In Silico Characterization of the RCC1 Family and the UVR8 Gene in Chenopodium quinoa Willd.
Abstract
1. Introduction
2. Results
2.1. Identification of the RCC1 Family in Chenopodium quinoa
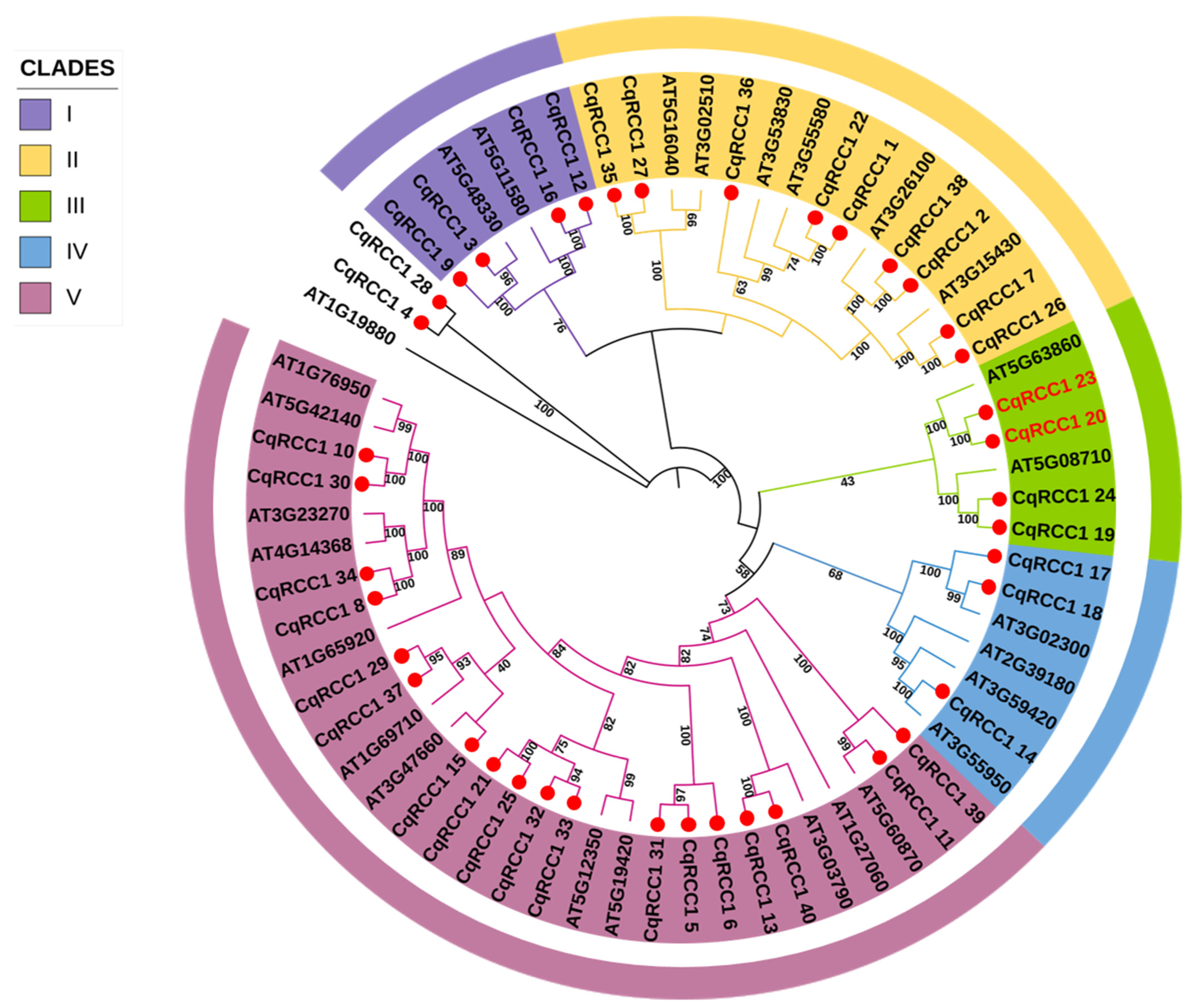
2.2. Phylogenetic Analysis of CqRCC1
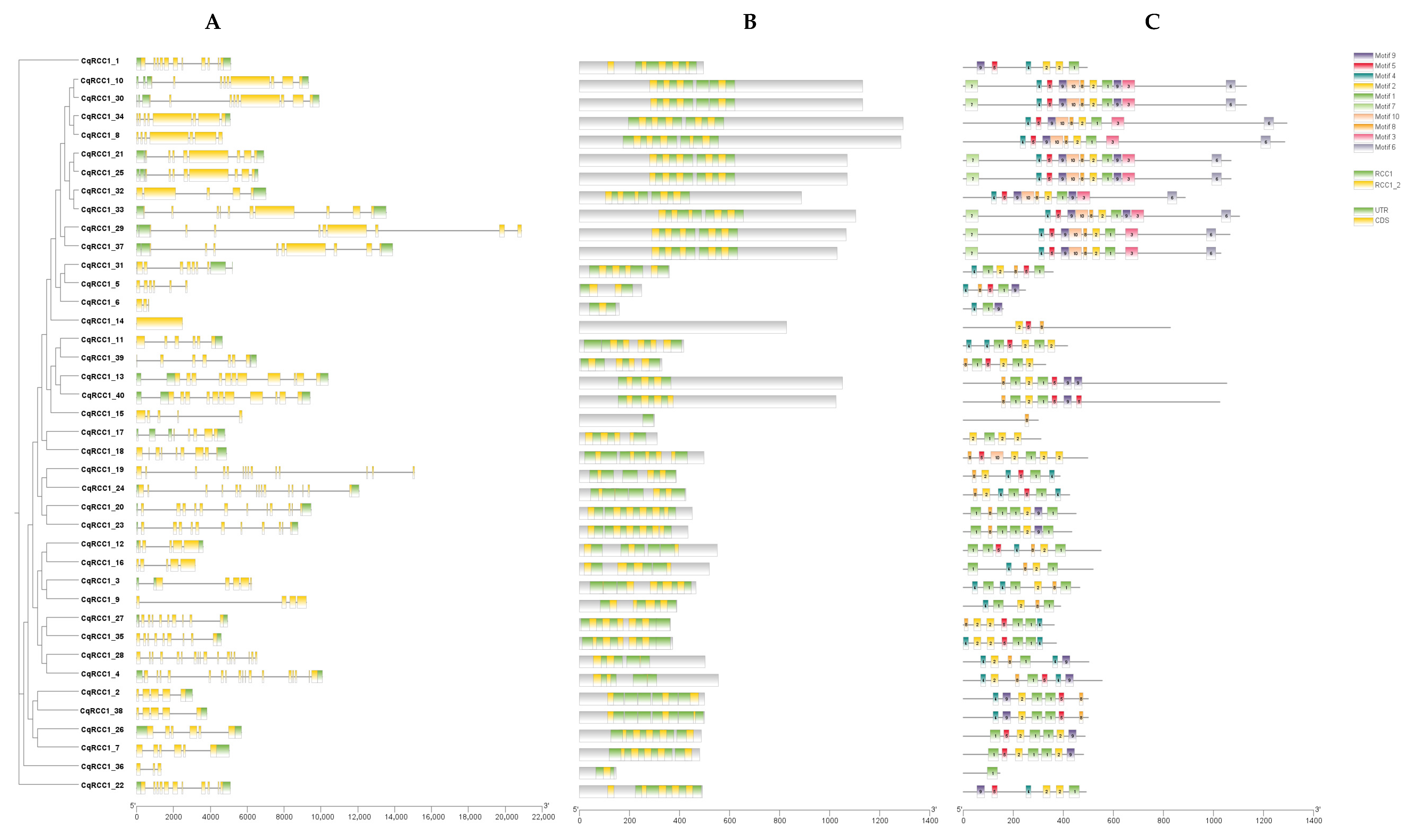
2.3. Structural and Genomic Analysis of CqRCC1
2.4. Multiple Sequence Alignment of CqRCC1
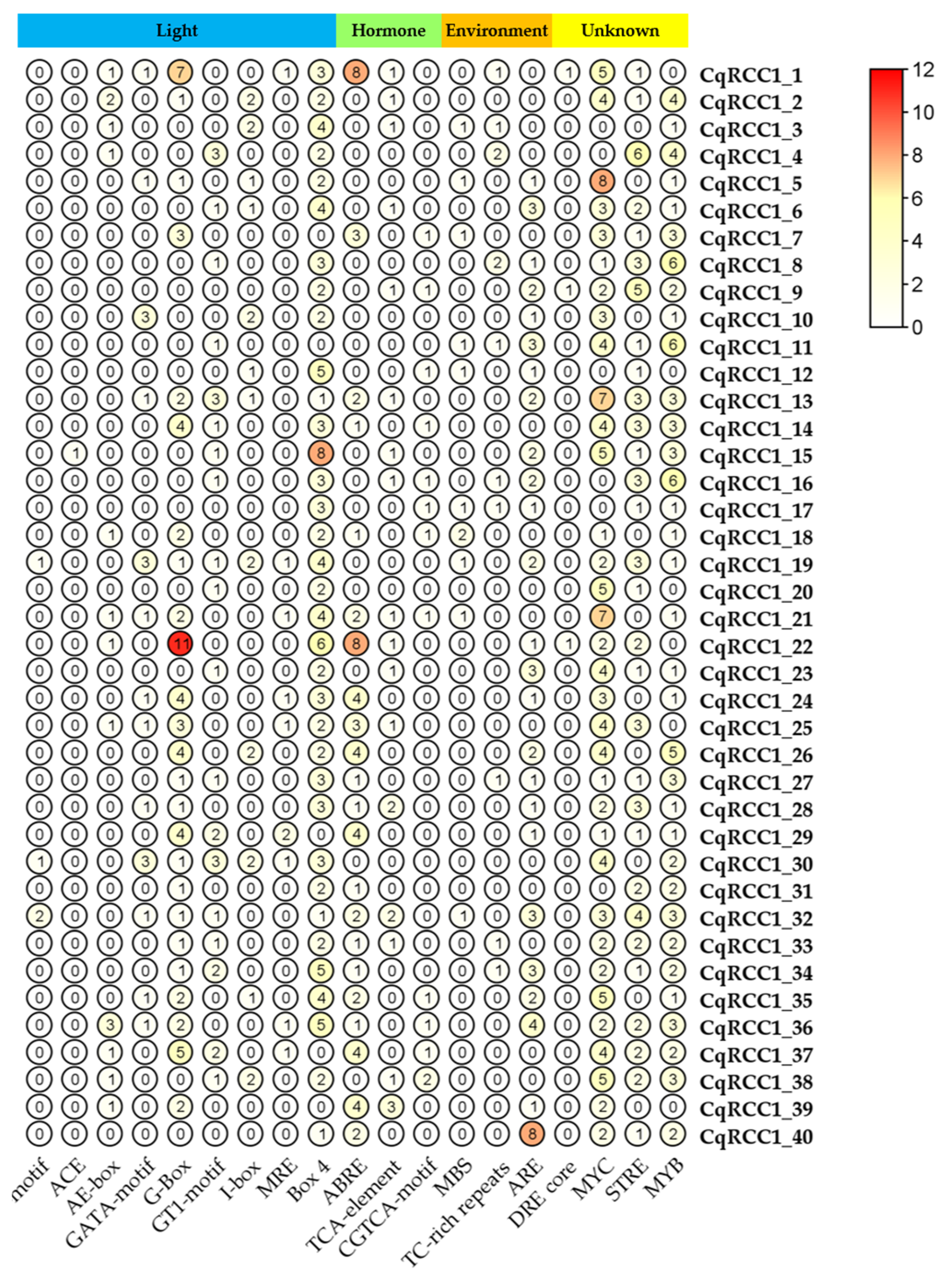
2.5. Cis-Regulatory Element (CRE) Analysis
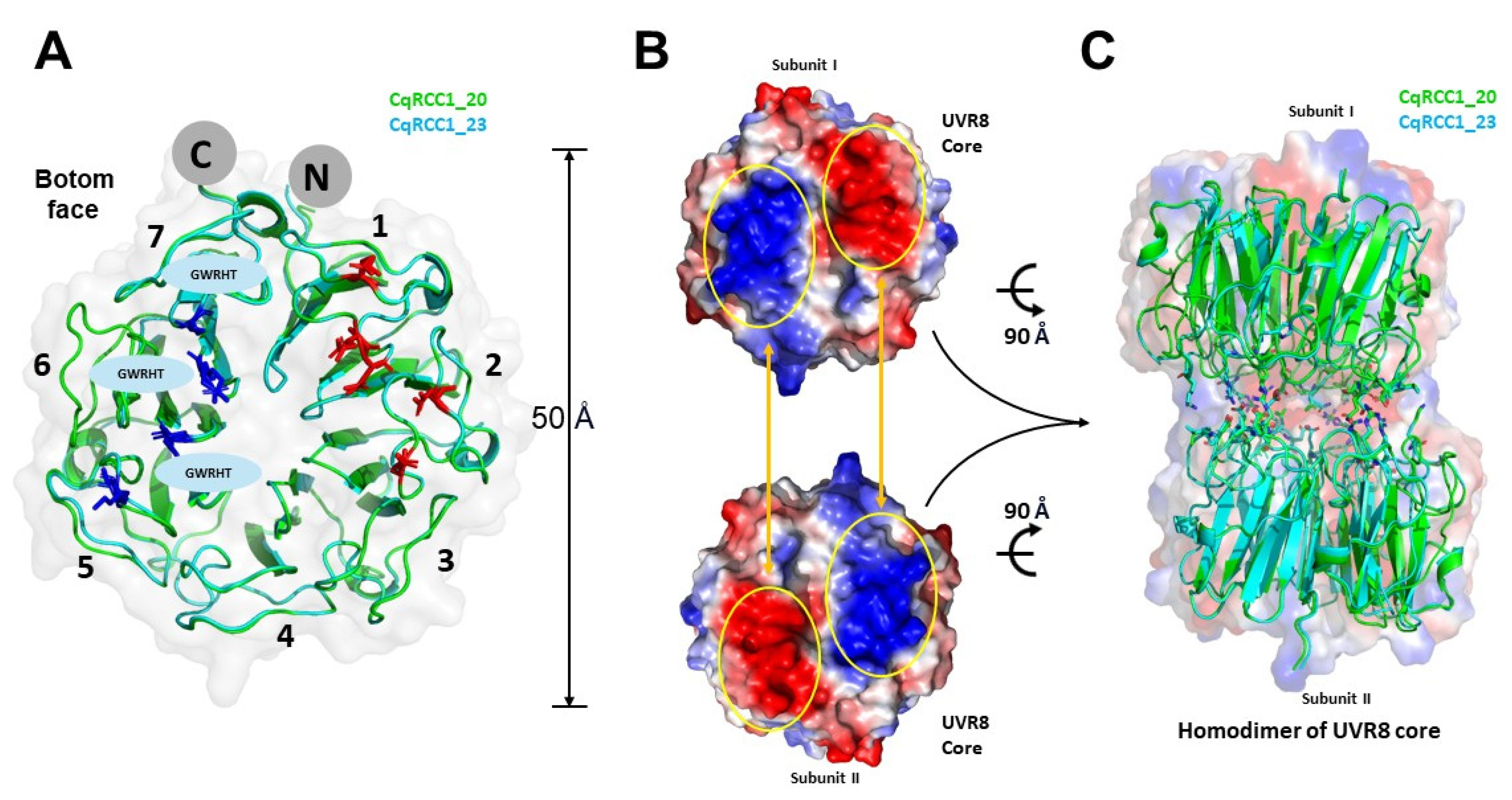
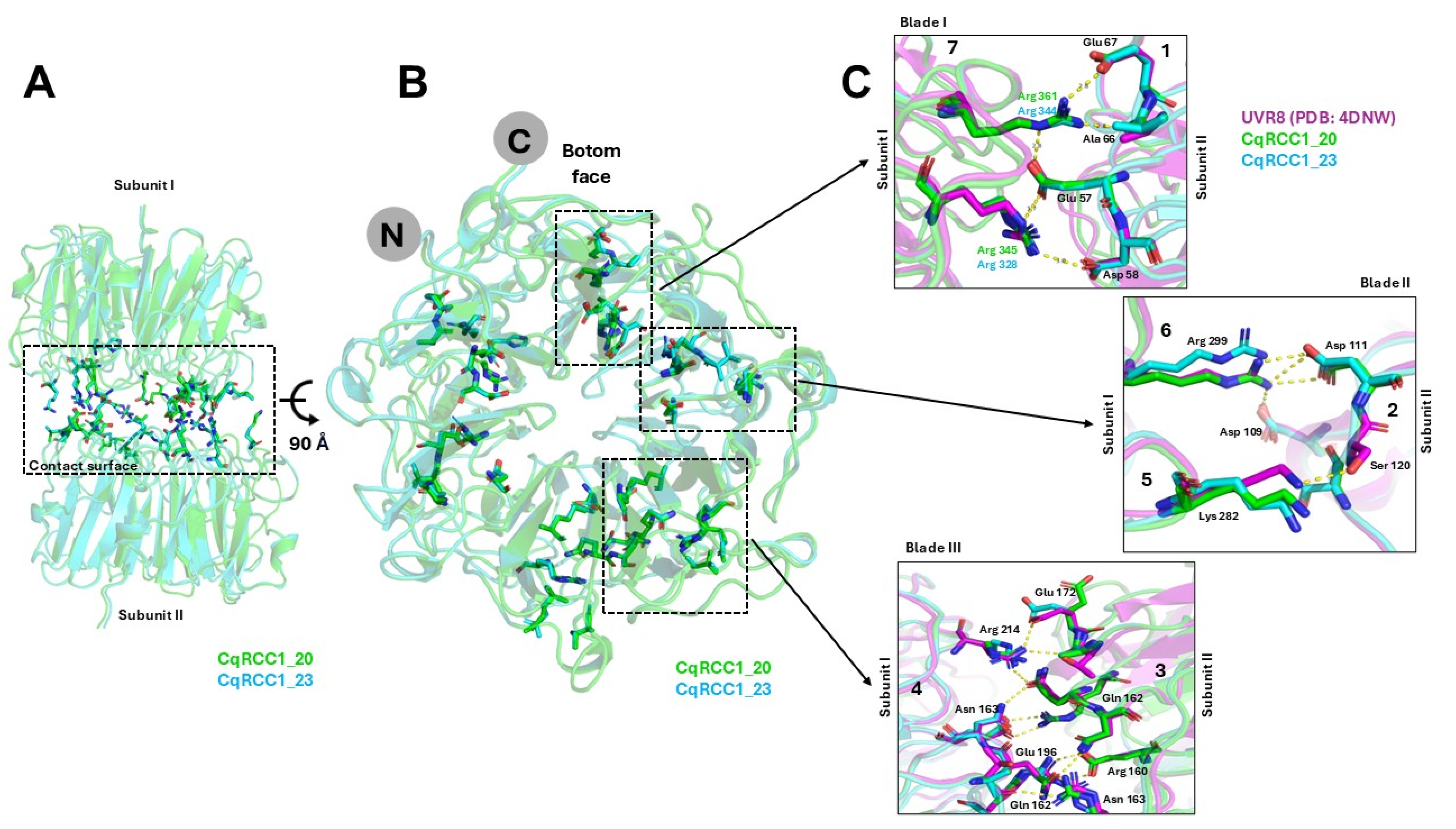
2.6. Structural Modeling of CqUVR8
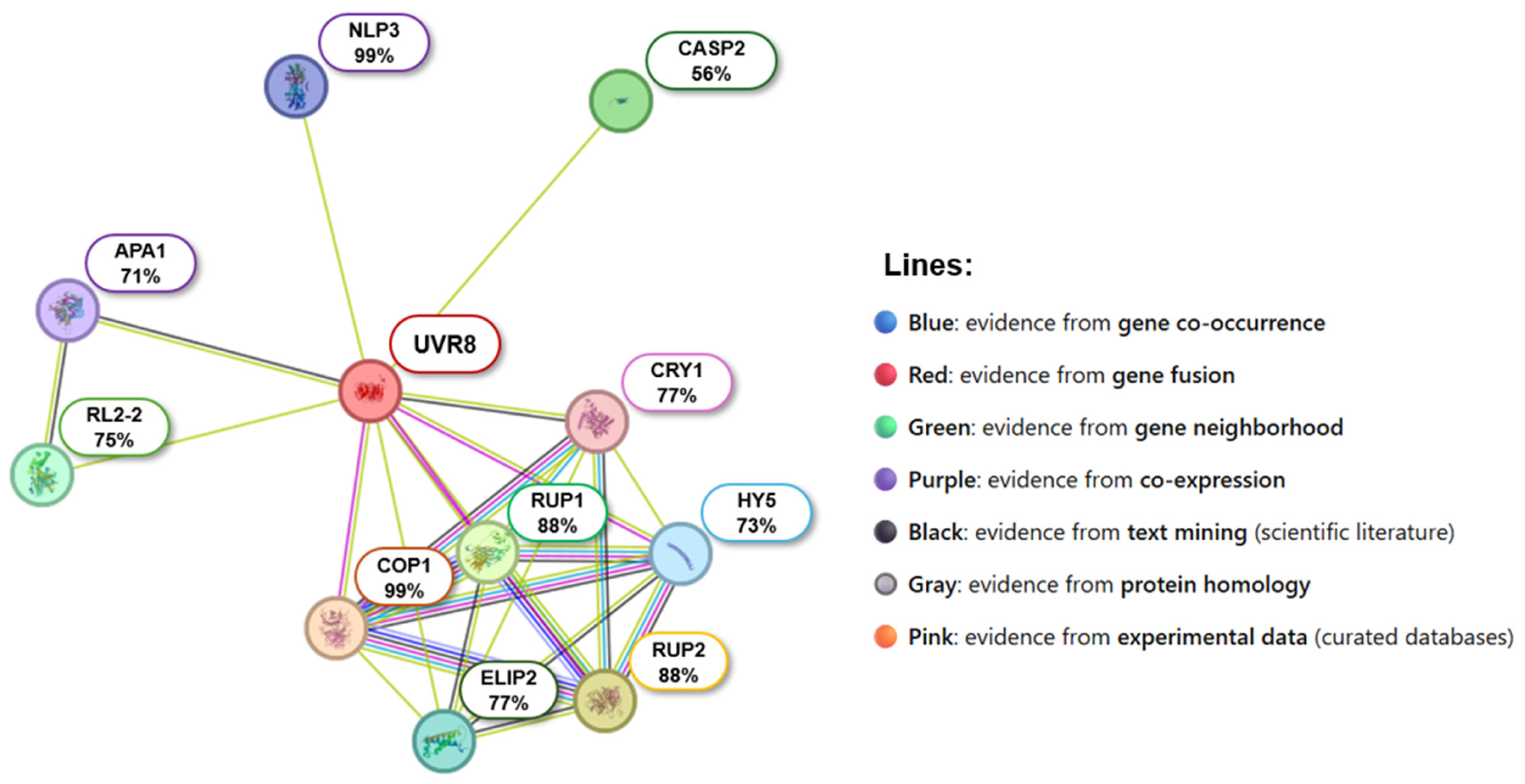
2.7. Predicted Protein–Protein Interactions of CqUVR8
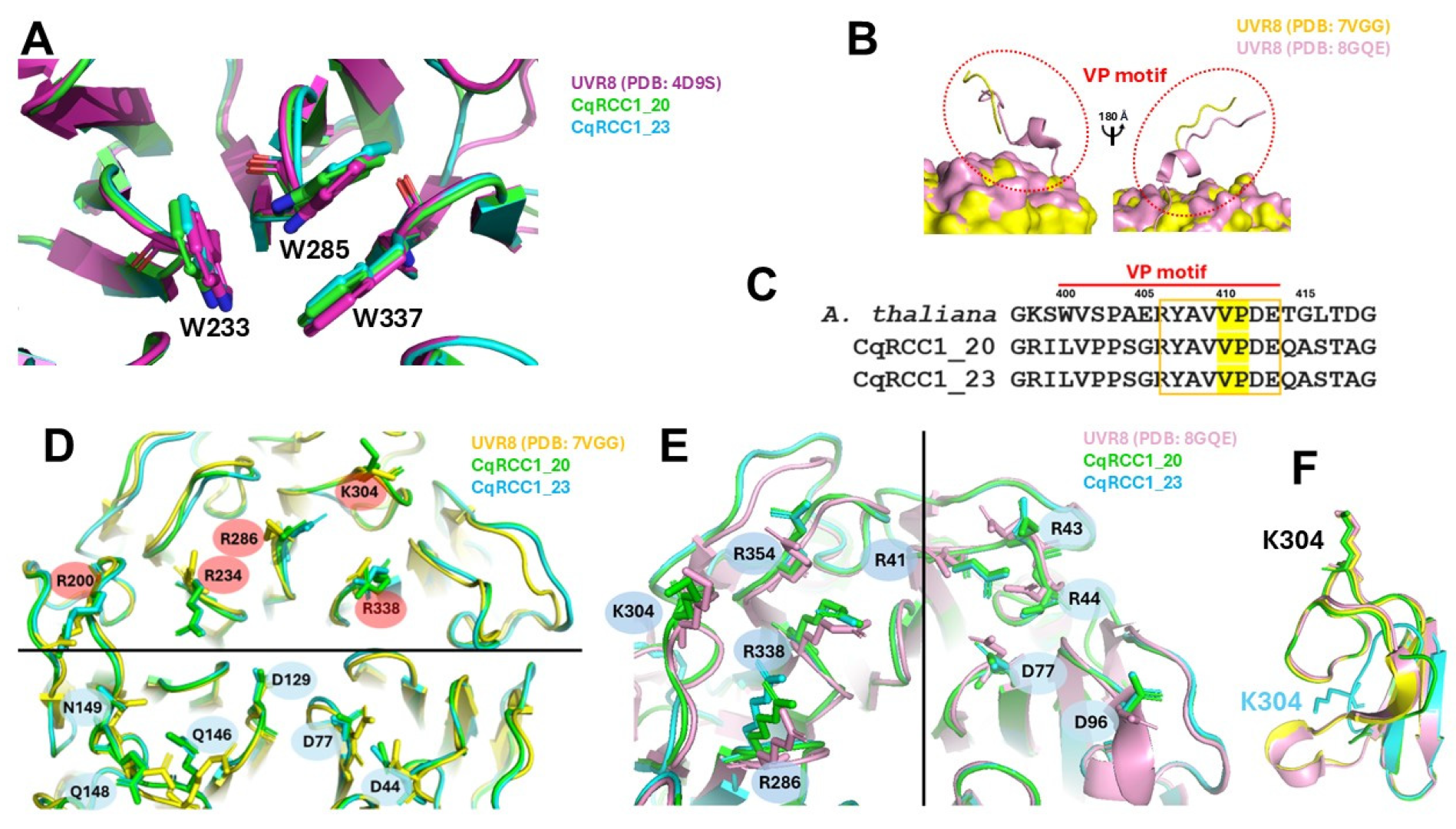
2.8. Critical Residues in the Photodynamics of CqUVR8
3. Discussion
3.1. Identification and Diversity of RCC1 Genes in Chenopodium quinoa
3.2. Physicochemical Properties and Subcellular Localization
3.3. Phylogenetic Relationship and Structural Conservation with UVR8
3.4. Cis-Regulatory Elements (CREs)
3.5. Structural and Functional Conservation of CqUVR8 Models
4. Materials and Methods
4.1. Identification of RCC1 Family Members in the Chenopodium quinoa Genome
4.2. Sequence Alignment and Phylogenetic Reconstruction
4.3. Gene Structure and Motif Analysis
4.4. Structural Modeling of CqUVR8 Proteins
4.5. Prediction of Protein Interactions
4.6. Analysis of Residues Involved in Photodynamics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ballaré, C.L.; Caldwell, M.M.; Flint, S.D.; Robinson, S.A.; Bornman, J.F. Effects of solar ultraviolet radiation on terrestrial ecosystems: Patterns, mechanisms, and interactions with climate change. Photochem. Photobiol. Sci. 2011, 10, 226–241. [Google Scholar] [CrossRef]
- Björn, L.O. History ultraviolet-A, B, and C. UV4Plants Bull. 2015, 1, 17–18. [Google Scholar] [CrossRef]
- Nowak, V.; Du, J.; Charrondière, U.R. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.). Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef]
- González, J.A.; Rosa, M.; Parrado, M.F.; Hilal, M.; Prado, F.E. Morphological and physiological responses of two varieties of a highland species (Chenopodium quinoa Willd.) growing under near-ambient and strongly reduced solar UV-B in a lowland location. J. Photochem. Photobiol. B 2009, 96, 144–151. [Google Scholar] [CrossRef]
- Hinojosa, L.; González, J.A.; Barrios-Masias, F.H.; Fuentes, F.; Murphy, K.M. Murphy. Quinoa abiotic stress responses: A review. Plants 2018, 7, 106. [Google Scholar] [CrossRef]
- Rizzini, L.; Favory, J.-J.; Cloix, C.; Faggionato, D.; O’hara, A.; Kaiserli, E.; Baumeister, R.; Schäfer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef]
- Jenkins, G.I. The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 2014, 26, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Heijde, M.; Ulm, R. UV-B photoreceptor-mediated signalling in plants. Trends Plant Sci. 2012, 17, 230–237. [Google Scholar] [CrossRef]
- Yin, R.; Ulm, R. How plants cope with UV-B: From perception to response. Curr. Opin. Plant Biol. 2017, 37, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Cloix, C.; Jiang, G.H.; Kaiserli, E.; Herzyk, P.; Kliebenstein, D.J.; Jenkins, G.I. A UV-B-Specific Signaling Component Orchestrates Plant UV Protection. Proc. Natl. Acad. Sci. USA 2005, 102, 18225–18230. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; He, K.; Stolc, V.; Lee, H.; Figueroa, P.; Gao, Y.; Tongprasit, W.; Zhao, H.; Lee, I.; Deng, X.W. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 2007, 19, 731–749. [Google Scholar] [CrossRef]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef]
- Christie, J.M.; Arvai, A.S.; Baxter, K.J.; Heilmann, M.; Pratt, A.J.; O’Hara, A.; Kelly, S.M.; Hohl, M.; Kovacs, L.; Toth, R.; et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 2012, 335, 1492–1496. [Google Scholar] [CrossRef]
- Wu, D.; Hu, Q.; Yan, Z.; Chen, W.; Yan, C.; Huang, X.; Zhang, J.; Yang, P.; Deng, H.; Wang, J.; et al. Structural basis of ultraviolet-B perception by UVR8. Nature 2012, 484, 214–219. [Google Scholar] [CrossRef]
- Ramsey, J.; Ramsey, T.S. Ecological studies of polyploidy in the 100 years following its discovery. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130352. [Google Scholar] [CrossRef]
- Adams, K.L. Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 2007, 98, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.-S.; Lightfoot, D.J.; Schmöckel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312, Erratum in Nature 2017, 545, 510. [Google Scholar] [CrossRef] [PubMed]
- Nene, T.; Yadav, M.; Yadav, H.S. Plant catalase in silico characterization and phylogenetic analysis with structural modeling. J. Genet. Eng. Biotechnol. 2022, 20, 125. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Zhao, H.; Zhao, Y.; Ji, H.; Wang, Y.; Zhi, L.; Li, X. RUG3 and ATM synergistically regulate the alternative splicing of mitochondrial nad2 and the DNA damage response in Arabidopsis thaliana. Sci. Rep. 2017, 7, 43897. [Google Scholar] [CrossRef]
- Kaiserli, E.; Jenkins, G.I. UV-B promotes rapid nuclear translocation of the Arabidopsis UVR8 and activates its function in the nucleus. Plant Cell 2007, 19, 2662–2673. [Google Scholar] [CrossRef]
- Tilbrook, K.; Dubois, M.; Crocco, C.D.; Yin, R.; Chappuis, R.; Allorent, G.; Schmid-Siegert, E.; Goldschmidt-Clermont, M.; Ulm, R. UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell 2016, 28, 966–983. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, B.; Hayes, S.; Kerner, K.; Höcker, U.; Jenkins, G.I.; Franklin, K.A. UVR8 disrupts stabilisation of PIF5 by COP1 to inhibit plant stem elongation in sunlight. Nat. Commun. 2019, 10, 3185. [Google Scholar] [CrossRef]
- Tissot, N.; Ulm, R. Cryptochrome-mediated blue-light signalling modulates UVR8 photoreceptor activity and contributes to UV-B tolerance in Arabidopsis. Nat. Commun. 2020, 11, 1323. [Google Scholar] [CrossRef]
- Zhao, C.; Mao, K.; You, C.; Zhao, X.; Wang, S.; Li, Y.; Hao, Y.-J. Molecular cloning and functional analysis of a UV-B photoreceptor gene, MdUVR8 (UV Resistance Locus 8), from apple. Plant Sci. 2016, 247, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Wang, L.; Li, Y.; Wu, R. Molecular cloning and functional analysis of UV RESISTANCE LOCUS 8 (PeUVR8) from Populus euphratica. PLoS ONE 2015, 10, e0132390. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, X.; Jang, Z.; Chen, Z.; Ruo, X.; Jin, W.; Xu, M. UV RESISTANCE LOCUS 8 from Chrysanthemum morifolium (CmUVR8) plays important roles in UV-B signal transduction and UV-B-induced accumulation of flavonoids. Front. Plant Sci. 2018, 9, 955. [Google Scholar] [CrossRef]
- Podolec, R.; Lau, K.; Wagnon, T.B.; Hothorn, M.; Ulm, R. A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023562118. [Google Scholar] [CrossRef]
- Fernández, M.B.; Lamattina, L.; Cassia, R. Functional analysis of the UVR8 photoreceptor from the monocotyledonous Zea mays. Plant Growth Regul. 2020, 92, 419–427. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Zhong, Y.-B.; Leung, D.W.-M.; Yan, X.-Y.; Ouyang, M.-N.; Ye, Y.-Z.; Li, S.-M.; Peng, X.-X.; Liu, E.-E. OsUVR8b, rather than OsUVR8a, plays a predominant role in rice UVR8-mediated UV-B response. Physiol. Plant. 2024, 176, e14471. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.; Podolec, R.; Chappuis, R.; Ulm, R.; Hothorn, M. Plant photoreceptors and their signaling components compete for COP1 binding via VP peptide motifs. EMBO J. 2019, 38, e102140. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, L.; Guan, Z.; Chang, H.; Ma, L.; Shen, C.; Qiu, L.; Yan, J.; Zhang, D.; Li, J.; et al. Structural insight into UV-B–activated UVR8 bound to COP1. Sci. Adv. 2022, 8, eabn3337. [Google Scholar] [CrossRef] [PubMed]
- Tossi, V.E.; Regalado, J.J.; Iannicelli, J.; Laino, L.E.; Burrieza, H.P.; Escandón, A.S.; Pitta-Álvarez, S.I. Beyond Arabidopsis: Differential UV-B response mediated by UVR8 in diverse species. Front. Plant Sci. 2019, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.B.; Tossi, V.; Lamattina, L.; Cassia, R. A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front. Plant Sci. 2016, 7, 1698. [Google Scholar] [CrossRef] [PubMed]
- Panchy, N.; Lehti-Shiu, M.; Shiu, S.-H. Evolution of gene duplication in plants. Plant Physiol. 2016, 171, 2294–2316. [Google Scholar] [CrossRef]

| Gene Name | Transcript ID | Localization | CDS (pb) | AA | Weight (kDa) | pI | Aliphatic Index | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| CqRCC1_1 | AUR62012141 | Scaffold_1995:925914..931012 f | 1494 | 497 | 52.14 | 5.76 | 72.23 | Me 1,2, C 1,2 |
| CqRCC1_2 | AUR62024206 | Scaffold_4119:1567432..1570457 f | 1506 | 501 | 53.79 | 6.09 | 80.4 | Me 1,2, C 1,2 |
| CqRCC1_3 | AUR62023679 | Scaffold_2187:562911..569141 r | 1401 | 466 | 49.81 | 5.75 | 88.91 | Cl 1, Mi 2,3 |
| CqRCC1_4 | AUR62019141 | Scaffold_3876:3492836..3502921 r | 1671 | 556 | 60.42 | 6.94 | 59.41 | Ex 1,2, N 1,2 |
| CqRCC1_5 | AUR62026458 | Scaffold_4244:6102177..6104925 f | 750 | 249 | 26.64 | 8.57 | 84.14 | Me 1, C 2 |
| CqRCC1_6 | AUR62026456 | Scaffold_4244:6090841..6091515 f | 483 | 160 | 17.38 | 6.42 | 81.06 | Me 1, N 2 |
| CqRCC1_7 | AUR62034255 | Scaffold_1337:1246314..1251339 r | 1446 | 481 | 51.94 | 6.99 | 84.28 | Me 1, C 1,2 |
| CqRCC1_8 | AUR62002142 | Scaffold_4480:2651092..2655743 f | 3861 | 1286 | 139.78 | 5.61 | 69.21 | Me 1, N 2 |
| CqRCC1_9 | AUR62002515 | Scaffold_4480:7952067..7961272 f | 1176 | 391 | 42.09 | 4.98 | 84.27 | Ex 1,2, M 1,2 |
| CqRCC1_10 | AUR62013542 | Scaffold_1611:4718188..4727510 r | 3402 | 1133 | 122.57 | 8.61 | 74.63 | Me 1,2, N 1,2 |
| CqRCC1_11 | AUR62003547 | Scaffold_2370:1986325..1990982 r | 1254 | 417 | 43.33 | 6.24 | 85.08 | P 1, M 1 |
| CqRCC1_12 | AUR62039706 | Scaffold_2419:242891..246491 r | 1656 | 551 | 59.37 | 4.9 | 77.66 | P 1,2, C 1,2 |
| CqRCC1_13 | AUR62039726 | Scaffold_2419:720240..730637 f | 3162 | 1053 | 114.57 | 9.23 | 80.85 | Me 1, N 2 |
| CqRCC1_14 | AUR62020143 | Scaffold_2465:1185020..1187510 r | 2490 | 829 | 90.09 | 7.65 | 90.05 | Me 1, N 2 |
| CqRCC1_15 | AUR62044490 | Scaffold_1723:8..5728 r | 903 | 301 | 33.74 | 4.73 | 84.95 | C 1,2 |
| CqRCC1_16 | AUR62009274 | Scaffold_3674:3391734..3394917 f | 1563 | 520 | 55.96 | 4.97 | 73.85 | P 1,2, C 1,2 |
| CqRCC1_17 | AUR62009482 | Scaffold_3674:5764320..5769103 f | 939 | 312 | 33.37 | 6.58 | 86.57 | Ex 1,2, N 1,2 |
| CqRCC1_18 | AUR62023161 | Scaffold_1189:2007096..2011966 f | 1497 | 498 | 53.66 | 6.24 | 83.03 | Me 1,2, C 1,2 |
| CqRCC1_19 | AUR62000563 | Scaffold_2088:6049517..6064591 r | 1161 | 387 | 41.43 | 6.35 | 79.84 | Me 3, N 1, M 2 |
| CqRCC1_20 | AUR62000528 | Scaffold_2088:5597182..5606640 f | 1356 | 451 | 47.85 | 5.77 | 73.1 | Ex 1,2, M 1,2 |
| CqRCC1_21 | AUR62000180 | Scaffold_2088:1942924..1949822 r | 3216 | 1071 | 115.86 | 9.09 | 67.63 | Me 1,2, N 1,2 |
| CqRCC1_22 | AUR62040694 | Scaffold_1465:99228..104321 r | 1482 | 493 | 51.33 | 5.47 | 71.26 | Me 1,2, C 1,2 |
| CqRCC1_23 | AUR62006875 | Scaffold_3429:5939170..5947925 f | 1305 | 434 | 46.23 | 6.06 | 74.15 | Ex 1,2, M 1,2 |
| CqRCC1_24 | AUR62006916 | Scaffold_3429:6412259..6424318 r | 1281 | 426 | 45.48 | 6.27 | 83.26 | Ex 1, M 2,3 |
| CqRCC1_25 | AUR62006531 | Scaffold_3429:2616822..2623416 r | 3216 | 1071 | 115.96 | 9.06 | 67.98 | Me 1,2, N 1,2 |
| CqRCC1_26 | AUR62029360 | Scaffold_2939:2226660..2232351 r | 1467 | 488 | 52.60 | 7.24 | 83.67 | Me 1,2, C 1,2 |
| CqRCC1_27 | AUR62014530 | Scaffold_1566:4165280..4170210 f | 1095 | 364 | 39.44 | 6.33 | 79.29 | P 1,2, C 1,2 |
| CqRCC1_28 | AUR62014404 | Scaffold_1566:2639482..2646019 r | 1512 | 503 | 54.57 | 8.94 | 65.88 | Me 1, Mi 2 |
| CqRCC1_29 | AUR62028803 | Scaffold_2837:713405..734287 f | 3204 | 1067 | 115.97 | 8.66 | 70.36 | Me 1,2, N 1,2 |
| CqRCC1_30 | AUR62026740 | Scaffold_2081:4343447..4353350 f | 3402 | 1133 | 122.53 | 8.58 | 74.97 | Me 1,2, N 1,2 |
| CqRCC1_31 | AUR62026265 | Scaffold_3298:5035900..5041104 f | 1083 | 360 | 38.23 | 6 | 78.56 | Ex 1,2, N 1,2 |
| CqRCC1_32 | AUR62037183 | Scaffold_1237:49943..56961 r | 2670 | 889 | 95.54 | 8.91 | 70.29 | Me 1,2, N 1,2 |
| CqRCC1_33 | AUR62026876 | Scaffold_2185:1349875..1363405 r | 3318 | 1105 | 119.45 | 9.05 | 73.13 | Me 1, N 2 |
| CqRCC1_34 | AUR62015566 | Scaffold_2751:8118037..8123109 r | 3888 | 1295 | 140.62 | 5.87 | 69.05 | Me 1, N 2 |
| CqRCC1_35 | AUR62008975 | Scaffold_1710:8511136..8515728 r | 1125 | 374 | 40.62 | 6.52 | 80.8 | P 1,2, N 1,2 |
| CqRCC1_36 | AUR62038079 | Scaffold_1613:1603591..1604910 f | 444 | 147 | 16.11 | 6.94 | 108.03 | C 1,2, Ex 1,2 |
| CqRCC1_37 | AUR62005755 | Scaffold_1214:7543923..7557818 r | 3096 | 1031 | 112.25 | 8.41 | 69.7 | Me 1,2, N 1,2 |
| CqRCC1_38 | AUR62005531 | Scaffold_1214:4144473..4148294 r | 1506 | 501 | 53.78 | 6.2 | 79.82 | Me 1,2, N 1,2 |
| CqRCC1_39 | AUR62017861 | Scaffold_3086:1094434..1100940 r | 993 | 330 | 34.44 | 6.45 | 82.12 | Me 1, Re 2 |
| CqRCC1_40 | AUR62036278 | Scaffold_2738:983326..992737 f | 3081 | 1026 | 111.84 | 9.32 | 82.11 | Me 1, N 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paredes Malca, J.C.; Fuentes Apaza, M.M.; Valderrama-Valencia, M.R.E.; Bardales Álvarez, R.; Condori Mamani, E.; Condori-Pacsi, S.J. In Silico Characterization of the RCC1 Family and the UVR8 Gene in Chenopodium quinoa Willd. Int. J. Mol. Sci. 2025, 26, 11657. https://doi.org/10.3390/ijms262311657
Paredes Malca JC, Fuentes Apaza MM, Valderrama-Valencia MRE, Bardales Álvarez R, Condori Mamani E, Condori-Pacsi SJ. In Silico Characterization of the RCC1 Family and the UVR8 Gene in Chenopodium quinoa Willd. International Journal of Molecular Sciences. 2025; 26(23):11657. https://doi.org/10.3390/ijms262311657
Chicago/Turabian StyleParedes Malca, Jean Carlo, Michell Maheba Fuentes Apaza, María Rosario Elsa Valderrama-Valencia, Roxana Bardales Álvarez, Eloy Condori Mamani, and Sandro Jhonatan Condori-Pacsi. 2025. "In Silico Characterization of the RCC1 Family and the UVR8 Gene in Chenopodium quinoa Willd." International Journal of Molecular Sciences 26, no. 23: 11657. https://doi.org/10.3390/ijms262311657
APA StyleParedes Malca, J. C., Fuentes Apaza, M. M., Valderrama-Valencia, M. R. E., Bardales Álvarez, R., Condori Mamani, E., & Condori-Pacsi, S. J. (2025). In Silico Characterization of the RCC1 Family and the UVR8 Gene in Chenopodium quinoa Willd. International Journal of Molecular Sciences, 26(23), 11657. https://doi.org/10.3390/ijms262311657






