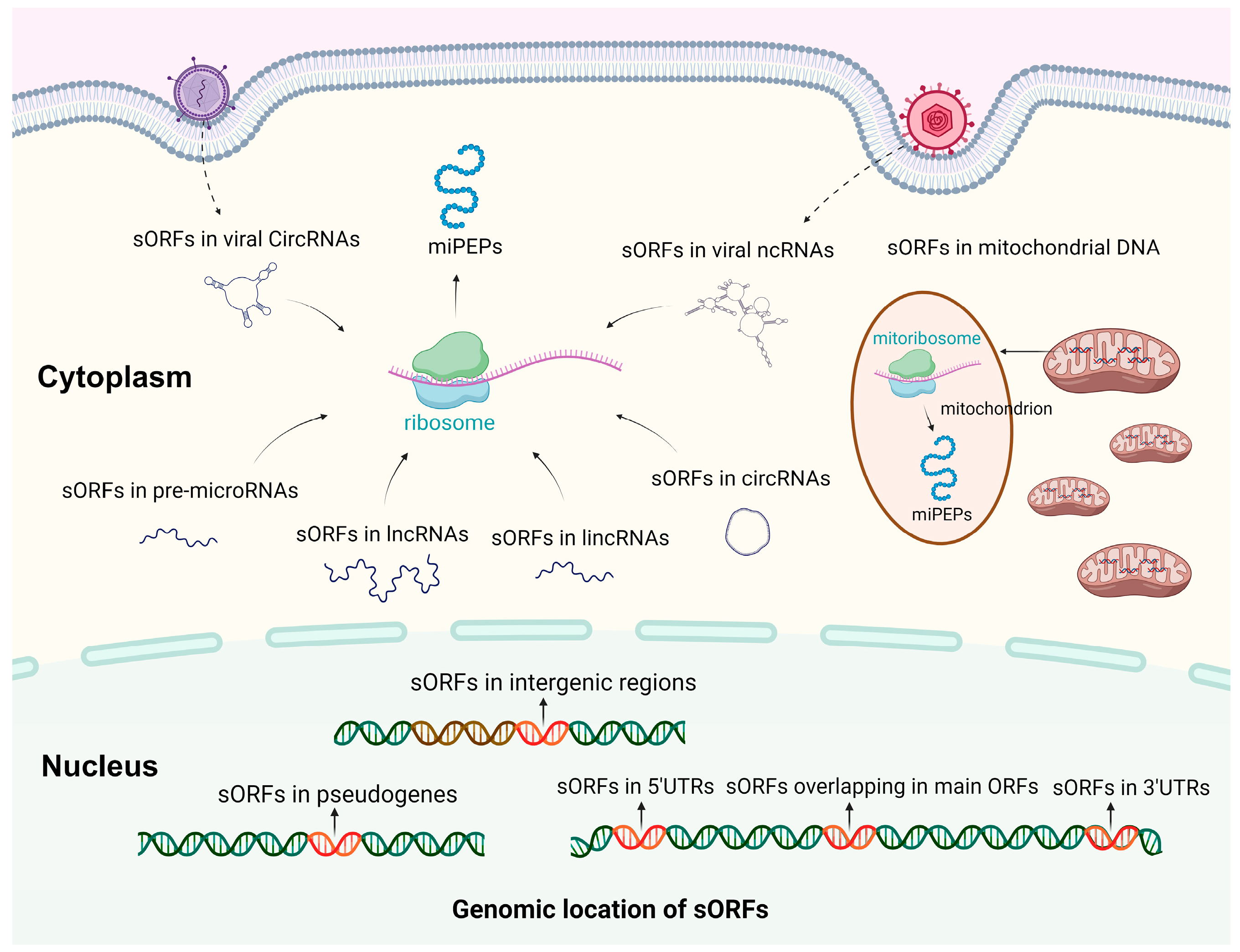
2. Biogenesis and Characterization of miPEPs
3. Roles of miPEPs in Virus–Host Interactions
3.1. Virus–Host Interactions
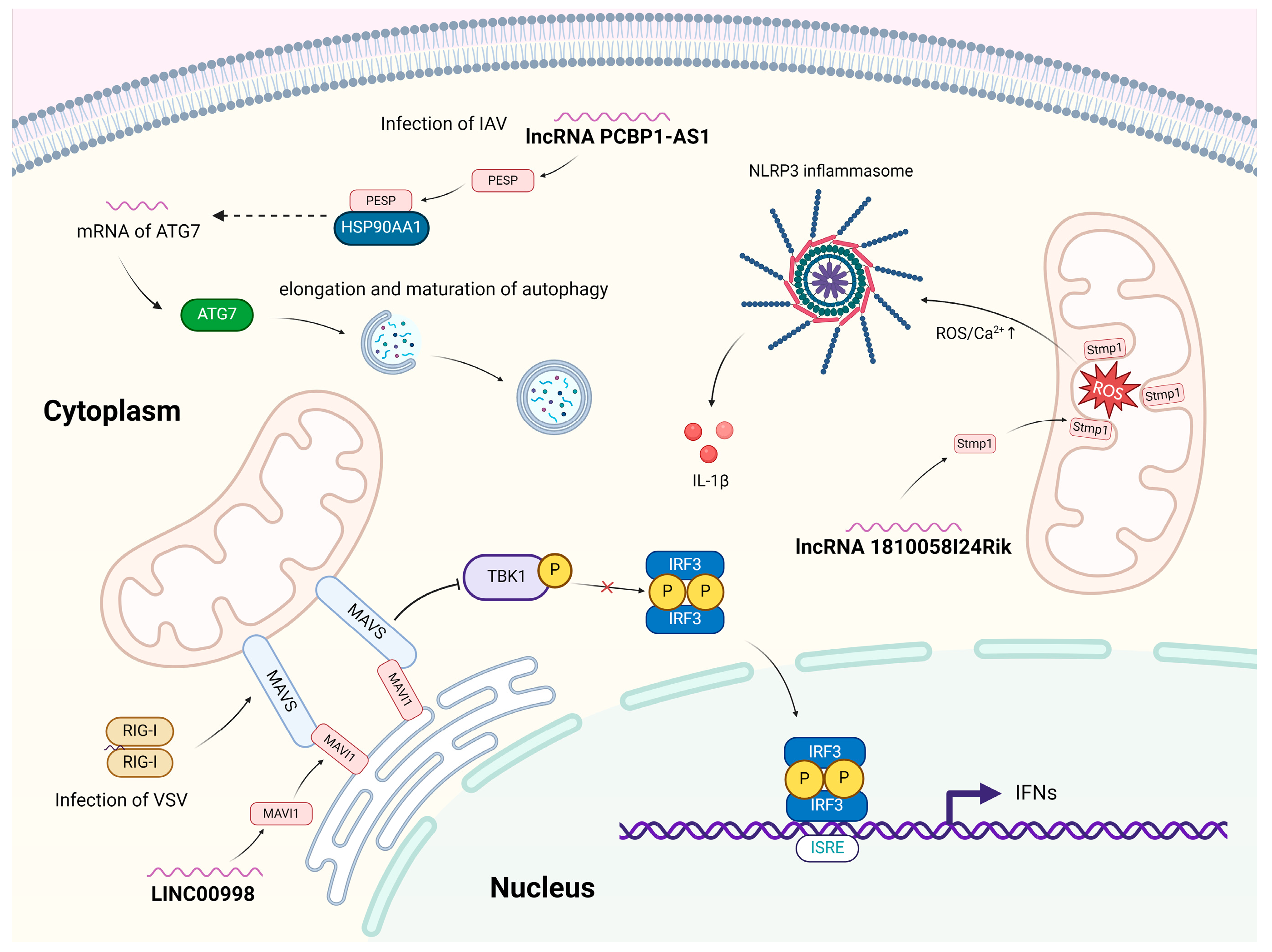
3.2. Roles of Host miPEPs in Virus–Host Interactions
3.3. Roles of Virus-Derived miPEPs in Virus–Host Interactions
| Name | Size (aa) | Origin/Source | Virus–Host Interactions | Function/Mechanism | Ref. |
|---|---|---|---|---|---|
| Host miPEPs involved in virus–host interactions | |||||
| ORF-674 | 71 | XR_001139971.3 (lnc557) | Bombyx mori nucleopolyhedrovirus (BmNPV) | Not Available | [56] |
| PESP | 110 | lncRNA PCBP1-AS1 (human) | IAV | Enhances the IAV-induced autophagy by increasing the expression of ATG7. | [2] |
| MIR22HG peptide | NA | lncRNA MIR22HG (human) | IAV | Not Available | [47] |
| SMIM30/ MAVI1 | 59 | LINC00998 | VSV | Endoplasmic reticulum–localized microprotein that suppresses antiviral innate immune response by targeting MAVS on mitochondria. | [50] |
| Virus-encoded miPEPs involved in virus–host interactions | |||||
| vsp21 | 21 | vcircRNA_000048 (silkworm) | BmCPV | Attenuates the viral replication. | [40] |
| vSP27 | NA | circular RNA (circRNA-vSP27) | BmCPV | Suppresses BmCPV infection. | [51] |
| VSP59 | 59 | S10 dsRNA genome | BmCPV | Negatively regulates of viral replication. | [57] |
| PB1-F2 | 87–90 | Influenza A/PR/8/34 virus | IAV | Mitochondria localized miPEP that induces apoptosis in host cells. | [58] |
| vSP-1 | 48 | T3.0 RNA from KSHV | KSHV | Precisely control of RTA abundance and activity in KSHV reactivation and initiates the establishment of latency of the KSHV. | [53,54] |
| vSP-2 | 27 | T3.0 RNA from KSHV | KSHV | Not Available | [53,54] |
3.4. Roles of Host-Derived miPEPs in Physiological and Pathological Processes: Potential Roles in Virus–Host Interactions
| Name | Size (aa) | Origin/Source | Function/Mechanism | Ref. |
|---|---|---|---|---|
| ASRPS | 60 | LINC00908 | miPEPs regulate innate or adaptive immunity. | [15,84,85,86,87] |
| P155 | 17 | lncRNA MIR155HG | ||
| miPEP31 | 44 | pri-miRNA-31 | ||
| Stmp1/Mm47 | 47 | lncRNA 1810058I24Rik | Activates the NLRP3 inflammasome pathway | [43,77,78,79,88] |
| SHLP2 | 26 | mitochondrial 16S rRNA gene | Regulate apoptosis. | [26,27,49,75,76] |
| PIGBOS | 54 | PIGB opposite strand 1 | ||
| FORCP | 79 | LINC00675 | ||
| YY1BM | 21 | LINC00278 | ||
| AC115619-22aa | 22 | lncRNA AC115619 | Regulates autophagy | [48] |
| PINT87aa | 87 | LINC-PINT | Regulates mitophagy | [89] |
| PACMP | 44 | lncRNA CTD-2256P15.2 | Modulates DNA damage response. | [90] |
4. Challenges and Future Perspectives of miPEP Study
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HSP90 | Heat shock protein 90 |
| NLRP3 | NOD-like receptor family Pyrin domain-containing protein 3 |
| LincRNAs | Long intronic non-coding RNAs |
| CRISPR-Cas9 | Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein 9 |
References
- Couso, J.P.; Patraquim, P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017, 18, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Chi, X.; Huang, G.; Wang, L.; Zhang, X.; Liu, J.; Yin, Z.; Guo, G.; Chen, Y.; Wang, S.; Chen, J.L. A small protein encoded by PCBP1-AS1 is identified as a key regulator of influenza virus replication via enhancing autophagy. PLoS Pathog. 2024, 20, e1012461. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhang, K.; Xun, C.; Chu, T.; Liang, S.; Zeng, Y.; Liu, Z. Small Open Reading Frame-Encoded Micro-Peptides: An Emerging Protein World. Int. J. Mol. Sci. 2023, 24, 10562. [Google Scholar] [CrossRef] [PubMed]
- Patraquim, P.; Magny, E.G.; Pueyo, J.I.; Platero, A.I.; Couso, J.P. Translation and natural selection of micropeptides from long non-canonical RNAs. Nat. Commun. 2022, 13, 6515. [Google Scholar] [CrossRef]
- Ryczek, N.; Łyś, A.; Makałowska, I. The Functional Meaning of 5′UTR in Protein-Coding Genes. Int. J. Mol. Sci. 2023, 24, 2976. [Google Scholar] [CrossRef]
- Tornesello, A.L.; Cerasuolo, A.; Starita, N.; Amiranda, S.; Cimmino, T.P.; Bonelli, P.; Tuccillo, F.M.; Buonaguro, F.M.; Buonaguro, L.; Tornesello, M.L. Emerging role of endogenous peptides encoded by non-coding RNAs in cancer biology. Noncoding RNA Res. 2025, 10, 231–241. [Google Scholar] [CrossRef]
- Kang, M.; Tang, B.; Li, J.; Zhou, Z.; Liu, K.; Wang, R.; Jiang, Z.; Bi, F.; Patrick, D.; Kim, D.; et al. Correction: Identification of miPEP133 as a novel tumor-suppressor microprotein encoded by miR-34a pri-miRNA. Mol. Cancer 2024, 23, 195. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, X.; Lai, K.; Zhou, R.; Chen, Z.; Yang, Z.; Gao, X. Discovery of the hidden coding information in cancers: Mechanisms and biological functions. Int. J. Cancer 2023, 153, 20–32. [Google Scholar] [CrossRef]
- Olexiouk, V.; Crappé, J.; Verbruggen, S.; Verhegen, K.; Martens, L.; Menschaert, G. sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016, 44, D324–D329. [Google Scholar] [CrossRef]
- Erokhina, T.N.; Ryazantsev, D.Y.; Zavriev, S.K.; Morozov, S.Y. Regulatory miPEP Open Reading Frames Contained in the Primary Transcripts of microRNAs. Int. J. Mol. Sci. 2023, 24, 2114. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Niikura, T.; Tajima, H.; Yasukawa, T.; Sudo, H.; Ito, Y.; Kita, Y.; Kawasumi, M.; Kouyama, K.; Doyu, M.; et al. A rescue factor abolishing neuronal cell death by a wide spectrum of familial Alzheimer’s disease genes and Abeta. Proc. Natl. Acad. Sci. USA 2001, 98, 6336–6341. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Gribskov, M. MiPepid: MicroPeptide identification tool using machine learning. BMC Bioinform. 2019, 20, 559. [Google Scholar] [CrossRef]
- Xiao, Y.; Ren, Y.; Hu, W.; Paliouras, A.R.; Zhang, W.; Zhong, L.; Yang, K.; Su, L.; Wang, P.; Li, Y.; et al. Long non-coding RNA-encoded micropeptides: Functions, mechanisms and implications. Cell Death Discov. 2024, 10, 450. [Google Scholar] [CrossRef]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef]
- Rai, K.R.; Shrestha, P.; Yang, B.; Chen, Y.; Liu, S.; Maarouf, M.; Chen, J.L. Acute Infection of Viral Pathogens and Their Innate Immune Escape. Front. Microbiol. 2021, 12, 672026. [Google Scholar] [CrossRef]
- Strumillo, S.T.; Kartavykh, D.; de Carvalho, F.F., Jr.; Cruz, N.C.; de Souza Teodoro, A.C.; Sobhie Diaz, R.; Curcio, M.F. Host-virus interaction and viral evasion. Cell Biol. Int. 2021, 45, 1124–1147. [Google Scholar] [CrossRef]
- Zhao, J.; Li, Y.; Wang, C.; Zhang, H.; Zhang, H.; Jiang, B.; Guo, X.; Song, X. IRESbase: A Comprehensive Database of Experimentally Validated Internal Ribosome Entry Sites. Genom. Proteom. Bioinform. 2020, 18, 129–139. [Google Scholar] [CrossRef]
- Tang, M.; Lv, Y. The Role of N(6)-Methyladenosine Modified Circular RNA in Pathophysiological Processes. Int. J. Biol. Sci. 2021, 17, 2262–2277. [Google Scholar] [CrossRef]
- Wright, B.W.; Yi, Z.; Weissman, J.S.; Chen, J. The dark proteome: Translation from noncanonical open reading frames. Trends Cell Biol. 2022, 32, 243–258. [Google Scholar] [CrossRef]
- Frei, Y.; Immarigeon, C.; Revel, M.; Karch, F.; Maeda, R.K. Upstream open reading frames repress the translation from the iab-8 RNA. PLoS Genet. 2024, 20, e1011214. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Zeng, J.; Drew, B.G.; Sallam, T.; Martin-Montalvo, A.; Wan, J.; Kim, S.J.; Mehta, H.; Hevener, A.L.; de Cabo, R.; et al. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab. 2015, 21, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Merry, T.L.; Chan, A.; Woodhead, J.S.T.; Reynolds, J.C.; Kumagai, H.; Kim, S.J.; Lee, C. Mitochondrial-derived peptides in energy metabolism. Am. J. Physiol. Endocrinol. Metab. 2020, 319, E659–E666. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yen, K.; Cohen, P. Humanin: A harbinger of mitochondrial-derived peptides? Trends Endocrinol. Metab. 2013, 24, 222–228. [Google Scholar] [CrossRef]
- Yen, K.; Mehta, H.H.; Kim, S.J.; Lue, Y.; Hoang, J.; Guerrero, N.; Port, J.; Bi, Q.; Navarrete, G.; Brandhorst, S.; et al. The mitochondrial derived peptide humanin is a regulator of lifespan and healthspan. Aging 2020, 12, 11185–11199. [Google Scholar] [CrossRef]
- Cobb, L.J.; Lee, C.; Xiao, J.; Yen, K.; Wong, R.G.; Nakamura, H.K.; Mehta, H.H.; Gao, Q.; Ashur, C.; Huffman, D.M.; et al. Naturally occurring mitochondrial-derived peptides are age-dependent regulators of apoptosis, insulin sensitivity, and inflammatory markers. Aging 2016, 8, 796–809. [Google Scholar] [CrossRef]
- Kim, S.K.; Tran, L.T.; NamKoong, C.; Choi, H.J.; Chun, H.J.; Lee, Y.H.; Cheon, M.; Chung, C.; Hwang, J.; Lim, H.H.; et al. Author Correction: Mitochondria-derived peptide SHLP2 regulates energy homeostasis through the activation of hypothalamic neurons. Nat. Commun. 2023, 14, 4995. [Google Scholar] [CrossRef]
- Olexiouk, V.; Van Criekinge, W.; Menschaert, G. An update on sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018, 46, D497–D502. [Google Scholar] [CrossRef]
- Hao, Y.; Zhang, L.; Niu, Y.; Cai, T.; Luo, J.; He, S.; Zhang, B.; Zhang, D.; Qin, Y.; Yang, F.; et al. SmProt: A database of small proteins encoded by annotated coding and non-coding RNA loci. Brief. Bioinform. 2018, 19, 636–643. [Google Scholar] [CrossRef]
- Brunet, M.A.; Lucier, J.F.; Levesque, M.; Leblanc, S.; Jacques, J.F.; Al-Saedi, H.R.H.; Guilloy, N.; Grenier, F.; Avino, M.; Fournier, I.; et al. OpenProt 2021: Deeper functional annotation of the coding potential of eukaryotic genomes. Nucleic Acids Res. 2021, 49, D380–D388. [Google Scholar] [CrossRef]
- Hazarika, R.R.; De Coninck, B.; Yamamoto, L.R.; Martin, L.R.; Cammue, B.P.; van Noort, V. ARA-PEPs: A repository of putative sORF-encoded peptides in Arabidopsis thaliana. BMC Bioinform. 2017, 18, 37. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Taniguchi, T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 2006, 6, 644–658. [Google Scholar] [CrossRef] [PubMed]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Park, E.S.; Dezhbord, M.; Lee, A.R.; Kim, K.H. The Roles of Ubiquitination in Pathogenesis of Influenza Virus Infection. Int. J. Mol. Sci. 2022, 23, 4593. [Google Scholar] [CrossRef]
- Resnik, R.; Lopez Mingorance, F.; Rivera, F.; Mitchell, F.; Gonzalez, C.D.; Vaccaro, M.I. Autophagy in Inflammatory Response against SARS-CoV-2. Int. J. Mol. Sci. 2023, 24, 4928. [Google Scholar] [CrossRef]
- Lamy-Besnier, Q.; Brancotte, B.; Ménager, H.; Debarbieux, L. Viral Host Range database, an online tool for recording, analyzing and disseminating virus-host interactions. Bioinformatics 2021, 37, 2798–2801. [Google Scholar] [CrossRef]
- Rai, K.R.; Liao, Y.; Cai, M.; Qiu, H.; Wen, F.; Peng, M.; Wang, S.; Liu, S.; Guo, G.; Chi, X.; et al. MIR155HG Plays a Bivalent Role in Regulating Innate Antiviral Immunity by Encoding Long Noncoding RNA-155 and microRNA-155-5p. mBio 2022, 13, e0251022. [Google Scholar] [CrossRef]
- Ouyang, J.; Zhu, X.; Chen, Y.; Wei, H.; Chen, Q.; Chi, X.; Qi, B.; Zhang, L.; Zhao, Y.; Gao, G.F.; et al. NRAV, a long noncoding RNA, modulates antiviral responses through suppression of interferon-stimulated gene transcription. Cell Host Microbe 2014, 16, 616–626. [Google Scholar] [CrossRef]
- Chen, B.; Guo, G.; Wang, G.; Zhu, Q.; Wang, L.; Shi, W.; Wang, S.; Chen, Y.; Chi, X.; Wen, F.; et al. ATG7/GAPLINC/IRF3 axis plays a critical role in regulating pathogenesis of influenza A virus. PLoS Pathog. 2024, 20, e1011958. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, M.; Zhang, X.; Dai, K.; Liang, Z.; Pan, J.; Zhang, Z.; Cao, M.; Xue, R.; Cao, G.; et al. Micropeptide vsp21 translated by Reovirus circular RNA 000048 attenuates viral replication. Int. J. Biol. Macromol. 2022, 209 Pt A, 1179–1187. [Google Scholar] [CrossRef]
- Maarouf, M.; Rai, K.R.; Goraya, M.U.; Chen, J.L. Immune Ecosystem of Virus-Infected Host Tissues. Int. J. Mol. Sci. 2018, 19, 1379. [Google Scholar] [CrossRef] [PubMed]
- Sakowski, E.G.; Arora-Williams, K.; Tian, F.; Zayed, A.A.; Zablocki, O.; Sullivan, M.B.; Preheim, S.P. Interaction dynamics and virus-host range for estuarine actinophages captured by epicPCR. Nat. Microbiol. 2021, 6, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Wang, M.; Liu, S.; Chen, H.; Li, Y.; Yuan, F.; Yang, L.; Qiu, S.; Wang, H.; Xie, Z.; et al. A lncRNA-encoded mitochondrial micropeptide exacerbates microglia-mediated neuroinflammation in retinal ischemia/reperfusion injury. Cell Death Dis. 2023, 14, 126. [Google Scholar] [CrossRef]
- Kane, M.; Zang, T.M.; Rihn, S.J.; Zhang, F.; Kueck, T.; Alim, M.; Schoggins, J.; Rice, C.M.; Wilson, S.J.; Bieniasz, P.D. Identification of Interferon-Stimulated Genes with Antiretroviral Activity. Cell Host Microbe 2016, 20, 392–405. [Google Scholar] [CrossRef]
- Rai, K.R.; Chen, B.; Zhao, Z.; Chen, Y.; Hu, J.; Liu, S.; Maarouf, M.; Li, Y.; Xiao, M.; Liao, Y.; et al. Robust expression of p27Kip1 induced by viral infection is critical for antiviral innate immunity. Cell Microbiol. 2020, 22, e13242. [Google Scholar] [CrossRef]
- Li, F.; Chen, Y.; Zhang, Z.; Ouyang, J.; Wang, Y.; Yan, R.; Huang, S.; Gao, G.F.; Guo, G.; Chen, J.L. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res. 2015, 43, 10321–10337. [Google Scholar] [CrossRef] [PubMed]
- Razooky, B.S.; Obermayer, B.; O’May, J.B.; Tarakhovsky, A. Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs. Genes 2017, 8, 206. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, T.; Yan, L.; Zhu, S.; Jin, W.; Bai, Y.; Zeng, Y.; Zhang, X.; Yin, Z.; Yang, J.; et al. Hypoxia-Responsive lncRNA AC115619 Encodes a Micropeptide That Suppresses m6A Modifications and Hepatocellular Carcinoma Progression. Cancer Res. 2023, 83, 2496–2512. [Google Scholar] [CrossRef]
- Chu, Q.; Martinez, T.F.; Novak, S.W.; Donaldson, C.J.; Tan, D.; Vaughan, J.M.; Chang, T.; Diedrich, J.K.; Andrade, L.; Kim, A.; et al. Regulation of the ER stress response by a mitochondrial microprotein. Nat. Commun. 2019, 10, 4883. [Google Scholar] [CrossRef]
- Shi, T.T.; Huang, Y.; Li, Y.; Dai, X.L.; He, Y.H.; Ding, J.C.; Ran, T.; Shi, Y.; Yuan, Q.; Li, W.J.; et al. MAVI1, an endoplasmic reticulum-localized microprotein, suppresses antiviral innate immune response by targeting MAVS on mitochondrion. Sci. Adv. 2023, 9, eadg7053. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.; Dai, K.; Zhu, M.; Liang, Z.; Pan, J.; Zhang, Z.; Xue, R.; Cao, G.; Hu, X.; et al. Bombyx mori Akirin hijacks a viral peptide vSP27 encoded by BmCPV circRNA and activates the ROS-NF-κB pathway against viral infection. Int. J. Biol. Macromol. 2022, 194, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, M.; Pan, J.; Qiu, Q.; Tong, X.; Hu, X.; Gong, C. BmCPV replication is suppressed by the activation of the NF-κB/autophagy pathway through the interaction of vsp21 translated by vcircRNA_000048 with ubiquitin carboxyl-terminal hydrolase. Insect Biochem. Mol. Biol. 2023, 156, 103947. [Google Scholar] [CrossRef] [PubMed]
- Jaber, T.; Yuan, Y. A virally encoded small peptide regulates RTA stability and facilitates Kaposi’s sarcoma-associated herpesvirus lytic replication. J. Virol. 2013, 87, 3461–3470. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yuan, Y. Two microPeptides are translated from a KSHV polycistronic RNA in human cells by leaky scanning mechanism. Biochem. Biophys. Res. Commun. 2020, 522, 568–573. [Google Scholar] [CrossRef]
- Yang, Z.; Cao, S.; Martens, C.A.; Porcella, S.F.; Xie, Z.; Ma, M.; Shen, B.; Moss, B. Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling. J. Virol. 2015, 89, 6874–6886. [Google Scholar] [CrossRef]
- Lin, S.; Shen, Z.Y.; Wang, M.D.; Zhou, X.M.; Xu, T.; Jiao, X.H.; Wang, L.L.; Guo, X.J.; Wu, P. Lnc557 promotes Bombyx mori nucleopolyhedrovirus replication by interacting with BmELAVL1 to enhance its stability and expression. Pestic. Biochem. Physiol. 2024, 204, 106046. [Google Scholar] [CrossRef]
- Cao, M.; Qiu, Q.; Zhang, X.; Zhang, W.; Shen, Z.; Ma, C.; Zhu, M.; Pan, J.; Tong, X.; Cao, G.; et al. Identification and characterization of a novel small viral peptide (VSP59) encoded by Bombyx mori cypovirus (BmCPV) that negatively regulates viral replication. Microbiol. Spectr. 2024, 12, e0082624. [Google Scholar] [CrossRef]
- Zamarin, D.; García-Sastre, A.; Xiao, X.; Wang, R.; Palese, P. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 2005, 1, e4. [Google Scholar] [CrossRef]
- Zheng, W.; Guo, Y.; Zhang, G.; Bai, J.; Song, Y.; Song, X.; Zhu, Q.; Bao, X.; Wu, G.; Zhang, C. Peptide encoded by lncRNA BVES-AS1 promotes cell viability, migration, and invasion in colorectal cancer cells via the SRC/mTOR signaling pathway. PLoS ONE 2023, 18, e0287133. [Google Scholar] [CrossRef]
- Li, M.; Li, X.; Zhang, Y.; Wu, H.; Zhou, H.; Ding, X.; Zhang, X.; Jin, X.; Wang, Y.; Yin, X.; et al. Micropeptide MIAC Inhibits HNSCC Progression by Interacting with Aquaporin 2. J. Am. Chem. Soc. 2020, 142, 6708–6716. [Google Scholar] [CrossRef]
- Huang, J.Z.; Chen, M.; Chen, D.; Gao, X.C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell 2017, 68, 171–184.e6. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Fan, J.; Han, L.; Qi, H.; Wang, Y.; Wang, H.; Chen, S.; Du, L.; Li, S.; Zhang, Y.; et al. The micropeptide LEMP plays an evolutionarily conserved role in myogenesis. Cell Death Dis. 2020, 11, 357. [Google Scholar] [CrossRef] [PubMed]
- Szafron, L.A.; Iwanicka-Nowicka, R.; Podgorska, A.; Bonna, A.M.; Sobiczewski, P.; Kupryjanczyk, J.; Szafron, L.M. The Clinical Significance of CRNDE Gene Methylation, Polymorphisms, and CRNDEP Micropeptide Expression in Ovarian Tumors. Int. J. Mol. Sci. 2024, 25, 7531. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Dai, Y.; Yu, Y.; Tang, J.; Cao, Z.; Zhang, Y.; Li, B.; Nie, J.; Hei, T.K.; Zhou, G. The Tumorigenic Effect of lncRNA AFAP1-AS1 is Mediated by Translated Peptide ATMLP Under the Control of m(6) A Methylation. Adv. Sci. 2023, 10, e2300314. [Google Scholar] [CrossRef]
- Xu, W.; Deng, B.; Lin, P.; Liu, C.; Li, B.; Huang, Q.; Zhou, H.; Yang, J.; Qu, L. Ribosome profiling analysis identified a KRAS-interacting microprotein that represses oncogenic signaling in hepatocellular carcinoma cells. Sci. China Life Sci. 2020, 63, 529–542. [Google Scholar] [CrossRef]
- Pan, J.; Liu, M.; Duan, X.; Wang, D. A short peptide LINC00665_18aa encoded by lncRNA LINC00665 suppresses the proliferation and migration of osteosarcoma cells through the regulation of the CREB1/RPS6KA3 interaction. PLoS ONE 2023, 18, e0286422. [Google Scholar] [CrossRef]
- Guo, B.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Wang, Y.; Zhang, S.; Wu, R.; Lu, J.; et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. Embo J. 2020, 39, e102190. [Google Scholar] [CrossRef]
- Polenkowski, M.; Burbano de Lara, S.; Allister, A.B.; Nguyen, T.N.Q.; Tamura, T.; Tran, D.D.H. Identification of Novel Micropeptides Derived from Hepatocellular Carcinoma-Specific Long Noncoding RNA. Int. J. Mol. Sci. 2021, 23, 58. [Google Scholar] [CrossRef]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef]
- Ge, Q.; Jia, D.; Cen, D.; Qi, Y.; Shi, C.; Li, J.; Sang, L.; Yang, L.J.; He, J.; Lin, A.; et al. Micropeptide ASAP encoded by LINC00467 promotes colorectal cancer progression by directly modulating ATP synthase activity. J. Clin. Investig. 2021, 131, e152911. [Google Scholar] [CrossRef]
- Meng, K.; Lu, S.; Li, Y.Y.; Hu, L.L.; Zhang, J.; Cao, Y.; Wang, Y.; Zhang, C.Z.; He, Q.Y. LINC00493-encoded microprotein SMIM26 exerts anti-metastatic activity in renal cell carcinoma. EMBO Rep. 2023, 24, e56282. [Google Scholar] [CrossRef] [PubMed]
- Meng, N.; Chen, M.; Chen, D.; Chen, X.H.; Wang, J.Z.; Zhu, S.; He, Y.T.; Zhang, X.L.; Lu, R.X.; Yan, G.R. Small Protein Hidden in lncRNA LOC90024 Promotes “Cancerous” RNA Splicing and Tumorigenesis. Adv. Sci. 2020, 7, 1903233. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Cai, Y.; Deng, S.; Yang, L.; Liu, N.; Chang, X.; Jing, L.; Zhou, Y.; Li, H. A peptide CORO1C-47aa encoded by the circular noncoding RNA circ-0000437 functions as a negative regulator in endometrium tumor angiogenesis. J. Biol. Chem. 2021, 297, 101182. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, L.; Zhou, Y.; Wang, Q.; Zheng, Z.; Xu, B.; Wu, C.; Zhou, Q.; Hu, W.; Wu, C.; et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol. Cancer 2019, 18, 47. [Google Scholar] [CrossRef]
- Li, X.L.; Pongor, L.; Tang, W.; Das, S.; Muys, B.R.; Jones, M.F.; Lazar, S.B.; Dangelmaier, E.A.; Hartford, C.C.; Grammatikakis, I.; et al. A small protein encoded by a putative lncRNA regulates apoptosis and tumorigenicity in human colorectal cancer cells. eLife 2020, 9, e53734. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, L.; Deng, J.; Guo, B.; Li, F.; Wang, Y.; Wu, R.; Zhang, S.; Lu, J.; Zhou, Y. A Novel Micropeptide Encoded by Y-Linked LINC00278 Links Cigarette Smoking and AR Signaling in Male Esophageal Squamous Cell Carcinoma. Cancer Res. 2020, 80, 2790–2803. [Google Scholar] [CrossRef]
- Xie, C.; Wang, F.-Y.; Sang, Y.; Chen, B.; Huang, J.-H.; He, F.-J.; Li, H.; Zhu, Y.; Liu, X.; Zhuang, S.-M.; et al. Mitochondrial Micropeptide STMP1 Enhances Mitochondrial Fission to Promote Tumor Metastasis. Cancer Res. 2022, 82, 2431–2443. [Google Scholar] [CrossRef]
- Zheng, X.; Guo, Y.; Zhang, R.; Chen, H.; Liu, S.; Qiu, S.; Xiang, M. The mitochondrial micropeptide Stmp1 promotes retinal cell differentiation. Biochem. Biophys. Res. Commun. 2022, 636 Pt 2, 79–86. [Google Scholar] [CrossRef]
- Bhatta, A.; Atianand, M.; Jiang, Z.; Crabtree, J.; Blin, J.; Fitzgerald, K.A. A Mitochondrial Micropeptide Is Required for Activation of the Nlrp3 Inflammasome. J. Immunol. 2020, 204, 428–437. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, S.; Liu, M.; Wei, Y.; Wang, Q.; Shen, W.; Lei, C.Q.; Zhu, Q. The nucleoprotein of influenza A virus inhibits the innate immune response by inducing mitophagy. Autophagy 2023, 19, 1916–1933. [Google Scholar] [CrossRef]
- Zeng, Y.; Xu, S.; Wei, Y.; Zhang, X.; Wang, Q.; Jia, Y.; Wang, W.; Han, L.; Chen, Z.; Wang, Z.; et al. The PB1 protein of influenza A virus inhibits the innate immune response by targeting MAVS for NBR1-mediated selective autophagic degradation. PLoS Pathog. 2021, 17, e1009300. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, A.; Chen, T.; Feng, Y.; Zou, J.; Tu, S.; Jiang, M.; Sun, H.; Zhou, H. The Hemagglutinin of Influenza A Virus Induces Ferroptosis to Facilitate Viral Replication. Adv. Sci. 2024, 11, e2404365. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.T.; Grant, A.; Manicassamy, B.; Palese, P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J. Virol. 2012, 86, 8359–8366. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2020, 217, jem.20190950. [Google Scholar] [CrossRef]
- Zhou, H.; Lou, F.; Bai, J.; Sun, Y.; Cai, W.; Sun, L.; Xu, Z.; Liu, Z.; Zhang, L.; Yin, Q.; et al. A peptide encoded by pri-miRNA-31 represses autoimmunity by promoting T(reg) differentiation. EMBO Rep. 2022, 23, e53475. [Google Scholar] [CrossRef]
- Li, X.; Zhou, H.; Lu, P.; Fang, Z.; Shi, G.; Tong, X.; Chen, W.; Jiang, G.; Zhang, P.; Tian, J.; et al. miPEP31 alleviates Ang II-induced hypertension in mice by occupying Cebpα binding sites in the pri-miR-31 promoter. Cardiovasc. Diabetol. 2024, 23, 249. [Google Scholar] [CrossRef]
- Zhou, Y.; Yuan, Y.; Yao, X.; Wang, L.; Yao, L.; Tang, D.; Chen, F.; Li, J. miPEP31 alleviates sepsis development by regulating Chi3l1-dependent macrophage polarization. Biol. Direct 2024, 19, 117. [Google Scholar] [CrossRef]
- Sang, Y.; Liu, J.-Y.; Wang, F.-Y.; Luo, X.-Y.; Chen, Z.-Q.; Zhuang, S.-M.; Zhu, Y. Mitochondrial micropeptide STMP1 promotes G1/S transition by enhancing mitochondrial complex IV activity. Mol. Ther. 2022, 30, 2844–2855. [Google Scholar] [CrossRef]
- Xiang, X.; Fu, Y.; Zhao, K.; Miao, R.; Zhang, X.; Ma, X.; Liu, C.; Zhang, N.; Qu, K. Cellular senescence in hepatocellular carcinoma induced by a long non-coding RNA-encoded peptide PINT87aa by blocking FOXM1-mediated PHB2. Theranostics 2021, 11, 4929–4944. [Google Scholar] [CrossRef]
- Zhang, C.; Zhou, B.; Gu, F.; Liu, H.; Wu, H.; Yao, F.; Zheng, H.; Fu, H.; Chong, W.; Cai, S.; et al. Micropeptide PACMP inhibition elicits synthetic lethal effects by decreasing CtIP and poly(ADP-ribosyl)ation. Mol. Cell 2022, 82, 1297–1312.e8. [Google Scholar] [CrossRef]
- Peeters, M.K.R.; Baggerman, G.; Gabriels, R.; Pepermans, E.; Menschaert, G.; Boonen, K. Ion Mobility Coupled to a Time-of-Flight Mass Analyzer Combined With Fragment Intensity Predictions Improves Identification of Classical Bioactive Peptides and Small Open Reading Frame-Encoded Peptides. Front. Cell Dev. Biol. 2021, 9, 720570. [Google Scholar] [CrossRef]
- Periasamy, P.; Rajandran, S.; Ziegman, R.; Casey, M.; Nakamura, K.; Kore, H.; Datta, K.; Gowda, H. A simple organic solvent precipitation method to improve detection of low molecular weight proteins. Proteomics 2021, 21, e2100152. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, L.; Helbig, A.O.; Kaulich, P.T.; Weidenbach, K.; Schmitz, R.A.; Tholey, A. Multidimensional separation schemes enhance the identification and molecular characterization of low molecular weight proteomes and short open reading frame-encoded peptides in top-down proteomics. J. Proteom. 2021, 230, 103988. [Google Scholar] [CrossRef] [PubMed]
- Valdivia-Francia, F.; Sendoel, A. No country for old methods: New tools for studying microproteins. iScience 2024, 27, 108972. [Google Scholar] [CrossRef] [PubMed]
- Brito Querido, J.; Díaz-López, I.; Ramakrishnan, V. The molecular basis of translation initiation and its regulation in eukaryotes. Nat. Rev. Mol. Cell Biol. 2024, 25, 168–186. [Google Scholar] [CrossRef]
- Kaulich, P.T.; Cassidy, L.; Bartel, J.; Schmitz, R.A.; Tholey, A. Multi-protease Approach for the Improved Identification and Molecular Characterization of Small Proteins and Short Open Reading Frame-Encoded Peptides. J. Proteome Res. 2021, 20, 2895–2903. [Google Scholar] [CrossRef]
- Cassidy, L.; Kaulich, P.T.; Tholey, A. Depletion of High-Molecular-Mass Proteins for the Identification of Small Proteins and Short Open Reading Frame Encoded Peptides in Cellular Proteomes. J. Proteome Res. 2019, 18, 1725–1734. [Google Scholar] [CrossRef]
- Peeters, M.K.R.; Menschaert, G. The hunt for sORFs: A multidisciplinary strategy. Exp. Cell Res. 2020, 391, 111923. [Google Scholar] [CrossRef]
- Yang, H.; Li, Q.; Stroup, E.K.; Wang, S.; Ji, Z. Widespread stable noncanonical peptides identified by integrated analyses of ribosome profiling and ORF features. Nat. Commun. 2024, 15, 1932. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, H.; Chen, X.; Zheng, Y.; Kang, Q.; Hao, D.; Zhang, L.; Song, T.; Luo, H.; Hao, Y.; et al. SmProt: A Reliable Repository with Comprehensive Annotation of Small Proteins Identified from Ribosome Profiling. Genom. Proteom. Bioinform. 2021, 19, 602–610. [Google Scholar] [CrossRef]
- Leblanc, S.; Yala, F.; Provencher, N.; Lucier, J.-F.; Levesque, M.; Lapointe, X.; Jacques, J.-F.; Fournier, I.; Salzet, M.; Ouangraoua, A.; et al. OpenProt 2.0 builds a path to the functional characterization of alternative proteins. Nucleic Acids Res. 2024, 52, D522–D528. [Google Scholar] [CrossRef]
- Machkovech, H.M.; Bloom, J.D.; Subramaniam, A.R. Comprehensive profiling of translation initiation in influenza virus infected cells. PLoS Pathog. 2019, 15, e1007518. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, H.; Gu, R.; Tang, T.; Rai, K.R.; Chen, J.-L. Current Perspectives on Functional Involvement of Micropeptides in Virus–Host Interactions. Int. J. Mol. Sci. 2025, 26, 3651. https://doi.org/10.3390/ijms26083651
Sun H, Gu R, Tang T, Rai KR, Chen J-L. Current Perspectives on Functional Involvement of Micropeptides in Virus–Host Interactions. International Journal of Molecular Sciences. 2025; 26(8):3651. https://doi.org/10.3390/ijms26083651
Chicago/Turabian StyleSun, Haowen, Rongrong Gu, Tingting Tang, Kul Raj Rai, and Ji-Long Chen. 2025. "Current Perspectives on Functional Involvement of Micropeptides in Virus–Host Interactions" International Journal of Molecular Sciences 26, no. 8: 3651. https://doi.org/10.3390/ijms26083651
APA StyleSun, H., Gu, R., Tang, T., Rai, K. R., & Chen, J.-L. (2025). Current Perspectives on Functional Involvement of Micropeptides in Virus–Host Interactions. International Journal of Molecular Sciences, 26(8), 3651. https://doi.org/10.3390/ijms26083651






