Comparative Analysis of Chromosome Repeat DNA Patterns in Four Amaranthus Species
Abstract
1. Introduction
2. Results
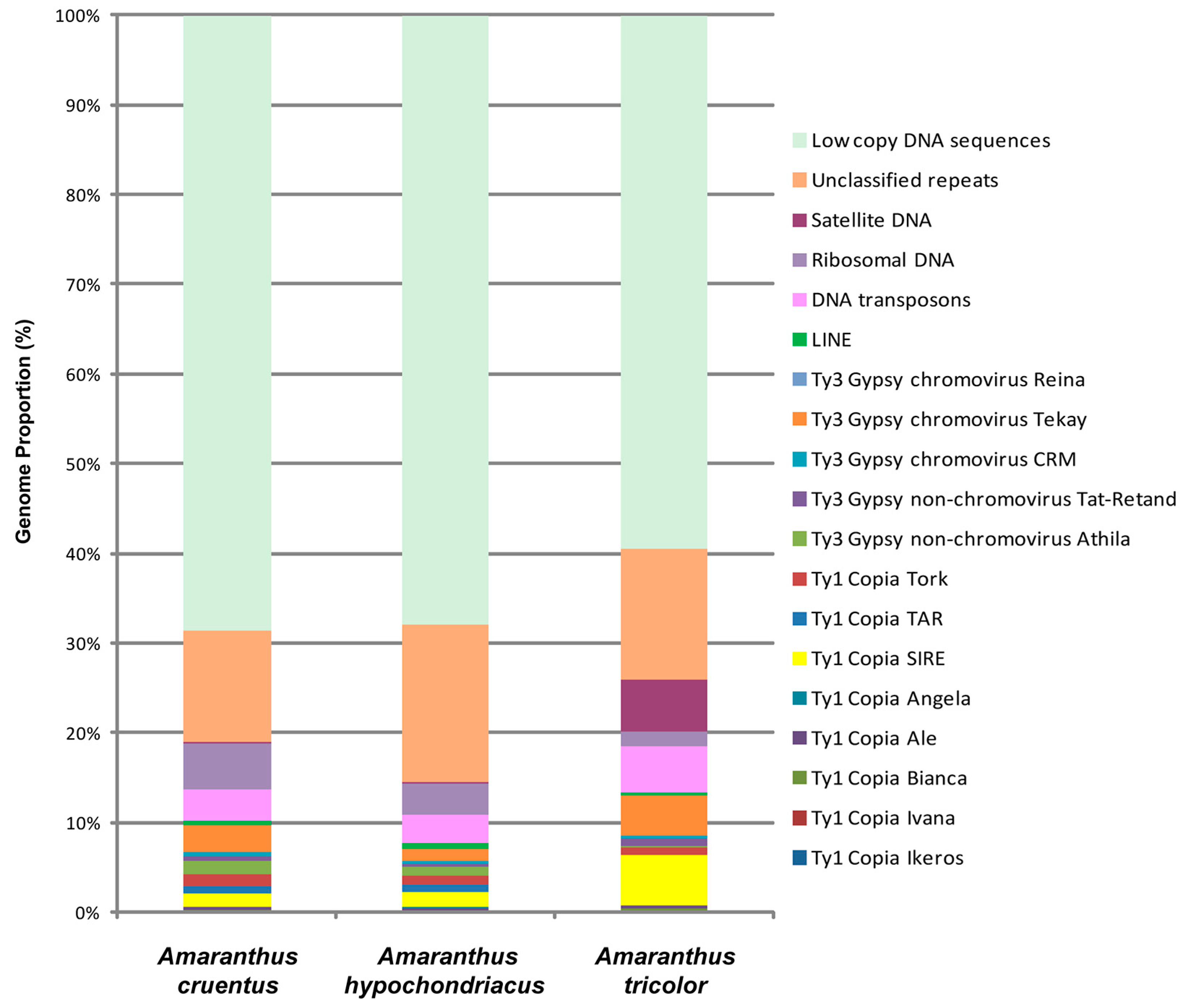
2.1. Comparative Analyses of DNA Repeats Identified in Genomes of A. tricolor, A. cruentus, and A. hypochondriacus
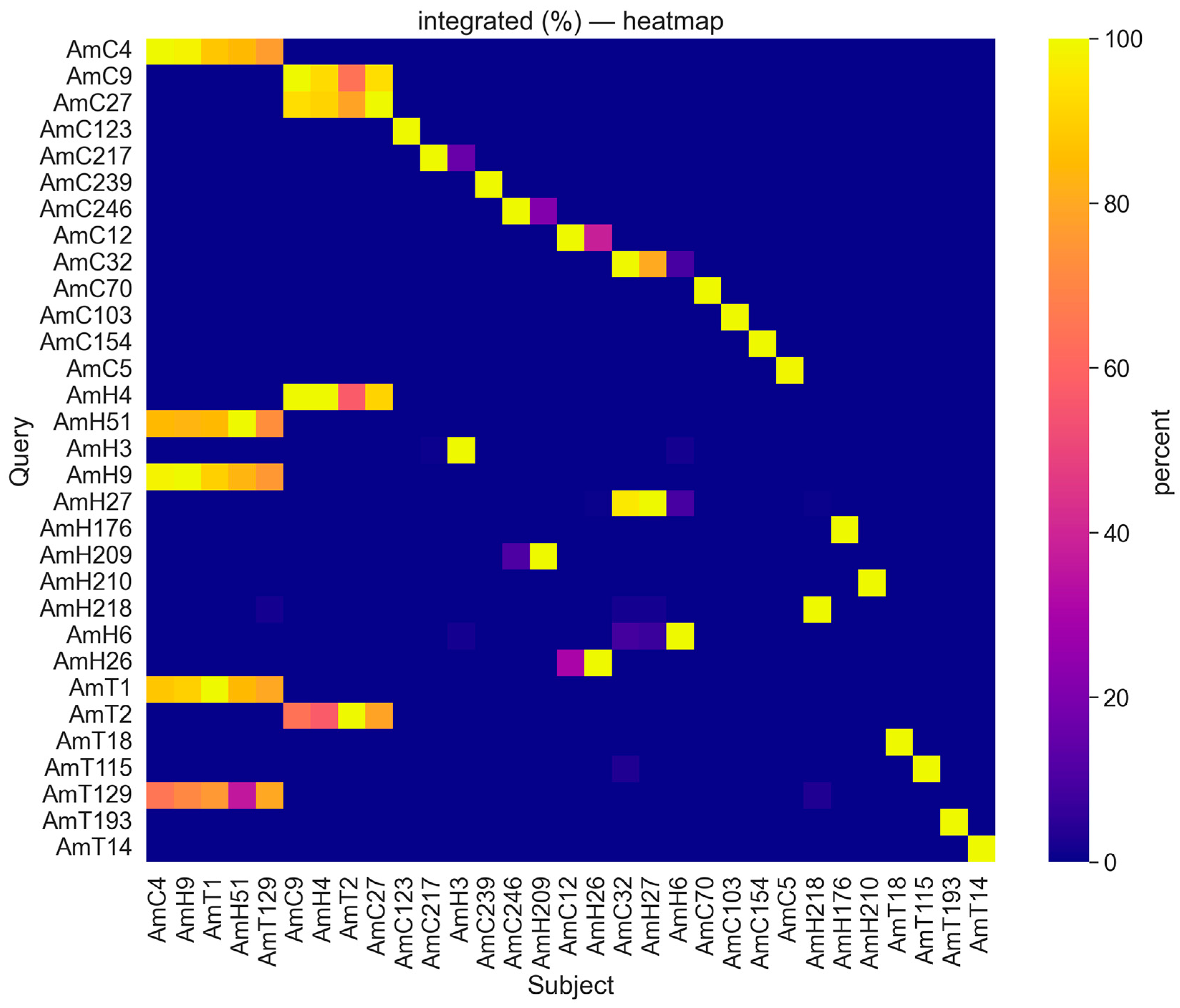
2.2. BLAST Analysis of the Identified Tandem DNAs
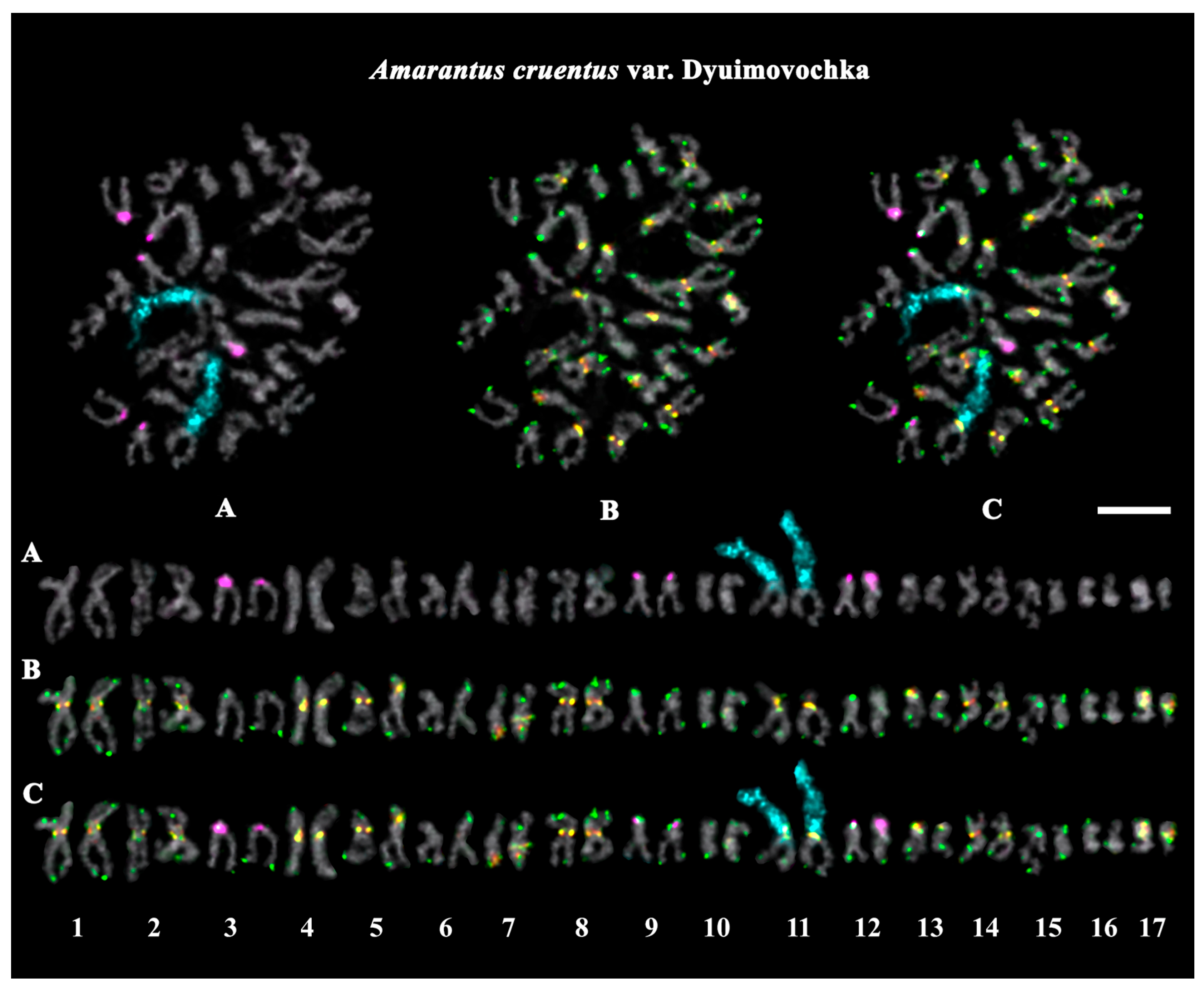
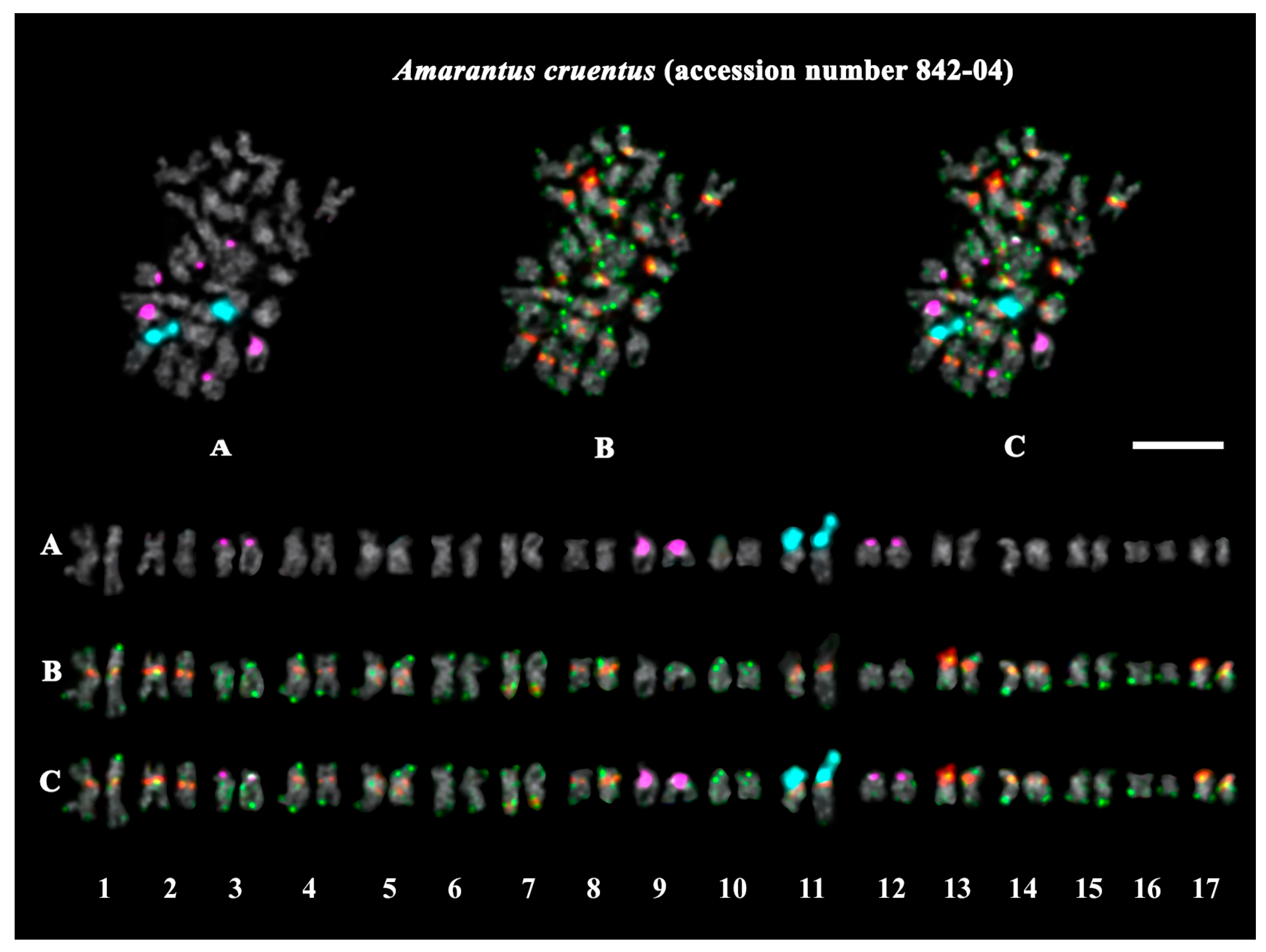
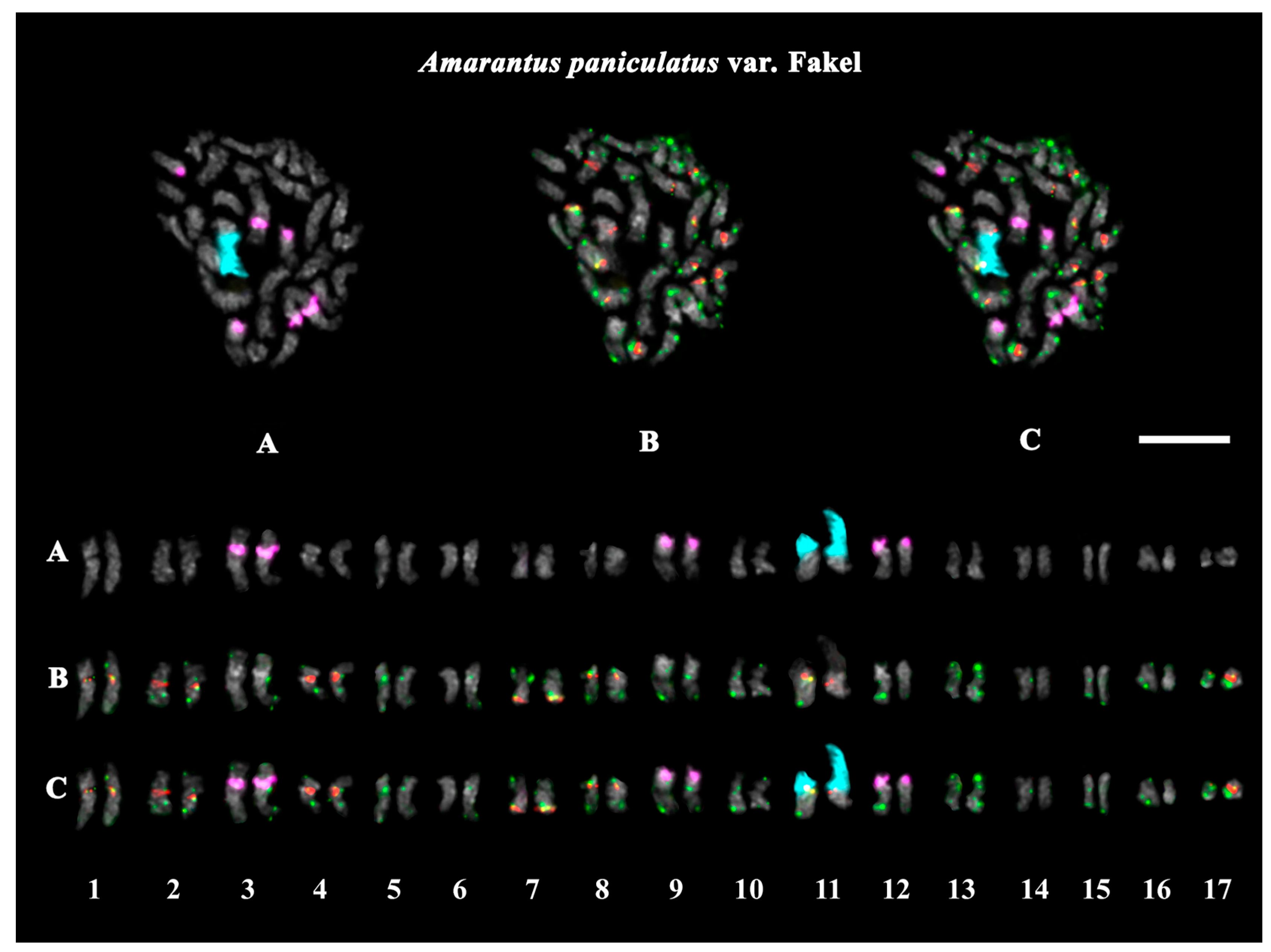
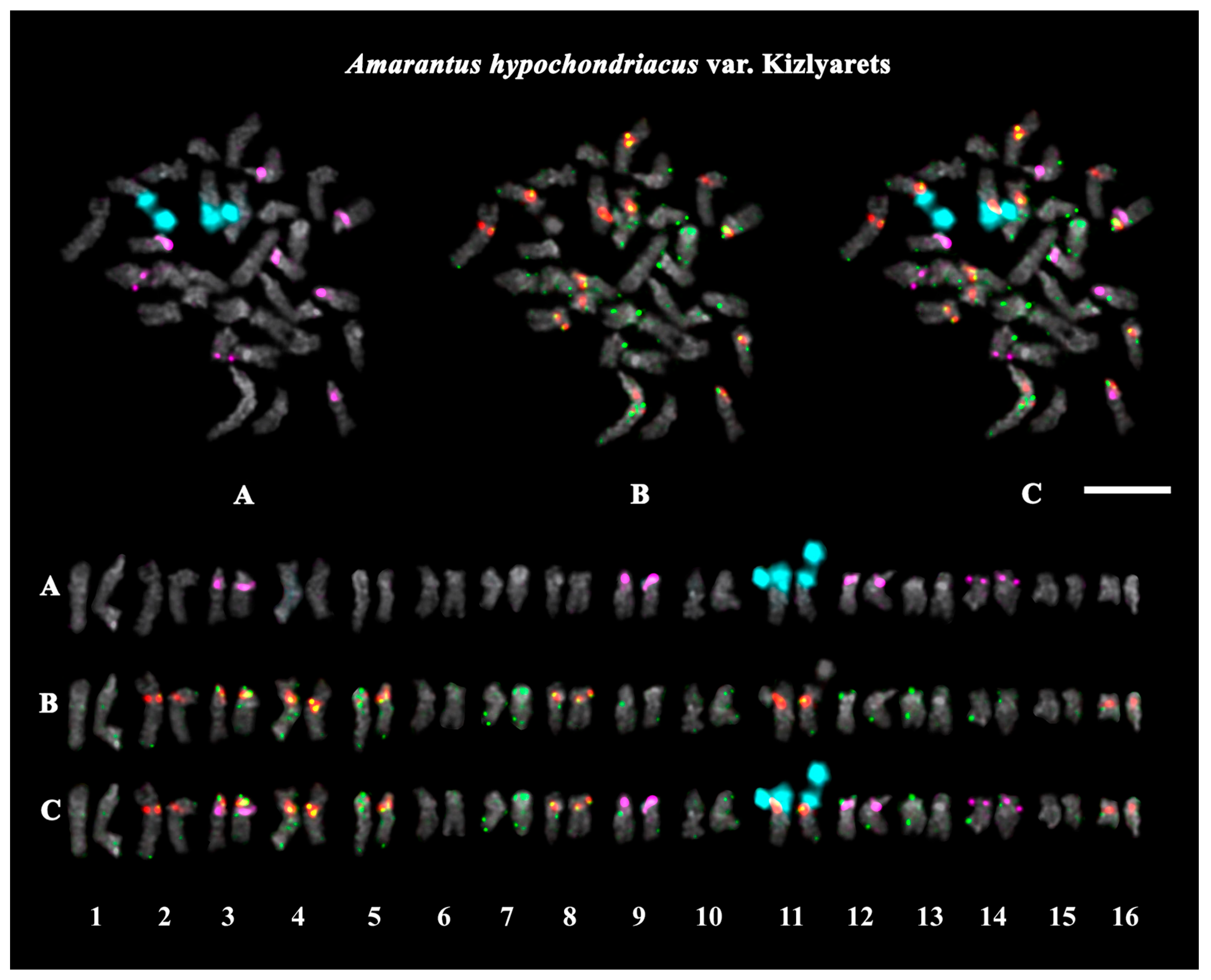
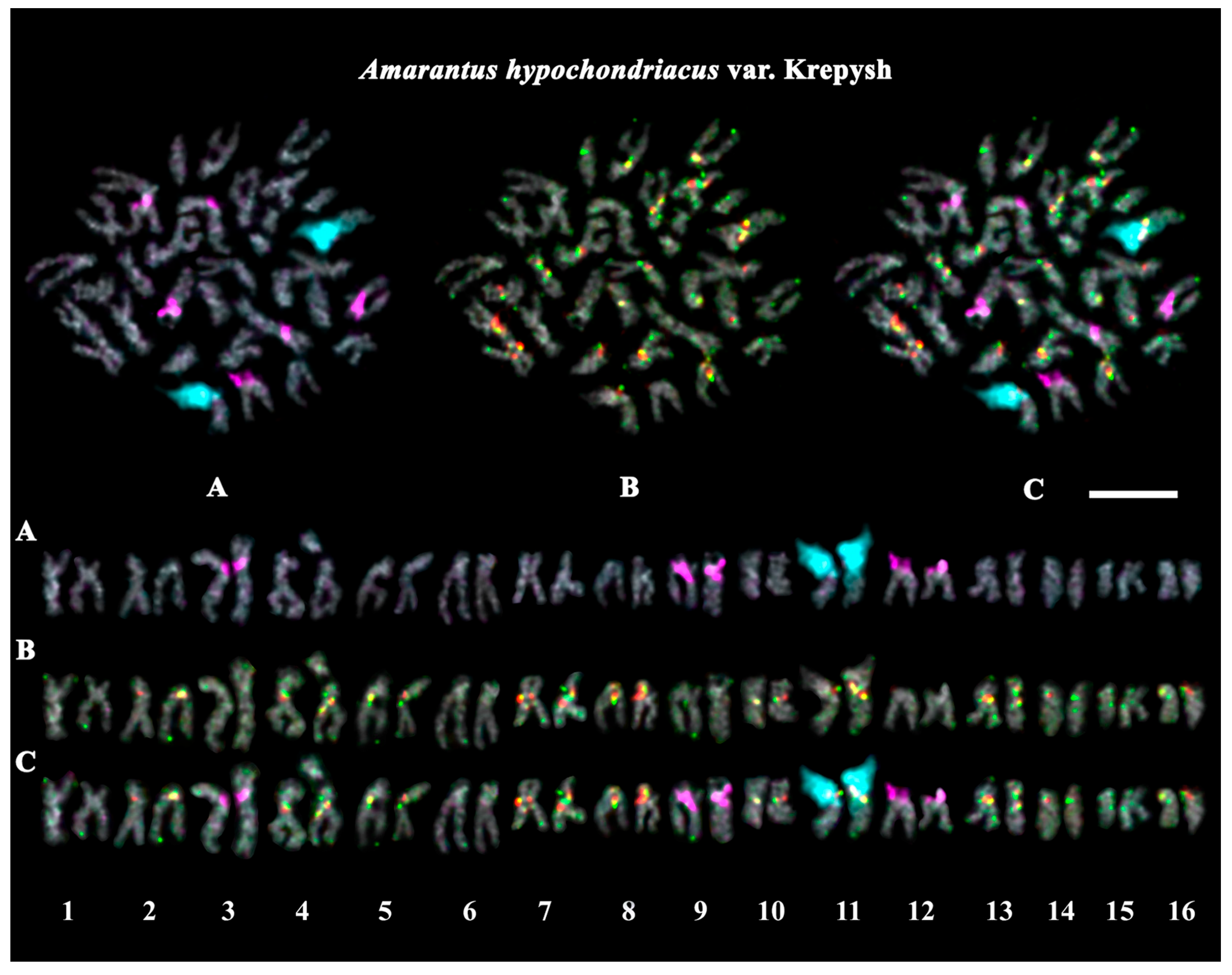
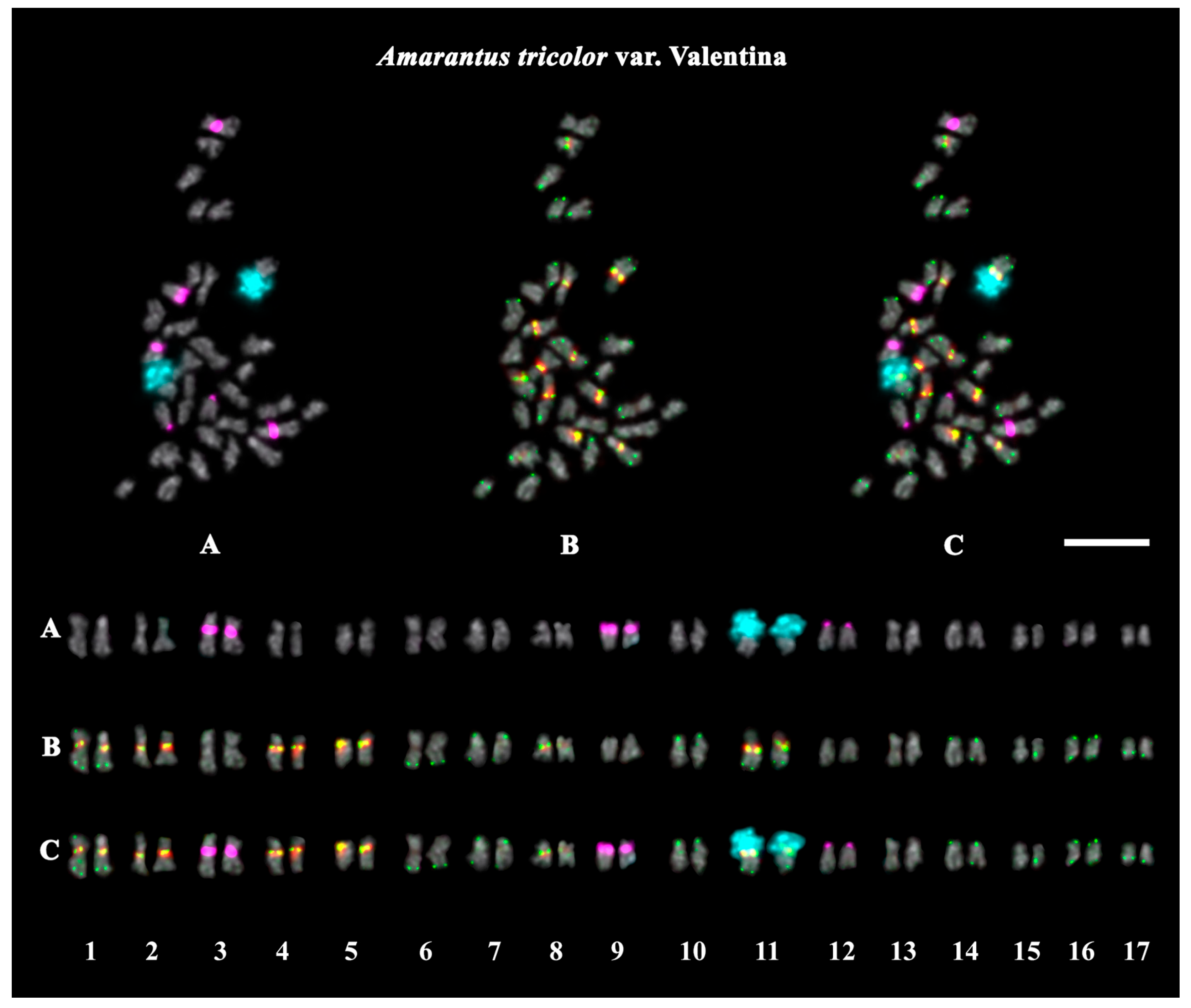
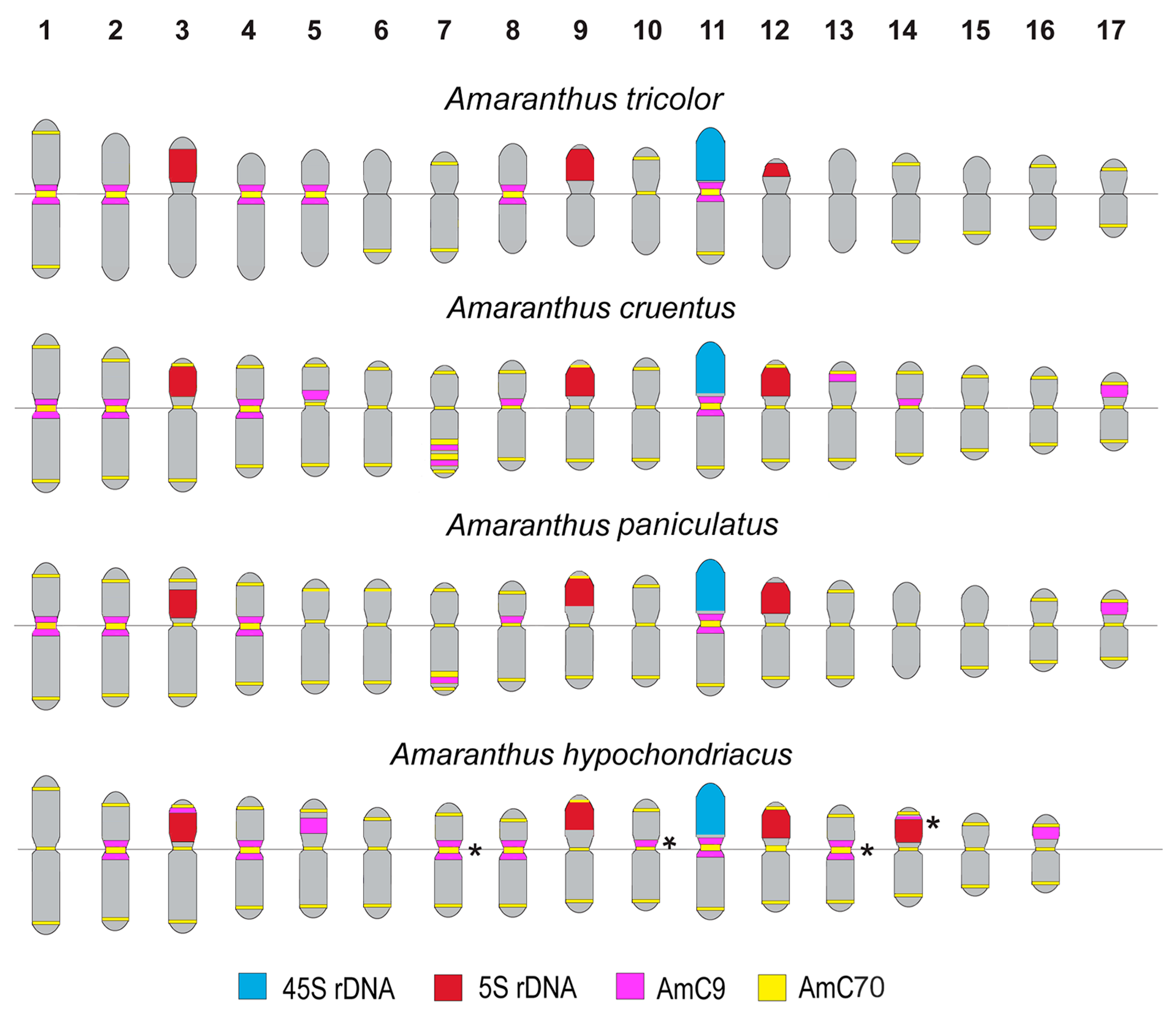
2.3. Structural Variations in Chromosomes Revealed by FISH
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Bioinformatic Sequence Analysis
4.3. Chromosome Spread Preparation
4.4. FISH
4.5. Analysis of Chromosome Slides
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Assad, R.; Reshi, Z.A.; Jan, S.; Rashid, I. Biology of Amaranths. Bot. Rev. 2017, 83, 382–436. [Google Scholar] [CrossRef]
- Hajyzade, M. Genome-wide identification and characterization of abiotic stress responsive mTERF gene family in Amaranthus hypochondriacus. Phyton-Int. J. Exp. Bot. 2023, 92, 1649–1664. [Google Scholar] [CrossRef]
- Chakraborty, S.; Chakraborty, N.; Agrawal, L.; Ghosh, S.; Narula, K.; Shekhar, S.; Naik, P.S.; Pande, P.C.; Chakrborti, S.K.; Datta, A. Next-generation protein-rich potato expressing the seed protein gene AmA1 is a result of proteome rebalancing I transgenic tuber. Proc. Natl. Acad. Sci. USA 2010, 107, 17533–17538. [Google Scholar] [CrossRef]
- Palmeros-Suarez, P.A.; MassangeSánchez, J.A.; Martínez-Gallardo, N.A.; Montero-Vargas, J.M.; Gomez-Leyva, J.F.; Delano-Frier, J.P. The overexpression of an Amaranthus hypochondriacus NF-YC gene modifies growth and confers water deficit stress resistance in Arabidopsis. Plant Sci. 2015, 240, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Massange-Sanchez, J.A.; Palmeros-Suarez, P.A.; Martinez-Gallardo, N.A.; Castrillon-Arbelaez, P.A.; Aviles-Arnaut, H.; Alatorre-Cobos, F.; Tiessen, A.; Delano-Frier, J.P. The novel and taxonomically restricted Ah24 gene from grain amaranth (Amaranthus hypochondriacus) has a dual role in development and defense. Front. Plant Sci. 2015, 6, 602. [Google Scholar] [CrossRef]
- Toimbayeva, D.; Saduakhasova, S.; Kamanova, S.; Kiykbay, A.; Tazhina, S.; Temirova, I.; Muratkhan, M.; Shaimenova, B.; Murat, L.; Khamitova, D.; et al. Prospects for the use of amaranth grain in the production of functional and specialized food products. Foods 2025, 14, 1603. [Google Scholar] [CrossRef]
- Szabóová, M.; Záhorský, M.; Gažo, J.; Geuens, J.; Vermoesen, A.; Hondt, E.D.; Hricov, A. Commercial Amaranth Varieties (Amaranthus spp.). Plants 2020, 9, 1412. [Google Scholar] [CrossRef]
- Tucker, J.B. Amaranth: The Once and Future Crop. BioScience 1986, 36, 9–13. [Google Scholar] [CrossRef]
- Martinez-Lopez, A.; Millan-Linares, M.C.; Noelia, M.; Rodriguez-Martin, N.M.; Francisco Millan, F.; Sergio Montserrat-de la Paz, S. Nutraceutical value of kiwicha (Amaranthus caudatus L.). J. Funct. Foods 2020, 65, 103735. [Google Scholar] [CrossRef]
- Cunha-Chiamolera, T.P.L.; Chileh-Chelh, T.; Urrestarazu, M.; Ezzaitouni, M.; López-Ruiz, R.; Gallón-Bedoya, M.; Rincón-Cervera, M.Á.; Guil-Guerrero, J.L. Crop productivity, phytochemicals, and bioactivities of wild and grown in controlled environment slender Amaranth (Amaranthus viridis L.). Agronomy 2024, 14, 2038. [Google Scholar] [CrossRef]
- Santiago, P.D.; Tenbergen, K.; Velez-Jimenez, E.; Cardador-Martínez, M.A. Functional attributes of Amaranth. Austin J. Nutr. Food Sci. 2014, 2, 1–6. [Google Scholar]
- Santiago, P.D.; Tenbergen, K.; Velez-Jimenez, E.; Cardador-Martínez, M.A.; Nazeer, S.; Yaman Firincioglu, S. Amaranth in animal nutrition. J. Agric. Food Environ. Anim. Sci. 2022, 3, 195–211. [Google Scholar]
- Maldonado-Cervantes, E.; Jeong, H.J.; Leon-Galvan, F.; Barrera-Pacheco, A.; De Leon-Rodriguez, A.; Gonzalez de Mejia, E.; de Lumen, B.O.; Barba de la Rosa, A.P. Amaranth lunasin-like peptide internalizes into the cell nucleus and inhibits chemical carcinogen-induced transformation of NIH-3T3 cells. Peptides 2010, 31, 1635–1642. [Google Scholar] [CrossRef]
- Tufts, H.R.; Harris, C.S.; Bukania, Z.N.; Johns, T. Antioxidant and anti-inflammatory activities of Kenyan leafy green vegetables, wild fruits, and medicinal plants with potential relevance for kwashiorkor. Evid. Based Complement. Altern. Med. 2015, 2015, 807158. [Google Scholar] [CrossRef]
- Lado, M.B.; Burini, J.; Rinaldi, G.; Anon, M.C.; Tironi, V.A. Effects of the dietary addition of Amaranth (Amaranthus mantegazzianus) protein isolate on antioxidant status, lipid profiles and blood pressure of rats. Plant Foods Hum. Nutr. 2015, 70, 371–379. [Google Scholar] [CrossRef]
- Sabbione, A.C.; Rinaldi, G.; Anon, M.C.; Scilingo, A.A. Antithrombotic effects of Amaranthus hypochondriacus proteins in rats. Plant Foods Hum. Nutr. 2016, 71, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Sammour, R.H.; Radwan, S.A.; Mira, M. Genetic diversity in genus Amaranthus: From morphology to genomic DNA. Res. Rev. Biosci. 2012, 6, 351–360. [Google Scholar]
- Yeshitila, M.; Gedebo, A.; Tesfaye, B.; Degu, H.D. Agro-morphological genetic diversity assessment of Amaranthus genotypes from Ethiopia based on qualitative traits. CABI Agric. Biosci. 2024, 5, 95. [Google Scholar] [CrossRef]
- Sammour, R.H.; Mira, M.; Radwan, S.A. Phenotypic and isoenzymatic variations in Amaranthus species. Int. J. Agric. Biol. 2021, 26, 587–596. [Google Scholar] [CrossRef]
- Iamonico, D. Nomenclature Survey of the Genus Amaranthus (Amaranthaceae): 12 Questions about Amaranthus caudatus. Plants 2023, 12, 1566. [Google Scholar] [CrossRef] [PubMed]
- Sauer, J.D. The grain amaranths and their relatives: A revised taxonomic and geographic survey. Ann. Mo. Bot. Gard. 1967, 54, 102–137. [Google Scholar] [CrossRef]
- Mosyakin, S.L.; Robertson, K.R. New infrageneric taxa and combinations in Amaranthus L. (Amaranthaceae). Ann. Bot. Fenn. 1996, 33, 275–281. [Google Scholar]
- Costea, M.; Sanders, A.; Waines, G. Preliminary results toward a revision of the Amaranthus hybridus complex (Amaranthaceae). Sida Contrib. Bot. 2001, 19, 931–974. [Google Scholar]
- Iamonico, D. Taxonomic revision of the genus Amaranthus (Amaranthaceae) in Italy. Phytotaxa 2015, 199, 1–84. [Google Scholar] [CrossRef]
- Hassan, W.A.; Al-shaye, N.A.; Alghamdi, S.; Korany, S.M.; Iamonico, D. Taxonomic revision of the genus Amaranthus (Amaranthaceae) in Saudi Arabia. Phytotaxa 2022, 576, 135–157. [Google Scholar] [CrossRef]
- Sammour, R.H.; Mira, M.; Radwan, S.; Fahmey, S. Genetic diversity and phylogenetic relationships between and within Amaranthus spp. using RAPD markers. Rev. Mex. Biodiv. 2020, 91, e913254. [Google Scholar] [CrossRef]
- Mallory, M.A.; Hall, R.V.; McNabb, A.R.; Pratt, D.B.; Jellen, E.N.; Maughan, P.J. Development and characterization of microsatellite markers for the grain amaranths. Crop Sci. 2008, 48, 1098–1106. [Google Scholar] [CrossRef]
- Maughan, P.J.; Smith, S.M.; Fairbanks, D.J.; Jellen, E.N. Development, characterization, and linkage mapping of single nucleotide polymorphisms in the grain amaranths (Amaranthus spp.). Plant Gen. 2011, 4, 92–101. [Google Scholar] [CrossRef]
- Thapa, R.; Edwards, M.; Blair, M.W. Relationship of cultivated grain amaranth species and wild relative accessions. Genes 2021, 12, 1849. [Google Scholar] [CrossRef]
- Maughan, P.J.; Sisneros, N.; Luo, M.; Kudrna, D.; Ammiraju, J.S.S.; Wing, R.A. Construction of an Amaranthus hypochondriacus bacterial artificial chromosome library and genomic sequencing of herbicide target genes. Crop Sci. 2008, 48, 85–94. [Google Scholar] [CrossRef]
- Waselkov, K.E.; Boleda, A.S.; Olsen, K.M. A Phylogeny of the genus Amaranthus (Amaranthaceae) based on several low-copy nuclear loci and chloroplast regions. Syst. Bot. 2018, 43, 439–458. [Google Scholar] [CrossRef]
- Lanoue, K.Z.; Wolf, P.G.; Browning, S.; Hood, E.E. Phylogenetic analysis of restriction-site variation in wild and cultivated Amaranthus species (Amaranthaceae). Theoret. Appl. Genet. 1996, 93, 722–732. [Google Scholar] [CrossRef]
- Han, J.; Lin, C.; Zhu, T.; Liu, Y.; Yan, J.; Qi, Z.; Yan, X. Comprehensive chloroplast genomic insights into Amaranthus: Resolving the phylogenetic and taxonomic status of A. powellii and A. bouchonii. Plants 2025, 14, 649. [Google Scholar] [CrossRef]
- Ma, X.; Vaistij, F.E.; Li, Y.; van Rensburg, W.S.J.; Harvey, S.; Bairu, M.W.; Venter, S.L.; Mavengahama, S.; Ning, Z.; Graham, I.A.; et al. A chromosome-level Amaranthus cruentus genome assembly highlights gene family evolution and biosynthetic gene clusters that may underpin the nutritional value of this traditional crop. Plant J. 2021, 107, 613–628, Erratum in Plant J. 2022, 110, 1829. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, D.; Wang, S.; Wang, A.; Lei, L.; Jiang, F.; Yang, B.; Yuan, L.; Chen, R.; Zhang, Y.; et al. Chromosome-scale Amaranthus tricolor genome provides insights into the evolution of the genus Amaranthus and the mechanism of betalain biosynthesis. DNA Res. 2022, 30, dsac050. [Google Scholar] [CrossRef] [PubMed]
- Deb, S.; Jayaprasad, S.; Ravi, S.; Rao, K.R.; Whadgar, S.; Hariharan, N.; Dixit, S.; Sunil, M.; Choudhary, B.; Stevanato, P.; et al. Classification of grain amaranths using chromosome-level genome assembly of Ramdana, A. hypochondriacus. Front. Plant Sci. 2020, 11, 579529. [Google Scholar] [CrossRef]
- Grant, F.W. Cytogenetic studies in Amaranthus I. Cytogenetical aspects of sex determination in dioecious species. Canad. J. Bot. 1959, 37, 413–417. [Google Scholar] [CrossRef]
- Grant, F.W. Cytogenetic studies in Amaranthus III. Chromosome numbers and phylogenetic aspects. Can. J. Genet. Cytol. 1959, 1, 313–318. [Google Scholar] [CrossRef]
- Greizerstein, E.J.; Poggio, L. Karyological studies in grain Amaranths. Cytology 1994, 59, 25–30. [Google Scholar] [CrossRef]
- Pal, M.; Ohri, D.; Subrahmanyam, G.V. A New Basic Chromosome Number for Amaranthus (Amaranthaceae). Cytologia 2000, 65, 13–16. [Google Scholar] [CrossRef][Green Version]
- Kolano, B.; Saracka, K.; Broda-Cnota, A.; Maluszynska, J. Localization of ribosomal DNA and CMA3/DAPI heterochromatin in cultivated and wild Amaranthus species. Sci. Hortic. 2013, 164, 249–255. [Google Scholar] [CrossRef]
- Prajitha, V.; Thoppil, J.E. Cytogenetic characterization of Amaranthus caudatus L. and Amaranthus hybridus subsp. cruentus (L.). Thell. Cytotechnology 2018, 70, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Toma, F.N.; Bonna, I.J.; Hossen, R.; Alam, S.S.; Sultana, S.S. Comparative chromosome analysis of three Amaranthus species. Cytologia 2019, 84, 147–151. [Google Scholar] [CrossRef]
- Sammour, R.H.; Radwan, S.; Mira, M.; Badar, S. Genetic variability and evolutionary relationships between Amaranthus spp. as revealed by karyotype analysis. Biochem. Indian J. 2014, 8, 129–137. [Google Scholar]
- Kolano, B.; Pando, L.G.; Małuszyńska, J. Molecular cytogenetic studies in Chenopodium quinoa and Amaranthus caudatus. Acta Soc. Bot. Pol. 2005, 70, 85–90. [Google Scholar] [CrossRef]
- Bonasora, M.G.; Poggio, L.; Greizerstein, E.J. Cytogenetic studies in four cultivated Amaranthus (Amaranthaceae) species. CompCytogen 2013, 7, 53–61. [Google Scholar] [CrossRef]
- Amosova, A.V.; Yurkevich, O.Y.; Semenov, A.R.; Samatadze, T.E.; Sokolova, D.V.; Artemyeva, A.M.; Zoshchuk, S.A.; Muravenko, O.V. Genome studies in Amaranthus cruentus L. and A. hypochondriacus L. based on repeatomic and cytogenetic data. Int. J. Mol. Sci. 2024, 25, 13575. [Google Scholar] [CrossRef]
- Kubis, S.; Schmidt, T.; Heslop-Harrison, J.S. Repetitive DNA elements as a major component of plant genomes. Ann. Bot. 1998, 82, 45–55. [Google Scholar] [CrossRef]
- Romano, N.C.; Laura Fanti, L. Transposable elements: Major players in shaping genomic and evolutionary patterns. Cells 2022, 11, 1048. [Google Scholar] [CrossRef]
- Lee, S.I.; Kim, N.S. Transposable elements and genome size variations in plants. Genom. Inform. 2014, 12, 87–97. [Google Scholar] [CrossRef]
- Sunil, M.; Hariharan, A.K.; Nayak, S.; Gupta, S.; Nambisan, S.R.; Gupta, R.P.; Panda, B.; Choudhary, B.; Srinivasan, S. The draft genome and transcriptome of Amaranthus hypochondriacus: A C4 dicot producing high-lysine edible pseudo-cereal. DNA Res. 2014, 21, 585–602. [Google Scholar] [CrossRef]
- Becher, H.; Powell, R.F.; Brown, M.R.; Metherell, C.; Pellicer, J.; Leitch, I.J.; Twyford, A.D. The nature of intraspecific and interspecific genome size variation in taxonomically complex eyebrights. Ann. Bot. 2021, 128, 639–651. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zheng, Z.; Li, Y.; Hu, H.; Wang, Z.; Du, X. Which factors contribute most to genome size variation within angiosperms? Ecol. Evol. 2021, 11, 2660–2668. [Google Scholar] [CrossRef]
- Zhang, Q.-J.; Gao, L.-I. Rapid and recent evolution of LTR retrotransposons drives rice genome evolution during the speciation of AA-genome Oryza species. G3 2017, 7, 1875–1885. [Google Scholar] [CrossRef]
- McCann, J.; Macas, J.; Novák, P.; Stuessy, T.F.; Villasenor, J.L.; Weiss-Schneweiss, H. Differential genome size and repetitive DNA evolution in diploid species of Melampodium sect Melampodium (Asteraceae). Front. Plant Sci. 2020, 11, 362. [Google Scholar] [CrossRef]
- Sáez-Vásquez, J.; Delseny, M. Ribosome Biogenesis in Plants: From Functional 45S Ribosomal DNA Organization to Ribosome Assembly Factors. Plant Cell. 2019, 31, 1945–1967. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Ganley, A.R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 2005, 309, 1581–1584. [Google Scholar] [CrossRef]
- Tsang, E.; Carr, A.M. Replication fork arrest, recombination and the maintenance of ribosomal DNA stability. DNA Repair. 2008, 7, 1613–1623. [Google Scholar] [CrossRef]
- Raskina, O.; Belyayev, A.; Nevo, E. Quantum speciation in Aegilops: Molecular cytogenetic evidence from rDNA cluster variability in natural populations. Proc. Natl. Acad. Sci. USA 2004, 101, 14818–14823. [Google Scholar] [CrossRef]
- Macas, J.; Novák, P.; Pellicer, J.; Čížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS ONE 2015, 10, e0143424. [Google Scholar] [CrossRef]
- Garrido-Ramos, M.A. Satellite DNA in plants: More than just rubbish. Cytogenet. Genome Res. 2015, 146, 153–170. [Google Scholar] [CrossRef]
- Plohl, M.; Meštrovic, N.; Mravinac, B. Satellite DNA evolution. In Repetitive DNA; Garrido-Ramos, M.A., Ed.; Karger: Granada, Spain, 2012; pp. 126–152. [Google Scholar]
- Ugarkovic, D. Functional elements residing within satellite DNAs. EMBO Rep. 2005, 6, 1035–1039. [Google Scholar] [CrossRef]
- Heslop-Harrison, J.S. Comparative genome organization in plants: From sequence and markers to chromatin and chromosomes. Plant Cell 2000, 12, 617–636. [Google Scholar] [CrossRef]
- Zang, Z.-T.; Yang, S.-Q.; Li, Z.-A.; Zhang, Y.-X.; Wang, Y.-Z.; Cheng, C.-Y.; Li, J.; Chen, J.-F.; Lou, Q.-F. Comparative chromosomal localization of 45S and 5S rDNAs and implications for genome evolution in Cucumis. Genome 2016, 59, 449–457. [Google Scholar] [CrossRef]
- Su, D.; Chen, L.; Sun, J.; Zhang, L.; Gao, R.; Li, Q.; Han, Y.; Li, Z. Comparative chromosomal localization of 45S and 5S rDNA sites in 76 purple-fleshed sweet potato cultivars. Plants 2020, 9, 865. [Google Scholar] [CrossRef]
- Pontes, O.; Neves, N.; Silva, M.; Lewis, M.S.; Madlung, A.; Comai, L.; Viegas, W.; Pikaard, C.S. Chromosomal locus rearrangements are a rapid response to formation of the allotetraploid Arabidopsis suecica genome. Proc. Natl. Acad. Sci. USA 2004, 101, 18240–18245. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa-Harand, A.; de Almeida, C.C.S.; Mosiolek, M.; Blair, M.W.; Schweizer, D.; Guerra, M. Extensive ribosomal DNA amplification during Andean common bean (Phaseolus vulgaris L.) evolution. Theor. Appl. Genet. 2006, 112, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Li, K.P.; Wu, Y.X.; Zhao, H.; Wang, Y.; Lu, X.M.; Wang, J.M.; Xu, Y.; Li, Z.Y.; Han, Y.H. Cytogenetic relationships among Citrullus species in comparison with some genera of the tribe Benincaseae (Cucurbitaceae) as inferred from rDNA distribution patterns. BMC Evol. Biol. 2016, 16, 85. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.; Kovařík, A.; Leitch, A.R.; Garnatje, T. Cytogenetic features of rRNA genes across land plants: Analysis of the plant rDNA database. Plant J. 2017, 89, 1020–1030. [Google Scholar] [CrossRef]
- Leitch, I.J.; Heslop-Harrison, J.S. Physical mapping of the 18S-5.8-26S rRNA genes in barley by in situ hybridization. Genome 1992, 35, 1013–1018. [Google Scholar] [CrossRef]
- Hanson, R.E.; Islam-Faridi, M.N.; Percival, E.A.; Crane, C.F.; Ji, Y.F.; McKnight, T.D.; Stelly, D.M.; Price, H.J. Distribution of 5S and 18S-28S rDNA loci in a tetraploid cotton (Gossypium hirsutum L.) and its putative diploid ancestors. Chromosoma 1996, 105, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Muravenko, O.V.; Yurkevich, O.Y.; Kalnyuk, J.V.; Samatadze, T.E.; Zoshchuk, S.A.; Amosova, A.V. Integration of repeatomic and cytogenetic data on satellite DNA for the genome analysis in the genus Salvia (Lamiaceae). Plants 2022, 11, 2244. [Google Scholar] [CrossRef]
- Yurkevich, O.Y.; Samatadze, T.E.; Zoshchuk, S.A.; Amosova, A.V.; Muravenko, O.V. Species of the Sections Hedysarum and Multicaulia of the Genus Hedysarum (Fabaceae): Taxonomy, Distribution, Chromosomes, Genomes, and Phylogeny. Int. J. Mol. Sci. 2024, 25, 8489. [Google Scholar] [CrossRef] [PubMed]
- Samatadze, T.E.; Yurkevich, O.Y.; Khazieva, F.M.; Basalaeva, I.V.; Savchenko, O.M.; Zoshchuk, S.A.; Morozov, A.I.; Amosova, A.V.; Muravenko, O.V. Genome studies in four species of Calendula L. (Asteraceae) using satellite DNAs as chromosome markers. Plants 2023, 12, 4056. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A galaxybased web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Robledillo, L.A.; Koblizkova, A.; Vrbova, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acid. Res. 2017, 45, e111. [Google Scholar] [CrossRef]
- Novák, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef]
- Novák, P.; Hoštáková, N.; Neumann, P.; Macas, J. DANTE and DANTE_LTR: Lineage-centric annotation pipelines for long terminal repeat retrotransposons in plant genomes. NAR Genom. Bioinform. 2024, 6, lqae113. [Google Scholar] [CrossRef]
- Untergasser, A.; Nijveen, H.; Rao, X.; Bisseling, T.; Geurts, R.; Leunissen, J.A.M. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007, 35, 71–74. [Google Scholar] [CrossRef]
- Gerlach, W.L.; Bedbrook, J.R. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res. 1979, 7, 1869–1885. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, W.L.; Dyer, T.A. Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res. 1980, 8, 4851–4855. [Google Scholar] [CrossRef] [PubMed]

| Variety | Leaf Color | Inflorescence Color | Stem Color | Vein Color | Seed Color | Usage |
|---|---|---|---|---|---|---|
| A. tricolor var. Valentina | purple | purple | purple | purple | black | leafy vegetables, grain crops |
| A. cruentus var. Dyuimovochka | green | green | green | green | black | leafy vegetables |
| A. paniculatus var. Fakel | green with red | purple | green with pink | pink | black | fodder and ornamental plants |
| A. hypochondriacus var. Kizlyarets | green | green with red | green | green | white | grain crops, fodder plants |
| A. hypochondriacus var. Krepysh | green | green with red | green | green | white | grain crops, fodder plants |
| Tandem Repeat/Genome Proportion [%] | Sequences of the Generated Oligonucleotide FISH Probes |
|---|---|
| CL9/0.55 | AmC9 CATTGTTCATTGATCATTGATCCTTGTTCATTGTTCATCGTT |
| CL70/0.1 | AmC70 AGGGTTTAGGGTTTAGGGTTT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amosova, A.V.; Yurkevich, O.Y.; Semenov, A.R.; Gins, M.S.; Kalnyuk, J.V.; Zemtsova, L.V.; Morozov, A.I.; Badaeva, E.D.; Zoshchuk, S.A.; Muravenko, O.V. Comparative Analysis of Chromosome Repeat DNA Patterns in Four Amaranthus Species. Int. J. Mol. Sci. 2025, 26, 11026. https://doi.org/10.3390/ijms262211026
Amosova AV, Yurkevich OY, Semenov AR, Gins MS, Kalnyuk JV, Zemtsova LV, Morozov AI, Badaeva ED, Zoshchuk SA, Muravenko OV. Comparative Analysis of Chromosome Repeat DNA Patterns in Four Amaranthus Species. International Journal of Molecular Sciences. 2025; 26(22):11026. https://doi.org/10.3390/ijms262211026
Chicago/Turabian StyleAmosova, Alexandra V., Olga Yu. Yurkevich, Alexey R. Semenov, Murat S. Gins, Julia V. Kalnyuk, Lyudmila V. Zemtsova, Alexander I. Morozov, Ekaterina D. Badaeva, Svyatoslav A. Zoshchuk, and Olga V. Muravenko. 2025. "Comparative Analysis of Chromosome Repeat DNA Patterns in Four Amaranthus Species" International Journal of Molecular Sciences 26, no. 22: 11026. https://doi.org/10.3390/ijms262211026
APA StyleAmosova, A. V., Yurkevich, O. Y., Semenov, A. R., Gins, M. S., Kalnyuk, J. V., Zemtsova, L. V., Morozov, A. I., Badaeva, E. D., Zoshchuk, S. A., & Muravenko, O. V. (2025). Comparative Analysis of Chromosome Repeat DNA Patterns in Four Amaranthus Species. International Journal of Molecular Sciences, 26(22), 11026. https://doi.org/10.3390/ijms262211026








