Prion-like Properties of Short Isoforms of Human Chromatin Modifier PHC3
Abstract
1. Introduction
2. Results
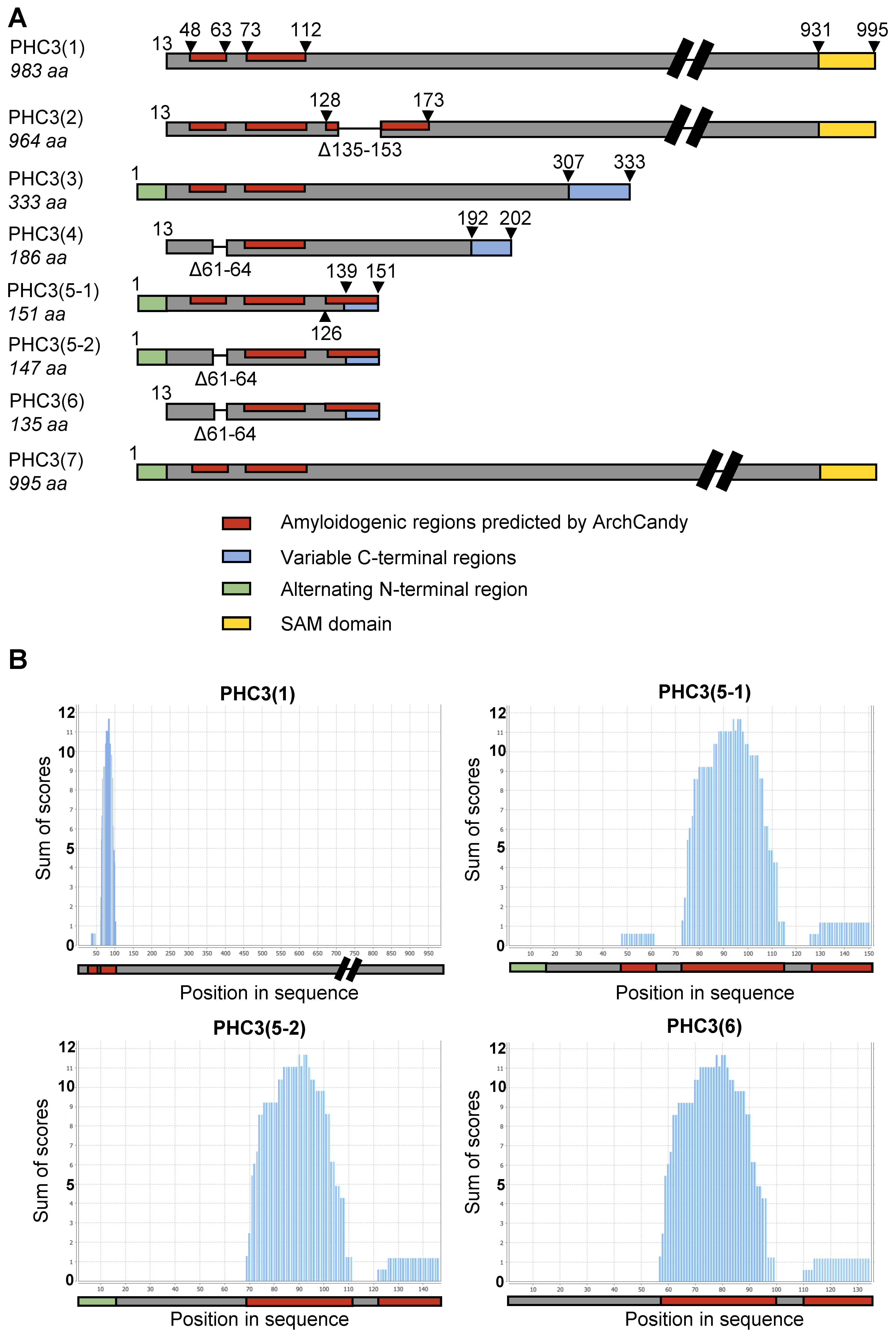
2.1. Multiple Isoforms of PHC3 Exist in Human Cells
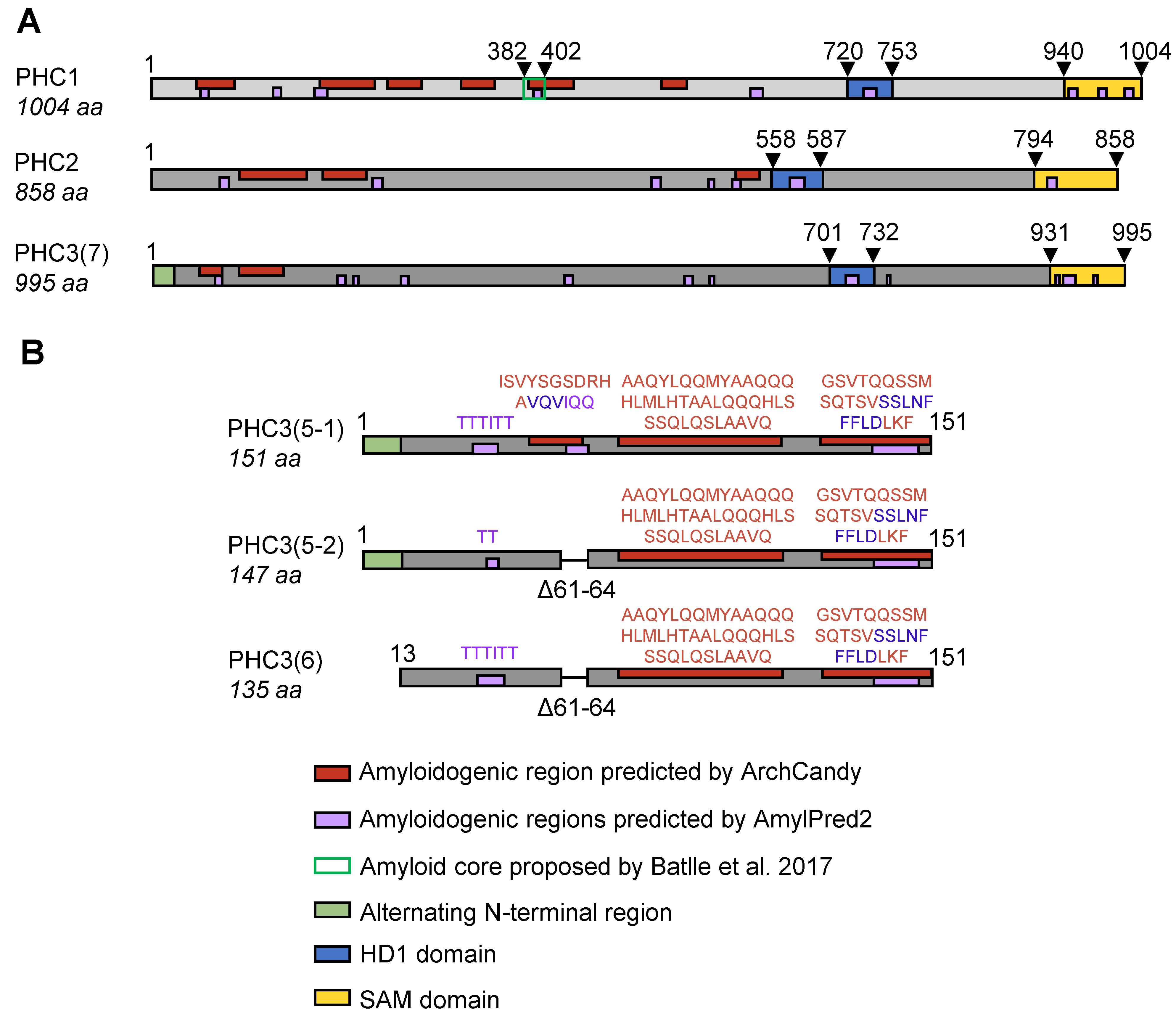
2.2. PHC3 Isoforms Contain Potentially Amyloidogenic Regions
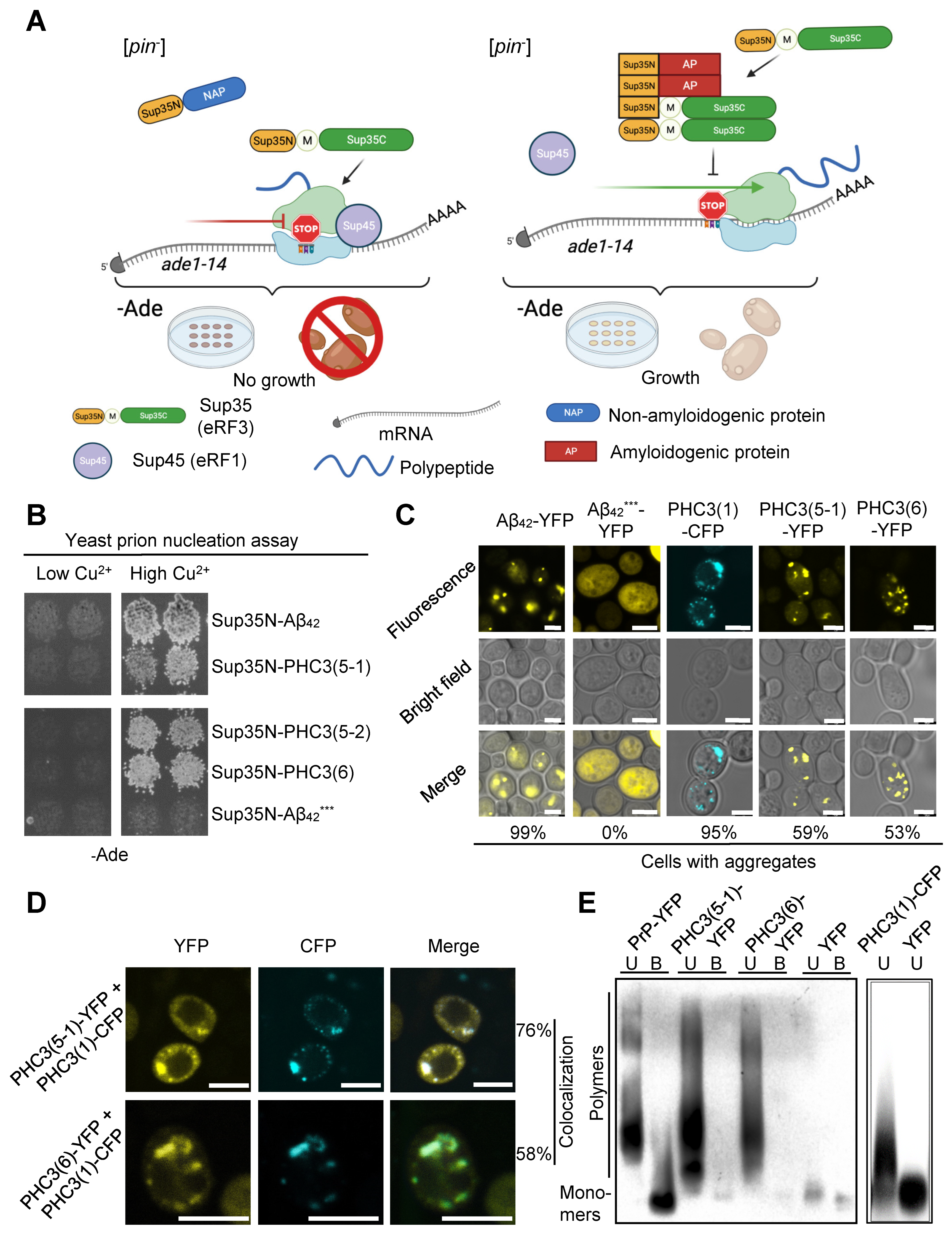
2.3. PHC3 Isoforms Possess Prion-Nucleating Properties in Yeast Cells
2.4. Short Isoforms of PHC3 Form Amyloid Fibrils in the C-DAG Assay
2.5. Short Isoforms of PHC3 Aggregate in Human Cells and Affect the Intracellular Localization of Full-Length PHC3
2.6. Overproduction of PHC3(5-1)-EGFP Alters Transcription in Human Cells
3. Discussion
4. Materials and Methods
4.1. Bacterial and Yeast Strains, and Cultivation Conditions
4.2. Human Cell Lines and Cultivation Conditions
4.3. Algorithms for Prediction of Amyloidogenic Sequence
4.4. Plasmid and Viral Vectors
4.4.1. Plasmids Used to Analyze Aggregation Properties of PHC3 Isoforms in Yeast
4.4.2. Plasmids for the Analysis of PHC3 Aggregation in the C-DAG Assay
4.4.3. Plasmids for the Analysis of PHC3 Aggregation in Human Cells
4.5. Yeast Assays
4.5.1. Phenotypic Detection of Sup35 Nucleation
4.5.2. Analysis of Aggregation by Fluorescence Microscopy
4.6. C-DAG Assay
4.7. Transfection and Viral Transduction of Human Cells
4.8. Analysis of Protein Aggregation by Semi-Denaturing Agarose Gel Electrophoresis (SDD-AGE)
4.9. Immunocytochemistry (ICC)
4.10. Analysis of Differential Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knowles, T.P.J.; Vendruscolo, M.; Dobson, C.M. The Amyloid State and Its Association with Protein Misfolding Diseases. Nat. Rev. Mol. Cell Biol. 2014, 15, 384–396. [Google Scholar] [CrossRef]
- Irvine, G.B.; El-Agnaf, O.M.; Shankar, G.M.; Walsh, D.M. Protein Aggregation in the Brain: The Molecular Basis for Alzheimer’s and Parkinson’s Diseases. Mol. Med. 2008, 14, 451–464. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid Nomenclature 2020: Update and Recommendations by the International Society of Amyloidosis (ISA) Nomenclature Committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef]
- Colby, D.W.; Prusiner, S.B. Prions. Cold Spring Harb. Perspect. Biol. 2011, 3, a006833. [Google Scholar] [CrossRef]
- Aguzzi, A.; Lakkaraju, A.K.K. Cell Biology of Prions and Prionoids: A Status Report. Trends Cell Biol. 2016, 26, 40–51. [Google Scholar] [CrossRef]
- Matiiv, A.B.; Trubitsina, N.P.; Matveenko, A.G.; Barbitoff, Y.A.; Zhouravleva, G.A.; Bondarev, S.A. Amyloid and Amyloid-Like Aggregates: Diversity and the Term Crisis. Biochemistry 2020, 85, 1011–1034. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Galkin, A.P. Functional Amyloids of Eukaryotes: Criteria, Classification, and Biological Significance. Curr. Genet. 2020, 66, 849–866. [Google Scholar] [CrossRef]
- Rubel, M.S.; Fedotov, S.A.; Grizel, A.V.; Sopova, J.V.; Malikova, O.A.; Chernoff, Y.O.; Rubel, A.A. Functional Mammalian Amyloids and Amyloid-Like Proteins. Life 2020, 10, 156. [Google Scholar] [CrossRef]
- Liebman, S.W.; Chernoff, Y.O. Prions in Yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Gorkovskiy, A.; Bezsonov, E.E.; Stroobant, E.E. Yeast and Fungal Prions. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2016; Volume 93, pp. 191–236. [Google Scholar]
- Chernova, T.A.; Wilkinson, K.D.; Chernoff, Y.O. Prions, Chaperones, and Proteostasis in Yeast. Cold Spring Harb. Perspect. Biol. 2017, 9, a023663. [Google Scholar] [CrossRef]
- Chakrabortee, S.; Byers, J.S.; Jones, S.; Garcia, D.M.; Bhullar, B.; Chang, A.; She, R.; Lee, L.; Fremin, B.; Lindquist, S.; et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell 2016, 167, 369–381.e12. [Google Scholar] [CrossRef]
- Michelitsch, M.D.; Weissman, J.S. A Census of Glutamine/Asparagine-Rich Regions: Implications for Their Conserved Function and the Prediction of Novel Prions. Proc. Natl. Acad. Sci. USA 2000, 97, 11910–11915. [Google Scholar] [CrossRef]
- Boeynaems, S.; Alberti, S.; Fawzi, N.L.; Mittag, T.; Polymenidou, M.; Rousseau, F.; Schymkowitz, J.; Shorter, J.; Wolozin, B.; Van Den Bosch, L.; et al. Protein Phase Separation: A New Phase in Cell Biology. Trends Cell Biol. 2018, 28, 420–435. [Google Scholar] [CrossRef]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct Stages in Stress Granule Assembly and Disassembly. eLife 2016, 5, e18413. [Google Scholar] [CrossRef]
- Franzmann, T.M.; Jahnel, M.; Pozniakovsky, A.; Mahamid, J.; Holehouse, A.S.; Nüske, E.; Richter, D.; Baumeister, W.; Grill, S.W.; Pappu, R.V.; et al. Phase Separation of a Yeast Prion Protein Promotes Cellular Fitness. Science 2018, 359, eaao5654. [Google Scholar] [CrossRef]
- Gorsheneva, N.A.; Sopova, J.V.; Azarov, V.V.; Grizel, A.V.; Rubel, A.A. Biomolecular Condensates: Structure, Functions, Methods of Research. Biochemistry 2024, 89, S205–S223. [Google Scholar] [CrossRef]
- Frieg, B.; Gremer, L.; Heise, H.; Willbold, D.; Gohlke, H. Binding Modes of Thioflavin T and Congo Red to the Fibril Structure of Amyloid-β(1–42). Chem. Commun. 2020, 56, 7589–7592. [Google Scholar] [CrossRef]
- Schütz, A.K.; Soragni, A.; Hornemann, S.; Aguzzi, A.; Ernst, M.; Böckmann, A.; Meier, B.H. The Amyloid-Congo Red Interface at Atomic Resolution. Angew. Chem. Int. Ed. Engl. 2011, 50, 5956–5960. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Alexandrov, I.M.; Mitkevich, O.V.; Shkundina, I.S.; Ter-Avanesyan, M.D. Purification and Analysis of Prion and Amyloid Aggregates. Methods 2006, 39, 50–55. [Google Scholar] [CrossRef]
- Howie, A.J. Origins of a Pervasive, Erroneous Idea: The “Green Birefringence” of Congo Red-stained Amyloid. Int. J. Exp. Pathol. 2019, 100, 208–221. [Google Scholar] [CrossRef]
- Yakupova, E.I.; Bobyleva, L.G.; Vikhlyantsev, I.M.; Bobylev, A.G. Congo Red and Amyloids: History and Relationship. Biosci. Rep. 2019, 39, BSR20181415. [Google Scholar] [CrossRef]
- Sivanathan, V.; Hochschild, A. A Bacterial Export System for Generating Extracellular Amyloid Aggregates. Nat. Protoc. 2013, 8, 1381–1390. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Grizel, A.V.; Rubel, A.A.; Zelinsky, A.A.; Chandramowlishwaran, P.; Chernova, T.A. Application of Yeast to Studying Amyloid and Prion Diseases. In Advances in Genetics; Elsevier: Amsterdam, The Netherlands, 2020; Volume 105, pp. 293–380. ISBN 978-0-12-821685-9. [Google Scholar]
- Ahmed, A.B.; Kajava, A.V. Breaking the Amyloidogenicity Code: Methods to Predict Amyloids from Amino Acid Sequence. FEBS Lett. 2013, 587, 1089–1095. [Google Scholar] [CrossRef]
- Ahmed, A.B.; Znassi, N.; Château, M.; Kajava, A.V. A Structure-based Approach to Predict Predisposition to Amyloidosis. Alzheimer’s Dement. 2015, 11, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Ito, S.; Koseki, H. Polycomb in Transcriptional Phase Transition of Developmental Genes. Trends Biochem. Sci. 2016, 41, 9–19. [Google Scholar] [CrossRef]
- Hernández-Muñoz, I.; Taghavi, P.; Kuijl, C.; Neefjes, J.; van Lohuizen, M. Association of BMI1 with Polycomb Bodies Is Dynamic and Requires PRC2/EZH2 and the Maintenance DNA Methyltransferase DNMT1. Mol. Cell. Biol. 2005, 25, 11047–11058. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef]
- Isono, K.; Endo, T.A.; Ku, M.; Yamada, D.; Suzuki, R.; Sharif, J.; Ishikura, T.; Toyoda, T.; Bernstein, B.E.; Koseki, H. SAM Domain Polymerization Links Subnuclear Clustering of PRC1 to Gene Silencing. Dev. Cell 2013, 26, 565–577. [Google Scholar] [CrossRef]
- Geng, Z.; Gao, Z. Mammalian PRC1 Complexes: Compositional Complexity and Diverse Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 8594. [Google Scholar] [CrossRef]
- Gunster, M.J.; Satijn, D.P.; Hamer, K.M.; den Blaauwen, J.L.; de Bruijn, D.; Alkema, M.J.; van Lohuizen, M.; van Driel, R.; Otte, A.P. Identification and Characterization of Interactions between the Vertebrate Polycomb-Group Protein BMI1 and Human Homologs of Polyhomeotic. Mol. Cell. Biol. 1997, 17, 2326–2335. [Google Scholar] [CrossRef]
- Tonkin, E.; Hagan, D.-M.; Li, W.; Strachan, T. Identification and Characterisation of Novel Mammalian Homologues of Drosophila Polyhomeoticpermits New Insights into Relationships between Members of the Polyhomeotic Family. Hum. Genet. 2002, 111, 435–442. [Google Scholar] [CrossRef]
- Robinson, A.K.; Leal, B.Z.; Nanyes, D.R.; Kaur, Y.; Ilangovan, U.; Schirf, V.; Hinck, A.P.; Demeler, B.; Kim, C.A. Human Polyhomeotic Homolog 3 (PHC3) Sterile Alpha Motif (SAM) Linker Allows Open-Ended Polymerization of PHC3 SAM. Biochemistry 2012, 51, 5379–5386. [Google Scholar] [CrossRef]
- Iwata, S.; Takenobu, H.; Kageyama, H.; Koseki, H.; Ishii, T.; Nakazawa, A.; Tatezaki, S.; Nakagawara, A.; Kamijo, T. Polycomb Group Molecule PHC3 Regulates Polycomb Complex Composition and Prognosis of Osteosarcoma. Cancer Sci. 2010, 101, 1646–1652. [Google Scholar] [CrossRef]
- Crea, F.; Sun, L.; Pikor, L.; Frumento, P.; Lam, W.L.; Helgason, C.D. Mutational Analysis of Polycomb Genes in Solid Tumours Identifies PHC3 Amplification as a Possible Cancer-Driving Genetic Alteration. Br. J. Cancer 2013, 109, 1699–1702. [Google Scholar] [CrossRef]
- Batlle, C.; de Groot, N.S.; Iglesias, V.; Navarro, S.; Ventura, S. Characterization of Soft Amyloid Cores in Human Prion-Like Proteins. Sci. Rep. 2017, 7, 12134. [Google Scholar] [CrossRef]
- Sabate, R.; Rousseau, F.; Schymkowitz, J.; Ventura, S. What Makes a Protein Sequence a Prion? PLoS Comput. Biol. 2015, 11, e1004013. [Google Scholar] [CrossRef]
- Frousios, K.K.; Iconomidou, V.A.; Karletidi, C.-M.; Hamodrakas, S.J. Amyloidogenic Determinants Are Usually Not Buried. BMC Struct. Biol. 2009, 9, 44. [Google Scholar] [CrossRef]
- Chandramowlishwaran, P.; Sun, M.; Casey, K.L.; Romanyuk, A.V.; Grizel, A.V.; Sopova, J.V.; Rubel, A.A.; Nussbaum-Krammer, C.; Vorberg, I.M.; Chernoff, Y.O. Mammalian Amyloidogenic Proteins Promote Prion Nucleation in Yeast. J. Biol. Chem. 2018, 293, 3436–3450. [Google Scholar] [CrossRef]
- Deshpande, A.M.; Akunowicz, J.D.; Reveles, X.T.; Patel, B.B.; Saria, E.A.; Gorlick, R.G.; Naylor, S.L.; Leach, R.J.; Hansen, M.F. PHC3, a Component of the hPRC-H Complex, Associates with E2F6 during G0 and Is Lost in Osteosarcoma Tumors. Oncogene 2007, 26, 1714–1722. [Google Scholar] [CrossRef]
- March, Z.M.; King, O.D.; Shorter, J. Prion-like Domains as Epigenetic Regulators, Scaffolds for Subcellular Organization, and Drivers of Neurodegenerative Disease. Brain Res. 2016, 1647, 9–18. [Google Scholar] [CrossRef]
- Du, Z.; Park, K.-W.; Yu, H.; Fan, Q.; Li, L. Newly Identified Prion Linked to the Chromatin-Remodeling Factor Swi1 in Saccharomyces Cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef]
- Harvey, Z.H.; Chakravarty, A.K.; Futia, R.A.; Jarosz, D.F. A Prion Epigenetic Switch Establishes an Active Chromatin State. Cell 2020, 180, 928–940.e14. [Google Scholar] [CrossRef]
- Jung, J.-H.; Barbosa, A.D.; Hutin, S.; Kumita, J.R.; Gao, M.; Derwort, D.; Silva, C.S.; Lai, X.; Pierre, E.; Geng, F.; et al. A Prion-like Domain in ELF3 Functions as a Thermosensor in Arabidopsis. Nature 2020, 585, 256–260. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Allen, K.D.; Chernova, T.A.; Tennant, E.P.; Wilkinson, K.D.; Chernoff, Y.O. Effects of Ubiquitin System Alterations on the Formation and Loss of a Yeast Prion. J. Biol. Chem. 2007, 282, 3004–3013. [Google Scholar] [CrossRef]
- Winston, F.; Dollard, C.; Ricupero-Hovasse, S.L. Construction of a Set of Convenient Saccharomyces Cerevisiae Strains That Are Isogenic to S288C. Yeast 1995, 11, 53–55. [Google Scholar] [CrossRef]
- Sherman, F. Getting Started with Yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar] [CrossRef]
- Gietz, R.D.; Schiestl, R.H. High-Efficiency Yeast Transformation Using the LiAc/SS Carrier DNA/PEG Method. Nat. Protoc. 2007, 2, 31–34. [Google Scholar] [CrossRef]
- Tsolis, A.C.; Papandreou, N.C.; Iconomidou, V.A.; Hamodrakas, S.J. A Consensus Method for the Prediction of ‘Aggregation-Prone’ Peptides in Globular Proteins. PLoS ONE 2013, 8, e54175. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A System of Shuttle Vectors and Yeast Host Strains Designed for Efficient Manipulation of DNA in Saccharomyces Cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Rubel, A.A.; Ryzhova, T.A.; Antonets, K.S.; Chernoff, Y.O.; Galkin, A.P. Identification of PrP Sequences Essential for the Interaction between the PrP Polymers and Aβ Peptide in a Yeast-Based Assay. Prion 2013, 7, 469–476. [Google Scholar] [CrossRef]
- Campeau, E.; Ruhl, V.E.; Rodier, F.; Smith, C.L.; Rahmberg, B.L.; Fuss, J.O.; Campisi, J.; Yaswen, P.; Cooper, P.K.; Kaufman, P.D. A Versatile Viral System for Expression and Depletion of Proteins in Mammalian Cells. PLoS ONE 2009, 4, e6529. [Google Scholar] [CrossRef]
- Kachkin, D.V.; Khorolskaya, J.I.; Ivanova, J.S.; Rubel, A.A. An Efficient Method for Isolation of Plasmid DNA for Transfection of Mammalian Cell Cultures. Methods Protoc. 2020, 3, 69. [Google Scholar] [CrossRef]
- Newnam, G.P.; Wegrzyn, R.D.; Lindquist, S.L.; Chernoff, Y.O. Antagonistic Interactions between Yeast Chaperones Hsp104 and Hsp70 in Prion Curing. Mol. Cell. Biol. 1999, 19, 1325–1333. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast [PSI+] Prion Aggregates Are Formed by Small Sup35 Polymers Fragmented by Hsp104. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar] [CrossRef]
- Walker, J.M. Protein Blotting by the Capillary Method. In The Protein Protocols Handbook; Humana Press: Totowa, NJ, USA, 2002; pp. 335–336. ISBN 978-1-59259-169-5. [Google Scholar]
- Hanna-Addams, S.; Wang, Z. Use of Two Dimensional Semi-Denaturing Detergent Agarose Gel Electrophoresis to Confirm Size Heterogeneity of Amyloid or Amyloid-like Fibers. J. Vis. Exp. 2018, 134, e57498. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 December 2024).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kachkin, D.; Zelinsky, A.A.; Romanova, N.V.; Kulichikhin, K.Y.; Zykin, P.A.; Khorolskaya, J.I.; Deckner, Z.J.; Kajava, A.V.; Rubel, A.A.; Chernoff, Y.O. Prion-like Properties of Short Isoforms of Human Chromatin Modifier PHC3. Int. J. Mol. Sci. 2025, 26, 1512. https://doi.org/10.3390/ijms26041512
Kachkin D, Zelinsky AA, Romanova NV, Kulichikhin KY, Zykin PA, Khorolskaya JI, Deckner ZJ, Kajava AV, Rubel AA, Chernoff YO. Prion-like Properties of Short Isoforms of Human Chromatin Modifier PHC3. International Journal of Molecular Sciences. 2025; 26(4):1512. https://doi.org/10.3390/ijms26041512
Chicago/Turabian StyleKachkin, Daniil, Andrew A. Zelinsky, Nina V. Romanova, Konstantin Y. Kulichikhin, Pavel A. Zykin, Julia I. Khorolskaya, Zachery J. Deckner, Andrey V. Kajava, Aleksandr A. Rubel, and Yury O. Chernoff. 2025. "Prion-like Properties of Short Isoforms of Human Chromatin Modifier PHC3" International Journal of Molecular Sciences 26, no. 4: 1512. https://doi.org/10.3390/ijms26041512
APA StyleKachkin, D., Zelinsky, A. A., Romanova, N. V., Kulichikhin, K. Y., Zykin, P. A., Khorolskaya, J. I., Deckner, Z. J., Kajava, A. V., Rubel, A. A., & Chernoff, Y. O. (2025). Prion-like Properties of Short Isoforms of Human Chromatin Modifier PHC3. International Journal of Molecular Sciences, 26(4), 1512. https://doi.org/10.3390/ijms26041512







