Generation of Induced Pluripotent Stem Cells and Neuroepithelial Stem Cells from a Family with the Pathogenic Variant p.Q337X in Progranulin
Abstract
1. Introduction
2. Results
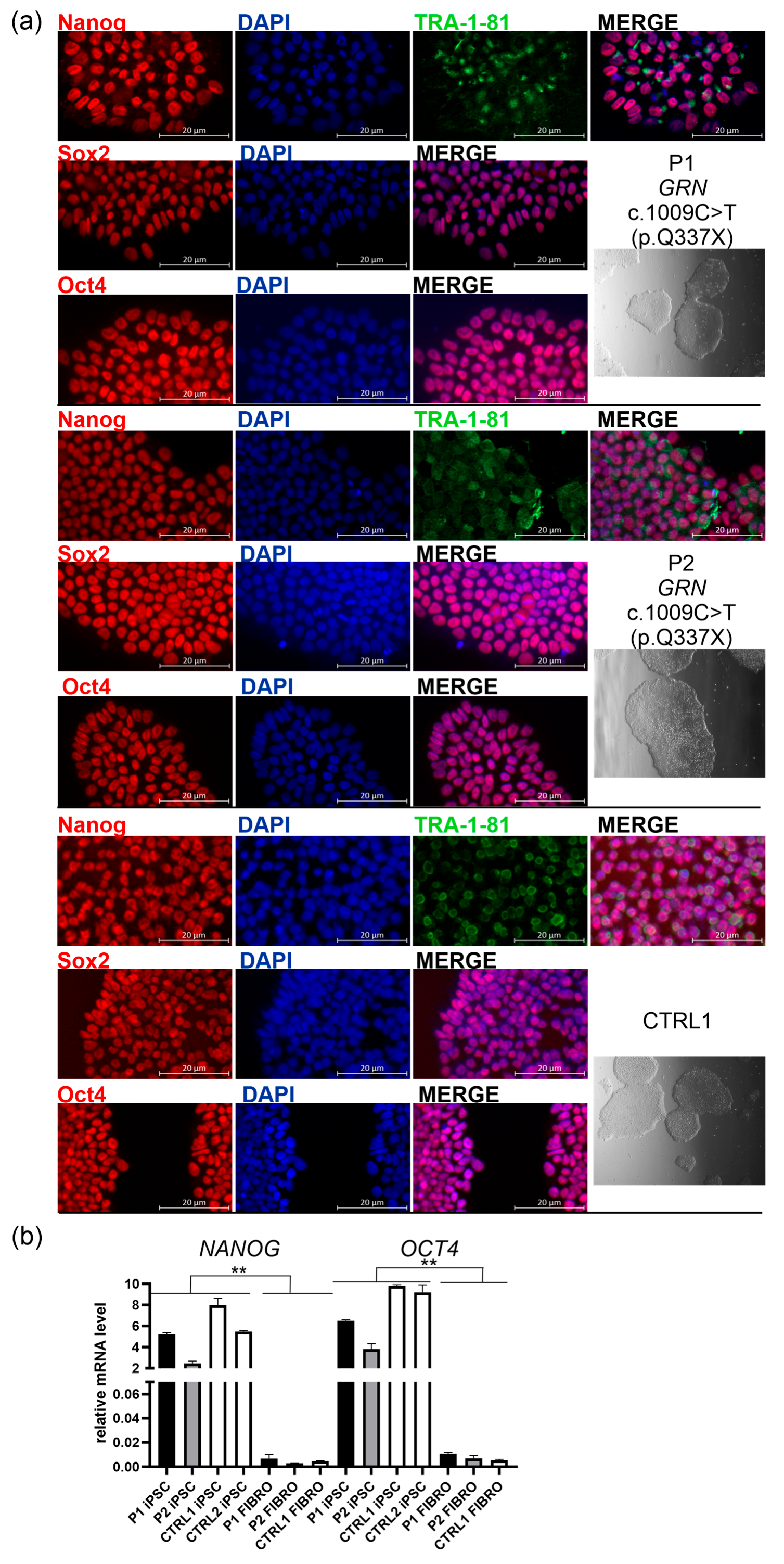
2.1. Characterization of iPSC Lines from a Family with GRN c.1009C>T (p.Q337X) Pathogenic Variant, and a Healthy First-Degree Relative


2.2. Characterization of Neuroepithelial Stem Cells (NES) Lines from a Symptomatic Patient with GRN c.1009C>T (p.Q337X) Pathogenic Variant, Asymptomatic Carrier (P2) of Pathogenic GRN Variant c.1009C>T (p.Q337X), and a Healthy First-Degree Relative (CTRL1)
3. Discussion
4. Materials and Methods
4.1. Reprogramming Fibroblasts into iPSCs
4.2. Real-Time-PCR (RT-PCR) Analysis
4.3. Karyotype Analysis
4.4. Sanger Sequencing and Authentication of iPSC Lines—Short Tandem Repeat (STR) Profiling
4.5. Mycoplasma Testing
4.6. Embryoid Body Formation
4.7. Generation of Neuroepithelial Stem Cells (NES) and Neuronal Differentiation
4.8. Immunofluorescence
4.9. Assessment of iPSC Line Purity
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bFGF | Basic Fibroblast Growth Factor |
| BMP4 | Bone Morphogenetic Protein 4 |
| CXCR4 | C-X-C chemokine receptor type 4 |
| EB | embryoid body |
| EGF | Epidermal Growth Factor |
| FOXA2 | Forkhead Box A2 |
| FTD | frontotemporal dementia |
| FTLD | frontotemporal lobar degeneration |
| FTLD-TDP | frontotemporal lobar degeneration with TDP-43–positive inclusions |
| GRN | Progranulin gene |
| GPCR | G protein-coupled receptor |
| GSK3β | Glycogen Synthase Kinase 3 Beta |
| hNoggin | Human Noggin |
| IMC | Integrated Modulation Contrast |
| iPSCs | induced pluripotent stem cells |
| KOSR | KnockOut Serum Replacement |
| MOI | multiplicity of infection |
| MAP2 | Microtubule-Associated Protein 2 |
| MKI67 | Marker Of Proliferation Ki-67 |
| Nanog | Nanog Homeobox |
| NCAM1 | Neural Cell Adhesion Molecule 1 |
| NCL11 | Neuronal Ceroid Lipofuscinosis type 11 |
| NES | neuroepithelial stem cells |
| NGS | normal goat serum |
| NMD | nonsense-mediated mRNA decay |
| Oct4 | Octamer-Binding Protein 4 |
| OSKM | Oct4, Sox2, Klf4, cMyc |
| PLAGL1 | PLAG1 Like Zinc Finger 1 |
| PGRN | progranulin |
| PODXL | Podocalyxin-like protein 1 |
| PTC | premature termination codon |
| RT-PCR | Real-time PCR |
| SOX1 | SRY-Box Transcription Factor 1 |
| Sox2 | SRY-Box Transcription Factor 2 |
| Sox17 | SRY-Box Transcription Factor 17 |
| STR | Short tandem repeat |
| TBXT | T Brachyury Transcription Factor, T-Box Transcription Factor T |
| TDP-43 | TAR DNA-binding protein 43 |
| TGFβ | Transforming Growth Factor-beta |
References
- Baker, M.; Mackenzie, I.R.; Pickering-Brown, S.M.; Gass, J.; Rademakers, R.; Lindholm, C.; Snowden, J.; Adamson, J.; Sadovnick, A.D.; Rollinson, S.; et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 2006, 442, 916–919. [Google Scholar] [CrossRef]
- Cruts, M.; Gijselinck, I.; van der Zee, J.; Engelborghs, S.; Wils, H.; Pirici, D.; Rademakers, R.; Vandenberghe, R.; Dermaut, B.; Martin, J.J.; et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature 2006, 442, 920–924. [Google Scholar] [CrossRef]
- Gaweda-Walerych, K.; Aragona, V.; Lodato, S.; Sitek, E.J.; Narozanska, E.; Buratti, E. Progranulin deficiency in the brain: The interplay between neuronal and non-neuronal cells. Transl. Neurodegener. 2025, 14, 18. [Google Scholar] [CrossRef]
- Nuytemans, K.; Franzen, S.; Broce, I.J.; Caramelli, P.; Ellajosyula, R.; Finger, E.; Gupta, V.; Illan-Gala, I.; Loi, S.M.; Morhardt, D.; et al. Gaps in biomedical research in frontotemporal dementia: A call for diversity and disparities focused research. Alzheimers Dement. 2024, 20, 9014–9036. [Google Scholar] [CrossRef]
- Pottier, C.; Ren, Y.; Perkerson, R.B., 3rd; Baker, M.; Jenkins, G.D.; van Blitterswijk, M.; DeJesus-Hernandez, M.; van Rooij, J.G.J.; Murray, M.E.; Christopher, E.; et al. Genome-wide analyses as part of the international FTLD-TDP whole-genome sequencing consortium reveals novel disease risk factors and increases support for immune dysfunction in FTLD. Acta Neuropathol. 2019, 137, 879–899. [Google Scholar] [CrossRef]
- Van Deerlin, V.M.; Wood, E.M.; Moore, P.; Yuan, W.; Forman, M.S.; Clark, C.M.; Neumann, M.; Kwong, L.K.; Trojanowski, J.Q.; Lee, V.M.; et al. Clinical, genetic, and pathologic characteristics of patients with frontotemporal dementia and progranulin mutations. Arch. Neurol. 2007, 64, 1148–1153. [Google Scholar] [CrossRef]
- Chen-Plotkin, A.S.; Martinez-Lage, M.; Sleiman, P.M.; Hu, W.; Greene, R.; Wood, E.M.; Bing, S.; Grossman, M.; Schellenberg, G.D.; Hatanpaa, K.J.; et al. Genetic and clinical features of progranulin-associated frontotemporal lobar degeneration. Arch. Neurol. 2011, 68, 488–497. [Google Scholar] [CrossRef]
- Yu, C.E.; Bird, T.D.; Bekris, L.M.; Montine, T.J.; Leverenz, J.B.; Steinbart, E.; Galloway, N.M.; Feldman, H.; Woltjer, R.; Miller, C.A.; et al. The spectrum of mutations in progranulin: A collaborative study screening 545 cases of neurodegeneration. Arch. Neurol. 2010, 67, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Gijselinck, I.; Van Broeckhoven, C.; Cruts, M. Granulin mutations associated with frontotemporal lobar degeneration and related disorders: An update. Hum. Mutat. 2008, 29, 1373–1386. [Google Scholar] [CrossRef] [PubMed]
- Gaweda-Walerych, K.; Walerych, D.; Berdynski, M.; Buratti, E.; Zekanowski, C. Parkin Levels Decrease in Fibroblasts With Progranulin (PGRN) Pathogenic Variants and in a Cellular Model of PGRN Deficiency. Front. Mol. Neurosci. 2021, 14, 676478. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Rodriguez, R.T.; Wang, J.; Ghodasara, A.; Kim, S.K. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell 2011, 8, 335–346. [Google Scholar] [CrossRef] [PubMed]
- Gil, C.H.; Chakraborty, D.; Vieira, C.P.; Prasain, N.; Li Calzi, S.; Fortmann, S.D.; Hu, P.; Banno, K.; Jamal, M.; Huang, C.; et al. Specific mesoderm subset derived from human pluripotent stem cells ameliorates microvascular pathology in type 2 diabetic mice. Sci. Adv. 2022, 8, eabm5559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Foudi, A.; Geay, J.F.; Berthebaud, M.; Buet, D.; Jarrier, P.; Jalil, A.; Vainchenker, W.; Louache, F. Intracellular localization and constitutive endocytosis of CXCR4 in human CD34+ hematopoietic progenitor cells. Stem Cells 2004, 22, 1015–1029. [Google Scholar] [CrossRef]
- Calvo-Garrido, J.; Winn, D.; Maffezzini, C.; Wedell, A.; Freyer, C.; Falk, A.; Wredenberg, A. Protocol for the derivation, culturing, and differentiation of human iPS-cell-derived neuroepithelial stem cells to study neural differentiation in vitro. STAR Protoc. 2021, 2, 100528. [Google Scholar] [CrossRef]
- Wężyk, M.; Szybinska, A.; Wojsiat, J.; Szczerba, M.; Day, K.; Ronnholm, H.; Kele, M.; Berdynski, M.; Peplonska, B.; Fichna, J.P.; et al. Overactive BRCA1 Affects Presenilin 1 in Induced Pluripotent Stem Cell-Derived Neurons in Alzheimer’s Disease. J. Alzheimers Dis. 2018, 62, 175–202. [Google Scholar] [CrossRef]
- Chung, S.H.; Marzban, H.; Aldinger, K.; Dixit, R.; Millen, K.; Schuurmans, C.; Hawkes, R. Zac1 plays a key role in the development of specific neuronal subsets in the mouse cerebellum. Neural Dev. 2011, 6, 25. [Google Scholar] [CrossRef]
- Sakai, H.; Fujii, Y.; Kuwayama, N.; Kawaji, K.; Gotoh, Y.; Kishi, Y. Plag1 regulates neuronal gene expression and neuronal differentiation of neocortical neural progenitor cells. Genes Cells 2019, 24, 650–666. [Google Scholar] [CrossRef]
- Valente, T.; Junyent, F.; Auladell, C. Zac1 is expressed in progenitor/stem cells of the neuroectoderm and mesoderm during embryogenesis: Differential phenotype of the Zac1-expressing cells during development. Dev. Dyn. 2005, 233, 667–679. [Google Scholar] [CrossRef]
- Rraklli, V.; Sodersten, E.; Nyman, U.; Hagey, D.W.; Holmberg, J. Elevated levels of ZAC1 disrupt neurogenesis and promote rapid in vivo reprogramming. Stem Cell Res. 2016, 16, 1–9. [Google Scholar] [CrossRef]
- Varrault, A.; Dantec, C.; Le Digarcher, A.; Chotard, L.; Bilanges, B.; Parrinello, H.; Dubois, E.; Rialle, S.; Severac, D.; Bouschet, T.; et al. Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res. 2017, 45, 10466–10480. [Google Scholar] [CrossRef] [PubMed]
- Liszewska, E.; Majchrowicz, L.; Krogulec, E.; Kotulska, K.; Kaczmarek, L.; Kalita, K.; Dobrzyn, A.; Jaworski, J. Establishment of two hiPSC lines (IIMCBi001-A and IIMCBi002-A) from dermal fibroblasts of healthy donors and characterization of their cell cycle. Stem Cell Res. 2021, 52, 102225. [Google Scholar] [CrossRef]
- Zhang, M.; Ngo, J.; Pirozzi, F.; Sun, Y.P.; Wynshaw-Boris, A. Highly efficient methods to obtain homogeneous dorsal neural progenitor cells from human and mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res. Ther. 2018, 9, 67. [Google Scholar] [CrossRef]
- Lee, D.H.; Lee, E.C.; Lee, J.Y.; Lee, M.R.; Shim, J.W.; Oh, J.S. Neuronal Cell Differentiation of iPSCs for the Clinical Treatment of Neurological Diseases. Biomedicines 2024, 12, 1350. [Google Scholar] [CrossRef]
- Li, W.; Sun, W.; Zhang, Y.; Wei, W.; Ambasudhan, R.; Xia, P.; Talantova, M.; Lin, T.; Kim, J.; Wang, X.; et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl. Acad. Sci. USA 2011, 108, 8299–8304. [Google Scholar] [CrossRef] [PubMed]
- Venere, M.; Han, Y.G.; Bell, R.; Song, J.S.; Alvarez-Buylla, A.; Blelloch, R. Sox1 marks an activated neural stem/progenitor cell in the hippocampus. Development 2012, 139, 3938–3949. [Google Scholar] [CrossRef] [PubMed]
- Gasperoni, J.G.; Tran, S.C.; Grommen, S.V.H.; De Groef, B.; Dworkin, S. The Role of PLAG1 in Mouse Brain Development and Neurogenesis. Mol. Neurobiol. 2024, 61, 5851–5867. [Google Scholar] [CrossRef]
- Almeida, S.; Zhang, Z.; Coppola, G.; Mao, W.; Futai, K.; Karydas, A.; Geschwind, M.D.; Tartaglia, M.C.; Gao, F.; Gianni, D.; et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012, 2, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Xu, Y.F.; Dickey, C.A.; Buratti, E.; Baralle, F.; Bailey, R.; Pickering-Brown, S.; Dickson, D.; Petrucelli, L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 2007, 27, 10530–10534. [Google Scholar] [CrossRef]
- Raitano, S.; Ordovas, L.; De Muynck, L.; Guo, W.; Espuny-Camacho, I.; Geraerts, M.; Khurana, S.; Vanuytsel, K.; Toth, B.I.; Voets, T.; et al. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Rep. 2015, 4, 16–24. [Google Scholar] [CrossRef]
- Lee, C.W.; Stankowski, J.N.; Chew, J.; Cook, C.N.; Lam, Y.W.; Almeida, S.; Carlomagno, Y.; Lau, K.F.; Prudencio, M.; Gao, F.B.; et al. The lysosomal protein cathepsin L is a progranulin protease. Mol. Neurodegener. 2017, 12, 55. [Google Scholar] [CrossRef]
- Valdez, C.; Wong, Y.C.; Schwake, M.; Bu, G.; Wszolek, Z.K.; Krainc, D. Progranulin-mediated deficiency of cathepsin D results in FTD and NCL-like phenotypes in neurons derived from FTD patients. Hum. Mol. Genet. 2017, 26, 4861–4872. [Google Scholar] [CrossRef]
- Lines, G.; Casey, J.M.; Preza, E.; Wray, S. Modelling frontotemporal dementia using patient-derived induced pluripotent stem cells. Mol. Cell Neurosci. 2020, 109, 103553. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.R.; Martins, S.; Cammarata, G.; Martins, M.; Cardoso, A.M.; Almeida, M.R.; do Carmo Macario, M.; Santana, I.; Peca, J.; Cardoso, A.L. Generation and Characterization of Novel iPSC Lines from a Portuguese Family Bearing Heterozygous and Homozygous GRN Mutations. Biomedicines 2022, 10, 1905. [Google Scholar] [CrossRef]
- Gaweda-Walerych, K.; Sitek, E.J.; Narozanska, E.; Wężyk, M.; Brockhuis, B.; Zekanowski, C.; Slawek, J. Functional characterization of a novel progranulin mutation in a patient with progressive nonfluent aphasia. Neurobiol. Aging 2018, 72, 186.e9–186.e12. [Google Scholar] [CrossRef] [PubMed]
- Gaweda-Walerych, K.; Mohagheghi, F.; Zekanowski, C.; Buratti, E. Parkinson’s disease-related gene variants influence pre-mRNA splicing processes. Neurobiol. Aging 2016, 47, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Vanderheijden, C.; Yakkioui, Y.; Vaessen, T.; Santegoeds, R.; Temel, Y.; Hoogland, G.; Hovinga, K. Developmental gene expression in skull-base chordomas and chondrosarcomas. J. Neurooncol. 2025, 172, 249–256. [Google Scholar] [CrossRef]
- Falk, A.; Koch, P.; Kesavan, J.; Takashima, Y.; Ladewig, J.; Alexander, M.; Wiskow, O.; Tailor, J.; Trotter, M.; Pollard, S.; et al. Capture of neuroepithelial-like stem cells from pluripotent stem cells provides a versatile system for in vitro production of human neurons. PLoS ONE 2012, 7, e29597. [Google Scholar] [CrossRef]

| Category | Method | Result | Data |
|---|---|---|---|
| iPSC morphology | phase contrast microscopy | normal human induced pluripotent stem cell morphology | Figure 1a—right panel |
| iPSC phenotype | immunocytochemistry | positive immunostaining of pluripotency markers: Nanog, Oct4, Sox2, and TRA1-1-81 | Figure 1a left panels, Figure S1 |
| iPSC phenotype | real-time PCR | mRNA expression of pluripotency markers: NANOG, OCT4 | Figure 1b |
| iPSC differentiation potential | Embroid body formation and STEMdiff ™ Trilineage Differentiation Kit | detection of the markers of the three germ layers, ectoderm (β3-Tubulin, Nestin), mesoderm (CXCR4), ectoderm/mesoderm (NCAM1), and endoderm (Sox17) by immunocytochemistry | Figure 2a, Figures S2 and S3 |
| iPSC differentiation potential | real-time PCR | mRNA expression of the markers of mesoderm (TBXT), and endoderm (FOXA2) | Figure 2b |
| karyotype | G-banding | a normal karyotype was confirmed for the established iPSC lines, P1 (passage: 22, resolution: 400, total counted: 20), P2 (passage: 20, resolution: 400, total counted: 20), and C1 (passage 23, resolution: 400, total counted: 20) | Figure 3a |
| phenotype related to the GRN pathogenic variant | real-time PCR | mRNA expression of GRN | Figure 3b |
| virology | real-time PCR | no residual expression of Sendai virus-derived reprogramming vectors was detected in the established iPSC lines | Figure S4 |
| mutation analysis | Sanger sequencing | confirmation of the presence of the pathogenic variant c.1009C>T in iPSC lines of carriers and the wild-type sequence in the non-carrier | Figure S5 |
| iPSC lines authentication | STR profiling | complete concordance of the respective fibroblast—iPSC pairs for P1, P2, and CTRL1 | not disclosed to protect the sensitive personal information of cell line donors |
| iPSC Line | Percentage of Nanog- and DAPI-Positive Cells (%) | Regions of Interests (ROI) 1 Count |
|---|---|---|
| P1 | 96.2 | 18,113 |
| P2 | 90.7 | 24,091 |
| CTRL1 | 93.3 | 42,501 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gaweda-Walerych, K.; Figarski, A.; Gawlik-Zawiślak, S.; Woźniak, M.; Chołoniewska, A.; Mierzwa, N.; Lutostańska, E.; Szymanowski, J.; Wężyk, M. Generation of Induced Pluripotent Stem Cells and Neuroepithelial Stem Cells from a Family with the Pathogenic Variant p.Q337X in Progranulin. Int. J. Mol. Sci. 2025, 26, 11242. https://doi.org/10.3390/ijms262311242
Gaweda-Walerych K, Figarski A, Gawlik-Zawiślak S, Woźniak M, Chołoniewska A, Mierzwa N, Lutostańska E, Szymanowski J, Wężyk M. Generation of Induced Pluripotent Stem Cells and Neuroepithelial Stem Cells from a Family with the Pathogenic Variant p.Q337X in Progranulin. International Journal of Molecular Sciences. 2025; 26(23):11242. https://doi.org/10.3390/ijms262311242
Chicago/Turabian StyleGaweda-Walerych, Katarzyna, Adam Figarski, Sylwia Gawlik-Zawiślak, Marta Woźniak, Anna Chołoniewska, Natalia Mierzwa, Eliza Lutostańska, Jakub Szymanowski, and Michalina Wężyk. 2025. "Generation of Induced Pluripotent Stem Cells and Neuroepithelial Stem Cells from a Family with the Pathogenic Variant p.Q337X in Progranulin" International Journal of Molecular Sciences 26, no. 23: 11242. https://doi.org/10.3390/ijms262311242
APA StyleGaweda-Walerych, K., Figarski, A., Gawlik-Zawiślak, S., Woźniak, M., Chołoniewska, A., Mierzwa, N., Lutostańska, E., Szymanowski, J., & Wężyk, M. (2025). Generation of Induced Pluripotent Stem Cells and Neuroepithelial Stem Cells from a Family with the Pathogenic Variant p.Q337X in Progranulin. International Journal of Molecular Sciences, 26(23), 11242. https://doi.org/10.3390/ijms262311242






