Molecular Interaction of Genes Related to Anthocyanin, Lipid and Wax Biosynthesis in Apple Red-Fleshed Fruits
Abstract
1. Introduction
2. Results
2.1. Significant Differences in Apple Genotypes with Regard to Total Anthocyanin Content
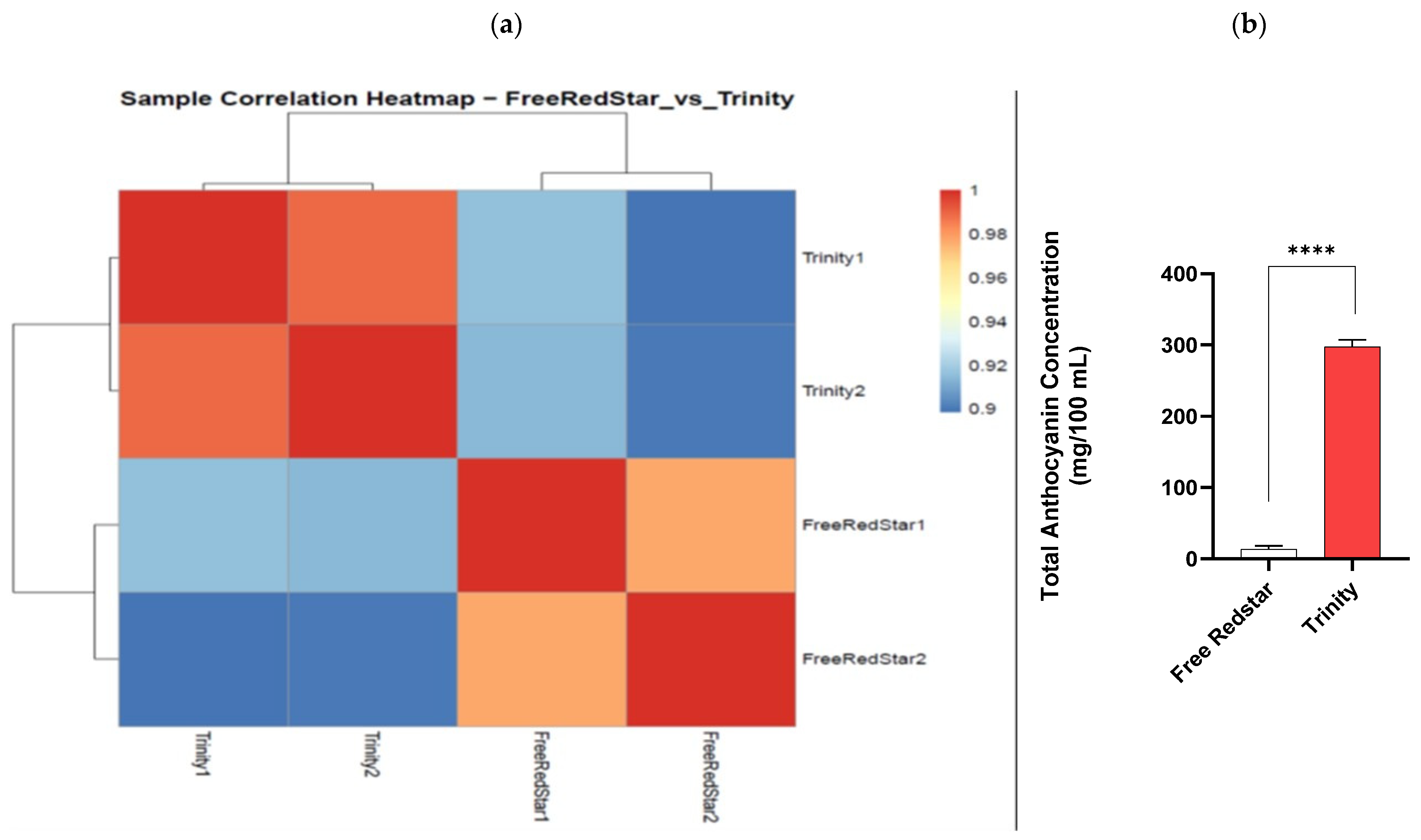
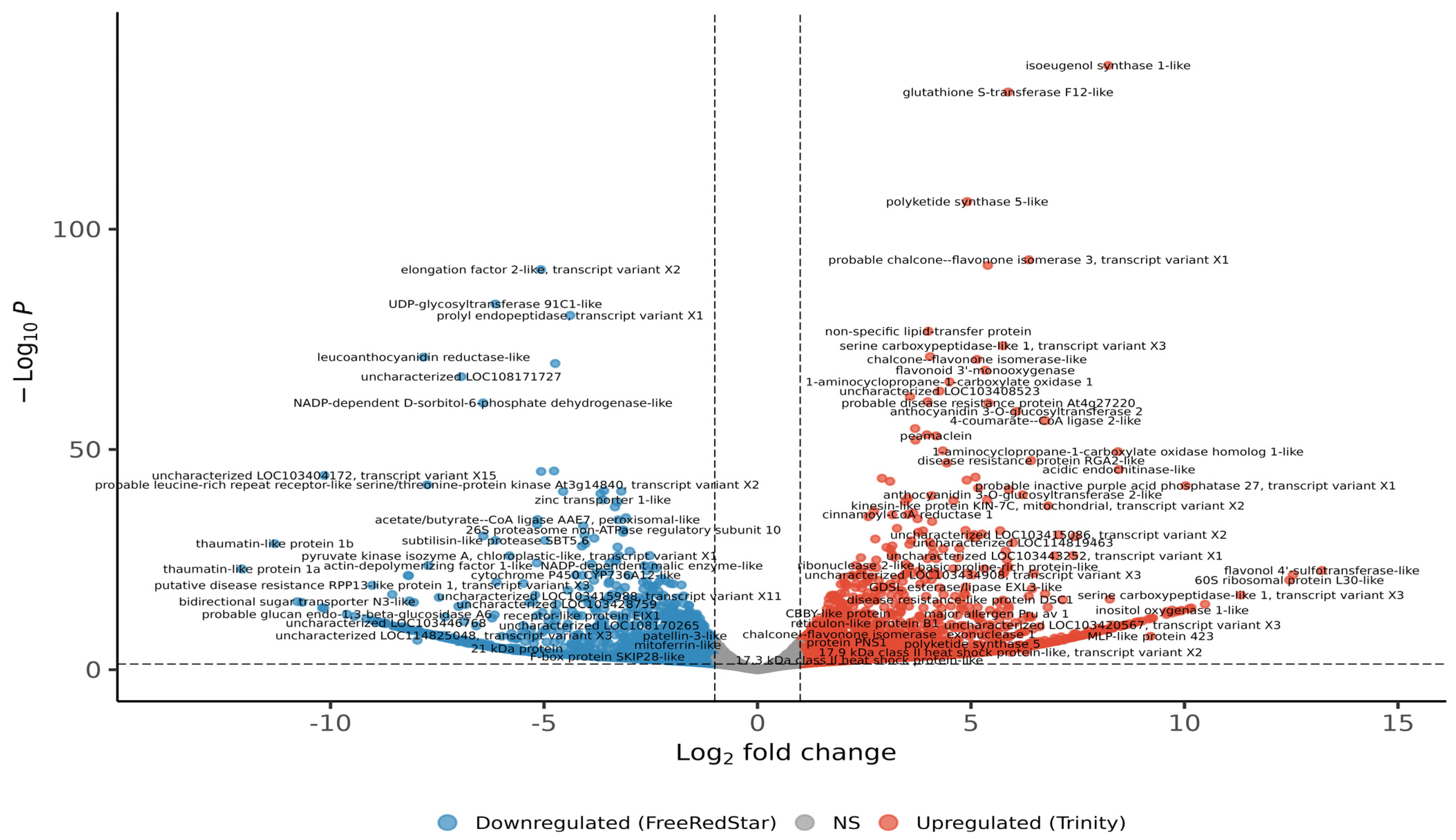
2.2. Comparative Analysis of Red and White Fruit Flesh Transcriptomes
2.3. Identification of Functional Categories of DEGs
2.3.1. Gene Ontology Enrichment of Differentially Expressed Genes (DEGs)
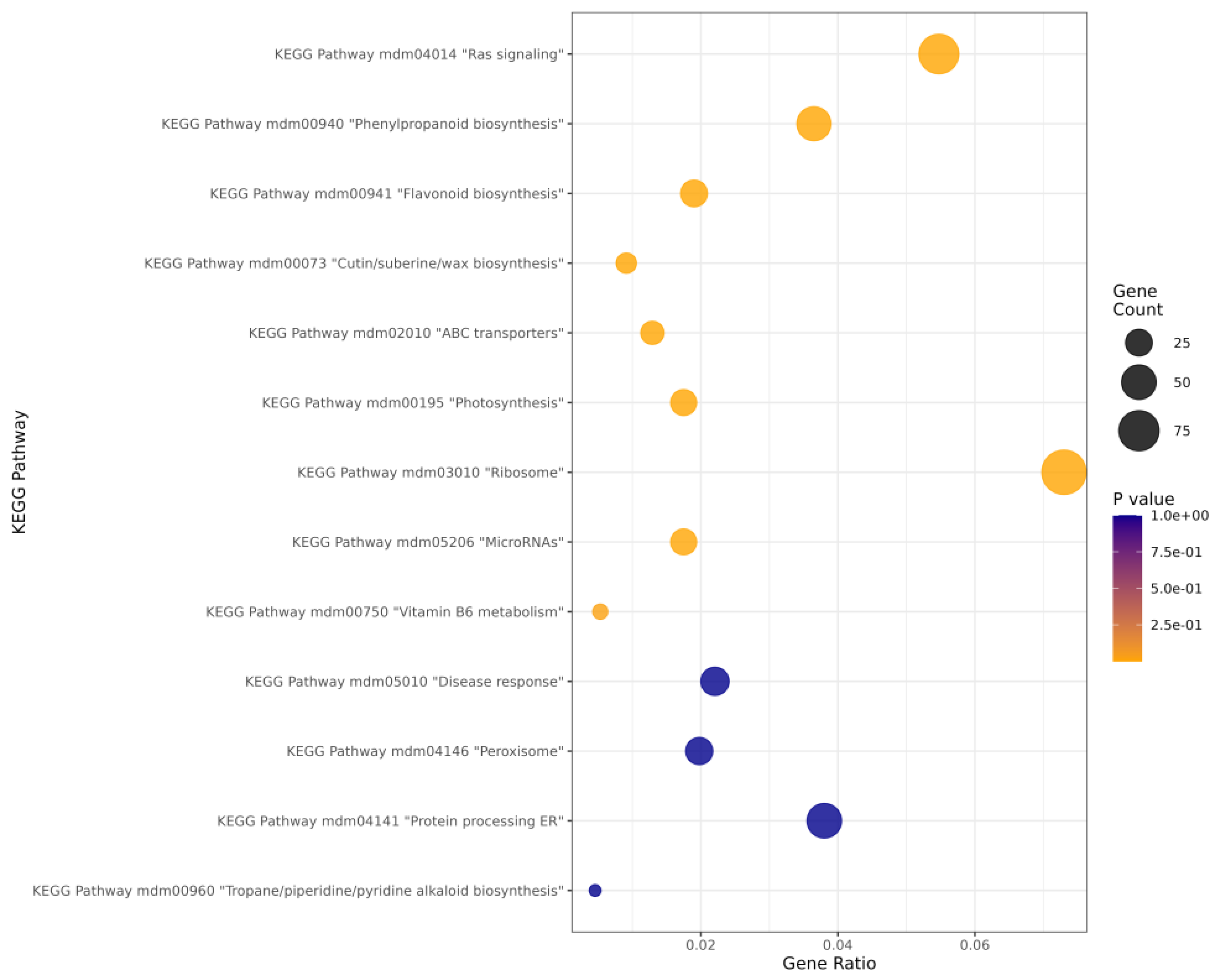
2.3.2. KEGG Enrichment Analysis
2.4. Activity of Selected Genes Depend on Anthocyanin Accumulation
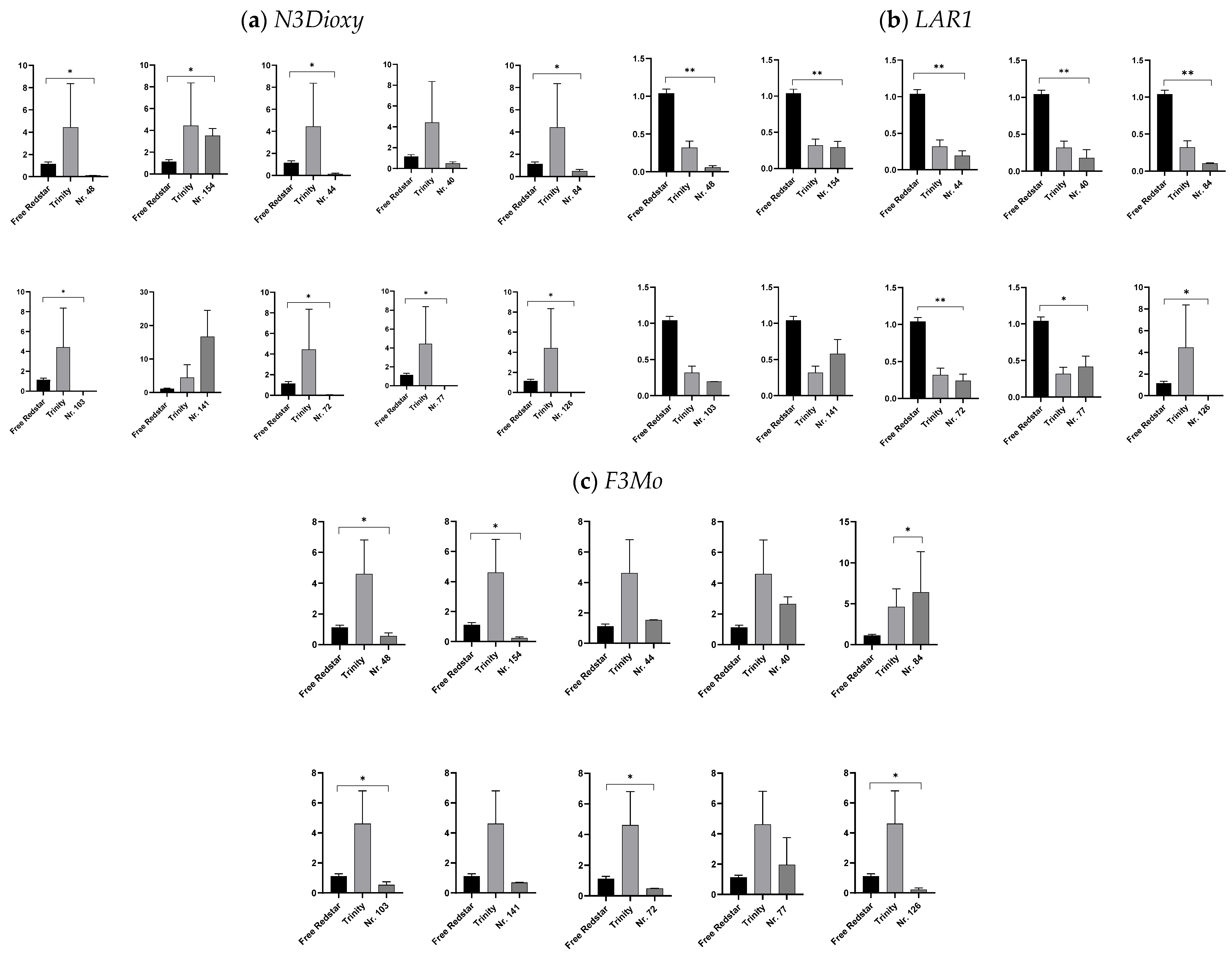
2.4.1. Expression Profiling of Genes Involved in Flavonoid Biosynthesis
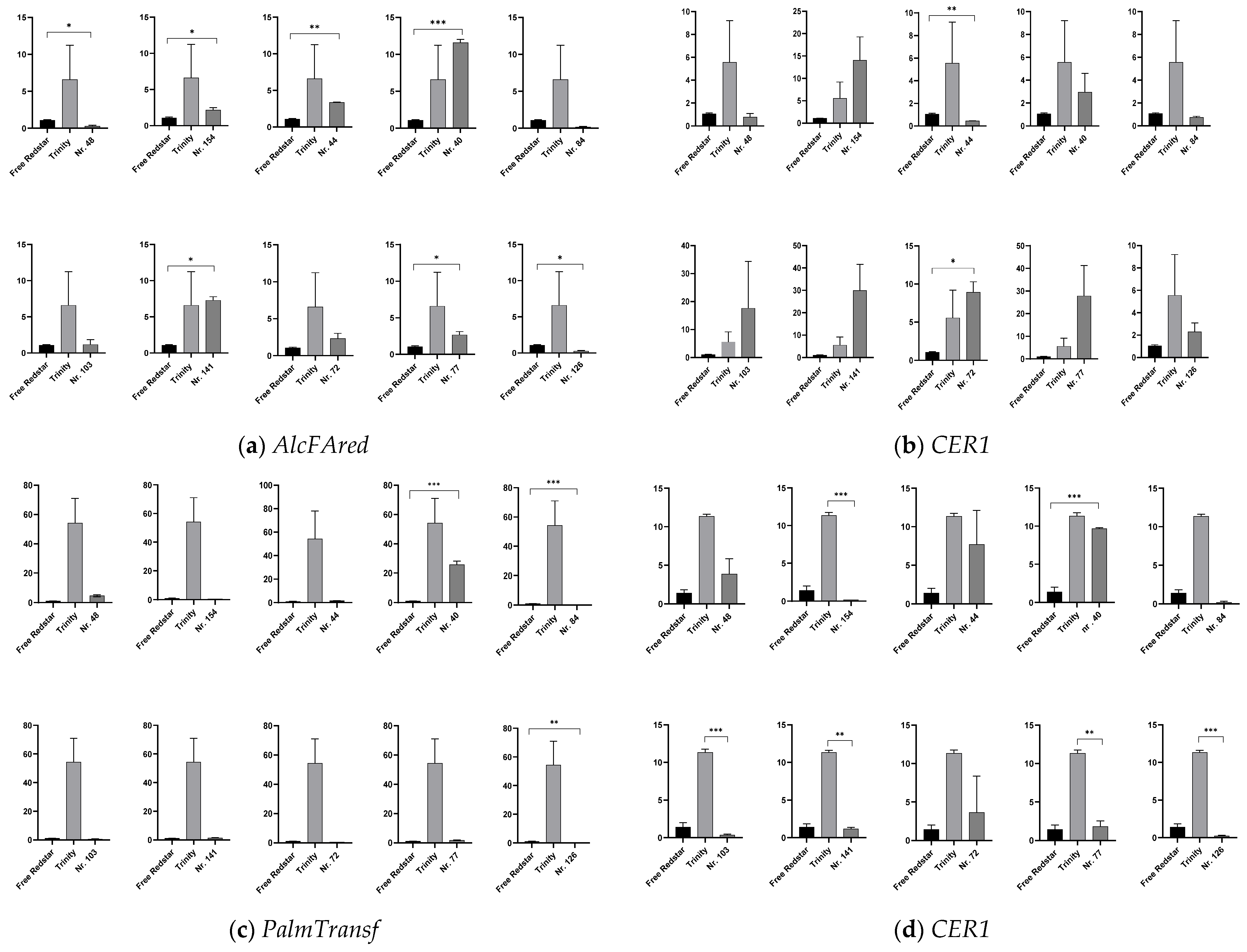
2.4.2. Expression Profiling of Genes Involved in Cutin and Wax Biosynthesis
2.4.3. Expression Profiling of Genes Involved in Tropane Piperidine and Pyridine Alkaloid Biosynthesis and Peroxisome
2.5. Validation of the Activity of Structural Genes from the Anthocyanin Biosynthesis Pathway in Accordance with Anthocyanin Accumulation
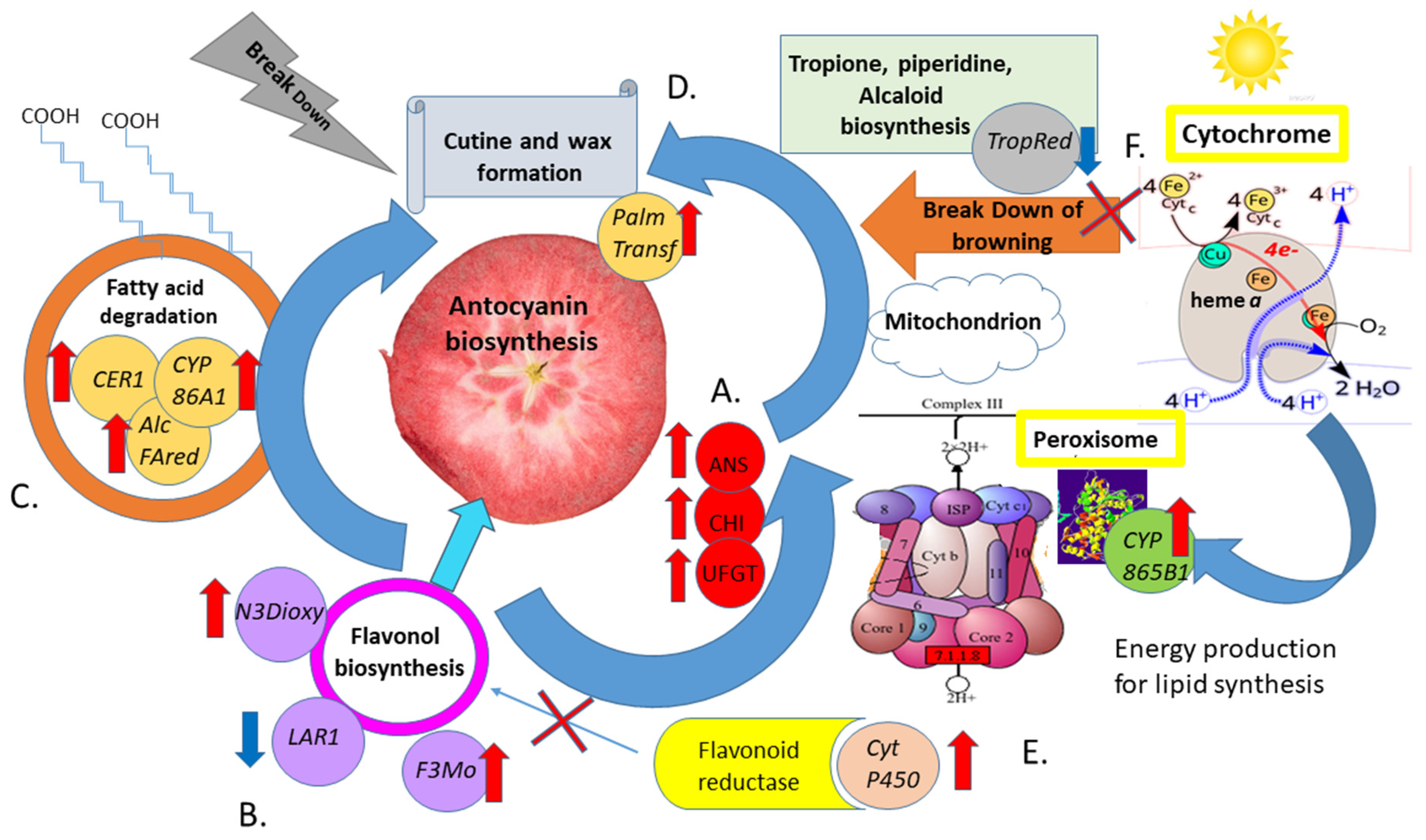
3. Discussion
3.1. Investigation of the Relationship of Uncovered Genes with Flavonoid and Pro-Anthocyanin Biosynthesis in Red-Fleshed Apple Fruits
3.2. Confirmation of Contribution of External Pathway Genes to Anthocyanin Biosynthesis
4. Materials and Methods
4.1. Plant Material
4.2. RNA Extraction
4.3. Transcriptome Sequencing and Differential Expression Analysis
4.4. Gene Annotation, GO and KEGG Enrichment Analyses
4.5. cDNA Synthesis and qPCR Tests
4.6. Anthocyanin Measurements and Fruit Phenotypical Assessment
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Available online: https://www.fao.org/faostat/en/#search/apple%20fruits (accessed on 3 November 2025).
- Keller-Przybylkowicz, S.; Oskiera, M.; Liu, X.; Song, L.; Zhao, L.; Du, X.; Kruczynska, D.; Walencik, A.; Kowara, N.; Bartoszewski, G. Transcriptome Analysis of White- and Red-Fleshed Apple Fruits Uncovered Novel Genes Related to the Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2024, 25, 1778. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Carlisle, C.M.; Blond, C.; Volz, R.K.; Whitworth, C.J.; Oraguzie, N.C.; Crowhurst, R.N.; Allan, A.C.; Espley, R.V.; Hellens, R.P.; et al. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genom. 2007, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Krieger, C.; Rassam, M.; Sullivan, M.; Fraser, J.; André, C.; Pindo, M.; Troggio, M.; Gardiner, S.E.; Henry, R.A.; et al. QTL and candidate gene mapping for polyphenolic composition in apple fruit. BMC Plant Biol. 2012, 12, 12. [Google Scholar] [CrossRef]
- Volz, R.K.; Oraguzie, N.; Whitworth, C.J.; How, N.; Chagne, D.; Carlisle, C.M.; Gardiner, S. Red flesh breeding in apple: Progress and challenges. Acta Hortic. 2009, 814, 304–309. [Google Scholar] [CrossRef]
- Available online: https://suttonelms.org.uk/apple43.html (accessed on 20 September 2025).
- Wang, N.; Xu, H.; Jiang, S.; Zhang, Z.; Lu, N.; Qiu, H.; Qu, C.; Wang, Y.; Wu, S.; Chen, X. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 2017, 90, 276–292. [Google Scholar] [CrossRef] [PubMed]
- Honda, C.; Moriya, S. Anthocyanin Biosynthesis in Apple Fruit. Hortic. J. 2018, 87, 305–314. [Google Scholar] [CrossRef]
- Yan, X.F.; Wang, Y.; Li, Y.M. Plant secondary metabolism and its response to environment. Acta Ecol. Sin. 2007, 6, 2554–2562. [Google Scholar]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red coloration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop Phenomics and High-Throughput Phenotyping: Past Decades, Current Challenges, and Future Perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, L.; Liu, W.; Zhang, J.; Wang, N.; Chen, X. Research progress of fruit color development in apple (Malus domestica Borkh.). Plant Physiol. Biochem. 2021, 162, 267–279. [Google Scholar] [CrossRef]
- Pinheiro, T.T.; Nishimura, D.S.; De Nadai, F.B.; Figueira, A.; Latado, R.R. Selection of reference genes for expression analyses of red-fleshed sweet orange (Citrus sinensis). Genet. Mol. Res. 2015, 14, 18440–18451. [Google Scholar] [CrossRef]
- Rumainum, I.; Worarad, K.; Yamaki, Y.; Yamane, K. Effects of Developmental Stages, Light, and an Auxin Polar Transport Inhibitor on the Skin and Flesh Pigmentation of Red-fleshed Peach Fruit. Hortic. J. 2016, 85, 141–147. [Google Scholar] [CrossRef]
- Brooks, R.M.; Olmo, H.P. The Brooks and Olmo Register of Fruit & Nut Varieties, 3rd ed.; ASHS Press: Alexandria, VA, USA, 1997. [Google Scholar]
- Sadilova, E.; Stintzing, F.C.; Carle, R. Anthocyanins, colour and antioxidant properties of eggplant (Solanum melongena L.) and violet pepper (Capsicum annuum L.) peel extracts. Naturforsch. C J. Biosci. 2006, 6, 527–535. [Google Scholar] [CrossRef]
- Sekido, K.; Hayashi, Y.; Yamada, K.; Shiratake, K.; Matsumoto, S.; Maejima, T.; Komatsu, H. Efficient Breeding System for Red-fleshed Apple Based on Linkage with S3-RNase Allele in ‘Pink Pearl’. HortScience 2010, 45, 534–537. [Google Scholar] [CrossRef]
- Wang, N.; Qu, C.; Jiang, S.; Chen, Z.; Xu, H.; Fang, H.; Su, M.; Zhang, J.; Wang, Y.; Liu, W.; et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018, 96, 39–55. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W.; Zhang, T.; Jiang, S.; Xu, H.; Wang, Y.; Zhang, Z.; Wang, C.; Chen, X. Transcriptomic Analysis of Red-Fleshed Apples Reveals the Novel Role of MdWRKY11 in Flavonoid and Anthocyanin Biosynthesis. J. Agric. Food Chem. 2018, 66, 7076–7086. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Jiang, S.; Zhang, Z.; Fang, H.; Xu, H.; Wang, Y.; Chen, X. Malus sieversii: The origin, flavonoid synthesis mechanism, and breeding of red-skinned and red-fleshed apples. Hortic. Res. 2018, 5, 70. [Google Scholar] [CrossRef]
- Wang, N.; Chen, X. Genetics and genomics of fruit color development in apple. In The Apple Genome; Korban, S.S., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 271–295. [Google Scholar]
- Duan, X.; Wang, K.; Tang, R.; Liu, J.; Cheng, K.; Gao, G.; Wang, Y.; Qin, G. Recent advances in biosynthesis and regulation of strawberry anthocyanins. Hortic. Res. 2025, 12, uhaf135. [Google Scholar] [CrossRef]
- Sun, P.; Yang, C.; Zhu, W.; Wu, J.; Lin, X.; Wang, Y.; Zhu, J.; Chen, C.; Zhou, K.; Qian, M.; et al. Metabolome, Plant Hormone, and Transcriptome Analyses Reveal the Mechanism of Spatial Accumulation Pattern of Anthocyanins in Peach Flesh. Foods 2023, 12, 2297. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, J.; Cherono, S.; An, J.P.; Allan, A.C.; Han, Y. Colorful hues: Insight into the mechanisms of anthocyanin pigmentation in fruit. Plant Physiol. 2023, 192, 1718–1732. [Google Scholar] [CrossRef] [PubMed]
- Heredia-Guerrero, J.A.; Domínguez, E. CHS silencing suggests a negative cross-talk between wax and flavonoid pathways in tomato fruit cuticle. Plant Signal. Behav. 2015, 10, 1019979. [Google Scholar] [CrossRef]
- Hu, Y.; Šmarda, P.; Liu, G.; Wang, B.; Gao, X.; Guo, Q. High-Depth Transcriptome Reveals Differences in Natural Haploid Ginkgo biloba L. Due to the Effect of Reduced Gene Dosage. Int. J. Mol. Sci. 2022, 23, 8958. [Google Scholar] [CrossRef]
- Honda, C.; Kotoda, N.; Wada, M.; Kondo, S.; Kobayashi, S.; Soejima, J.; Zhang, Z.; Tsuda, T.; Moriguchi, T. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biochem. 2002, 40, 955–962. [Google Scholar] [CrossRef]
- Bouillon, P.; Belin, E.; Fanciullino, A.L.; Balzergue, S.; Hanteville, S.; Letekoma, Y.; Cournol, M.; Faris, F.; Bouanich, A.; Bréard, D.; et al. Fade into you: Genetic control of pigmentation patterns in red-flesh apple (Malus domestica). Front. Plant Sci. 2025, 15, 1462545. [Google Scholar] [CrossRef]
- Zhao, D.; Tao, J.; Han, C.; Ge, J. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.). Mol. Biol. Rep. 2012, 39, 11263–11275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Li, B.; Wang, Z.; Hu, D.; Zhang, X.; Zhao, B.; Wang, J. Green biosynthesis of rare DHA-phospholipids by lipase-catalyzed transesterification with edible algal oil in solvent-free system and catalytic mechanism study. Front. Bioeng. Biotechnol. 2023, 11, 1158348. [Google Scholar] [CrossRef]
- Espley, R.V.; Bovy, A.; Bava, C.; Jaeger, S.R.; Tomes, S.; Norling, C.; Crawford, J.; Rowan, D.; McGhie, T.K.; Brendolise, C.; et al. Analysis of genetically modified red-fleshed apples reveals effects on growth and consumer attributes. Plant Biotechnol. J. 2013, 11, 408–419. [Google Scholar] [CrossRef]
- Chen, C.; Li, H.; Zhang, D.; Li, P.; Ma, F. The role of anthocyanin in photoprotection and its relationship with the xanthophyll cycle and the antioxidant system in apple peel depends on the light conditions. Physiol. Plant. 2013, 149, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, J.M.; Heine, G.F.; Irani, N.G.; Feller, A.; Kim, M.G.; Matulnik, T.; Chandler, V.L.; Grotewold, E. Different mechanisms participate in the R-dependent activity of the R2R3 MYB transcription factor C1. J. Biol. Chem. 2004, 279, 48205–48213. [Google Scholar] [CrossRef] [PubMed]
- Baudry, A.; Heim, M.A.; Dubreucq, B.; Caboche, M.; Weisshaar, B.; Lepiniec, L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004, 39, 366–380. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef]
- Davies, K.M.; Schwinn, K.E.; Deroles, S.C.; Manson, D.G.; Lewis, D.H.; Bloor, S.J.; Bradley, J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica 2003, 131, 259–268. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002, 5, 218–223. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Smeekens, S. Sugar-induced signal transduction in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 49–81. [Google Scholar] [CrossRef]
- Supriya, L.; Deepika, D.; Nyanthanglo, W.; Prodosh, G.; Kodetham, G.; Gudipalli, P.; Mehanathan, M. Sugar sensors in plants: Orchestrators of growth, stress tolerance, and hormonal crosstalk. J. Plant Physiol. 2025, 307, 154471. [Google Scholar] [CrossRef]
- Dooner, H.K.; Robbins, T.P.; Jorgensen, R.A. Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 1991, 25, 173–199. [Google Scholar] [CrossRef] [PubMed]
- Holton, T.A.; Cornish, E.C. Genetics and Biochemistry of Anthocyanin Biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T.; Jones, P. Glycosyltransferases in plant natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Lloyd, J.C.; Zakhleniuk, O.V. Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J. Exp. Bot. 2004, 55, 1221–1230. [Google Scholar] [CrossRef]
- Teng, S.; Keurentjes, J.; Bentsink, L.; Koornneef, M.; Smeekens, S. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 2005, 139, 1840–1852. [Google Scholar] [CrossRef]
- Solfanelli, C.; Poggi, A.; Loreti, E.; Alpi, A.; Perata, P. Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol. 2006, 140, 637–646. [Google Scholar] [CrossRef]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hülskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef]
- Sivitz, A.B.; Reinders, A.; Ward, J.M. Arabidopsis sucrose transporter AtSUC1 is important for pollen germination and sucrose-induced anthocyanin accumulation. Plant Physiol. 2008, 147, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.G.; Sun, C.H.; Zhang, Q.Y.; An, J.P.; You, C.X.; Hao, Y.J. Glucose Sensor MdHXK1 Phosphorylates and Stabilizes MdbHLH3 to Promote Anthocyanin Biosynthesis in Apple. PLoS Genet. 2016, 12, e1006273. [Google Scholar] [CrossRef] [PubMed]
- Mieszczakowska-Frąc, M.; Buczek, M.; Kruczyńska, D.; Markowski, J. Cloudy red-fleshed apple juice production and quality. Pol. J. Nat. Sci. 2015, 30, 59–72. [Google Scholar]
- Trivedi, P.; Nguyen, N.; Hykkerud, A.L.; Häggman, H.; Martinussen, I.; Jaakola Land Karppinen, K. Developmental and Environmental Regulation of Cuticular Wax Biosynthesis in Fleshy. Fruits Front. Plant Sci. 2019, 10, 431. [Google Scholar] [CrossRef]
- Arrieta-Baez, D.; Perea-Flores, M.J.; Méndez-Méndez, J.; Mendoza, H.; Gómez-Patiño, M. Structural Studies of the Cutin from Two Apple Varieties: Golden Delicious and Red Delicious (Malus domestica). Molecules 2020, 25, 5955. [Google Scholar] [CrossRef]
- Fatland, B.L.; Nikolau, B.J.; Wurtele, E.S. Reverse Genetic Characterization of Cytosolic Acetyl-CoA Generation by ATP-Citrate Lyase in Arabidopsis. Plant Cell 2005, 17, 182–203. [Google Scholar] [CrossRef]
- Schwender, J.; Shachar-Hill, Y.; Ohlrogge, J.B. Mitochondrial metabolism in developing embryos of Brassica napus. J. Biol. Chem. 2006, 281, 34040–34047. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef]
- Akram, M. Citric Acid Cycle and Role of its Intermediates in Metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef]
- Zhang, Y.; Fernie, A.R. On the role of the tricarboxylic acid cycle in plant productivity. J. Integr. Plant Biol. 2018, 60, 1199–1216. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, R.; Gu, H.; Zhang, E.; Qu, J.; Cao, W.; Huang, X.; Yan, H.; He, J.; Cai, Z. Metabolic reprogramming in macrophage responses. Biomark. Res. 2021, 9, 1. [Google Scholar] [CrossRef]
- Proels, R.K.; Hückelhoven, R. Cell-wall invertases, key enzymes in the modulation of plant metabolism during defence responses. Mol. Plant Pathol. 2014, 15, 858–864. [Google Scholar] [CrossRef]
- Vallarino, J.G.; Yeats, T.H.; Maximova, E.; Rose, J.K.; Fernie, A.R.; Osorio, S. Postharvest changes in LIN5-down-regulated plants suggest a role for sugar deficiency in cuticle metabolism during ripening. Phytochemistry 2017, 142, 11–20. [Google Scholar] [CrossRef]
- Min, D.; Li, F.; Wang, J.; Fu, X.; Ali, M.; Song, Y.; Ding, J.; Li, X.; Li, M.; Yang, K.; et al. Transcriptome reveals insights into the regulatory mechanism of cuticular wax synthesis in developing apple fruit. Sci. Hortic. 2024, 328, 112891. [Google Scholar] [CrossRef]
- Zhao, Y.-W.; Wang, C.-K.; Huang, X.-Y.; Hu, D.-G. Genome-Wide Analysis of the Glutathione S-Transferase (GST) Genes and Functional Identification of MdGSTU12 Reveals the Involvement in the Regulation of Anthocyanin Accumulation in Apple. Genes 2021, 12, 1733. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.H.; Cao, H.H.; Pan, L.; Niu, L.; Wei, B.; Cui, G.C.; Wang, L.W.; Yao, J.L.; Zeng, W.F.; Wang, Z.Q. Two loss-of-function alleles of the glutathione S-transferase (GST) gene cause anthocyanin deficiency in flower and fruit skin of peach (Prunus persica). Plant J. 2021, 107, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Huang, X.Y.; Zhao, Y.W.; Wang, C.K.; Sun, Q.; Hu, D.G. Role of an ATP-binding cassette (ABC) transporter MdABCI17 in the anthocyanin accumulation of apple. Sci. Hortic. 2024, 323, 112502. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Ayaz, A.; Zhao, H.; Lü, S. The Plant Fatty Acyl Reductases. Int. J. Mol. Sci. 2022, 23, 16156. [Google Scholar] [CrossRef]
- Buhrman, K.; Aravena-Calvo, J.; Ross Zaulich, C.; Hinz, K.; Laursen, T. Anthocyanic Vacuolar Inclusions: From Biosynthesis to Storage and Possible Applications. Front. Chem. 2022, 10, 913324. [Google Scholar] [CrossRef]
- Kondo, S.; Hiraoka, K.; Kobayashi, S.; Honda, C.; Terahara, N. Changes in the expression of anthocyanin biosynthetic genes during apple development. J. Am. Soc. Hortic. Sci. 2002, 127, 971–997. [Google Scholar] [CrossRef]
- Bogs, J.; Ebadi, A.; McDavid, D.; Robinson, S.P. Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 2006, 140, 279–291. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic expression of apple F3′H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef]
- Huo, W.; Liu, S.; Chen, X.; Gu, T.; Wang, Z.; Xu, X.; Liu, D.; Zhang, Y.; Jiang, S. Combined analysis of lncRNAs and mRNAs associated with coloration and wax formation during ‘Fumei’ Apple development. BMC Plant Biol. 2025, 25, 498. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, S.; Lee, E.; Walker, A.R.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003, 35, 624–636. [Google Scholar] [CrossRef]
- Tanner, G.J.; Francki, K.T.; Abrahams, S.; Watson, J.M.; Larkin, P.J.; Ashton, A. Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol. Chem. 2003, 278, 31647–33156. [Google Scholar] [CrossRef]
- Liao, L.; Vimolmangkang, S.; Wei, G.; Zhou, H.; Korban, S.S.; Han, Y. Molecular characterization of genes encoding leucoanthocyanidin reductase involved in proanthocyanidin biosynthesis in apple. Front. Plant Sci. 2015, 6, 243. [Google Scholar] [CrossRef]
- Li, H.; Tian, J.; Yao, Y.Y.; Zhang, J.; Song, T.T.; Li, K.T.; Yao, Y.C. Identification of leucoanthocyanidin reductase and anthocyanidin reductase genes involved in proanthocyanidin biosynthesis in Malus crabapple plants. Plant Physiol. Biochem. 2019, 139, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Samuels, L.; Kunst, L.; Jetter, R. Sealing plant surfaces: Cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 2008, 59, 683–707. [Google Scholar] [CrossRef]
- Wu, W.; Jiang, B.; Liu, R.; Han, Y.; Fang, X.; Mu, H.; Farag, M.A.; Simal-Gandara, J.; Prieto, M.A.; Chen, H.; et al. Structures and Functions of Cuticular Wax in Postharvest Fruit and Its Regulation: A Comprehensive Review with Future Perspectives. Engineering 2023, 23, 118–129. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, T.; Zhu, S.; Wang, D.; Sun, S.; Xin, L. Short-term hypobaric treatment alleviates chilling injury by regulating membrane fatty acids metabolism in peach fruit. J. Food Biochem. 2022, 46, 14113. [Google Scholar] [CrossRef]
- Höfer, R.; Briesen, I.; Beck, M.; Pinot, F.; Schreiber, L.; Franke, R. The Arabidopsis cytochrome P450 CYP86A1 encodes a fatty acid omega-hydroxylase involved in suberin monomer biosynthesis. J. Exp. Bot. 2008, 59, 2347–2360. [Google Scholar] [CrossRef]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef]
- Pascal, S.; Bernard, A.; Sorel, M.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Domergue, F.; Joubès, J. The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J. 2013, 73, 733–746. [Google Scholar] [CrossRef]
- Leide, J.; Hildebrandt, U.; Reussing, K.; Riederer, M.; Vogg, G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: Effects of a deficiency in a beta-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol. 2007, 144, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Alkio, M.; Jonas, U.; Sprink, T.; van Nocker, S.; Knoche, M. Identification of putative candidate genes involved in cuticle formation in Prunus avium (sweet cherry) fruit. Ann. Bot. 2012, 110, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Hao, H.; Liu, R.; Ma, Q.; Xu, J.; Chen, F.; Cheng, Y.; Deng, X. Comparative analysis of surface wax in mature fruits between Satsuma mandarin (Citrus unshiu) and ‘Newhall’ navel orange (Citrus sinensis) from the perspective of crystal morphology, chemical composition and key gene expression. Food Chem. 2014, 153, 177–185. [Google Scholar] [CrossRef]
- Rowland, O.; Domergue, F. Plant fatty acyl reductases: Enzymes generating fatty alcohols for protective layers with potential for industrial applications. Plant Sci. 2012, 193, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Li, C.; Xu, F.; Li, Y.; Shi, X.; Wang, Y.; Wang, Z. Three endoplasmic reticulum-associated fatty acyl-coenzyme a reductases were involved in the production of primary alcohols in hexaploid wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 18, 41. [Google Scholar] [CrossRef]
- Liu, H.; Liu, Z.; Wu, Y.; Zheng, L.; Zhang, G. Regulatory Mechanisms of Anthocyanin Biosynthesis in Apple and Pear. Int. J. Mol. Sci. 2021, 22, 8441. [Google Scholar] [CrossRef]
- Deng, Y.; Lu, S. Biosynthesis and Regulation of Phenylpropanoids in Plants. Crit. Rev. Plant Sci. 2017, 36, 257–290. [Google Scholar] [CrossRef]
- Tang, C.; Yang, M.; Fang, Y.; Luo, Y.; Gao, S.; Xiao, X.; An, Z.; Zhou, B.; Zhang, B.; Tan, X.; et al. The rubber tree genome reveals new insights into rubber production and species adaptation. Nat. Plants 2016, 2, 16073. [Google Scholar] [CrossRef]
- Schuler, M.A.; Werck-Reichhart, D. Functional genomics of P450s. Annu. Rev. Plant Biol. 2003, 54, 629–667. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, P.; Biswas, A.; Dey, S.; Bhattacharjee, T.; Chakrabarty, S. Cytochrome P450 Gene Families: Role in Plant Secondary Metabolites Production and Plant Defense. J. Xenobiot. 2023, 13, 402–423. [Google Scholar] [CrossRef]
- Ayabe, S.; Akashi, T. Cytochrome P450s in flavonoid metabolism. Phytochem. Rev. 2006, 5, 271–282. [Google Scholar] [CrossRef]
- Hu, X.; Liu, W.; Yan, Y.; Deng, H.; Cai, Y. Tropinone reductase: A comprehensive review on its role as the key enzyme in tropane alkaloids biosynthesis. Int. J. Biol. Macromol. 2023, 253, 127377. [Google Scholar] [CrossRef] [PubMed]
- Bedewitz, M.A.; Jones, A.D.; D’Auria, J.C.; Barry, C.S. Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization. Nat. Commun. 2018, 9, 5281. [Google Scholar] [CrossRef] [PubMed]
- Dräger, B. Tropinone reductases, enzymes at the branch point of tropane alkaloid metabolism. Phytochemistry 2006, 67, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)). Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef]
- Taghavi, T.; Patel, H.; Rafie, R. Comparing pH differential and methanol-based methods for anthocyanin assessments of strawberries. Food Sci. Nutr. 2021, 10, 2123–2131. [Google Scholar] [CrossRef] [PubMed]
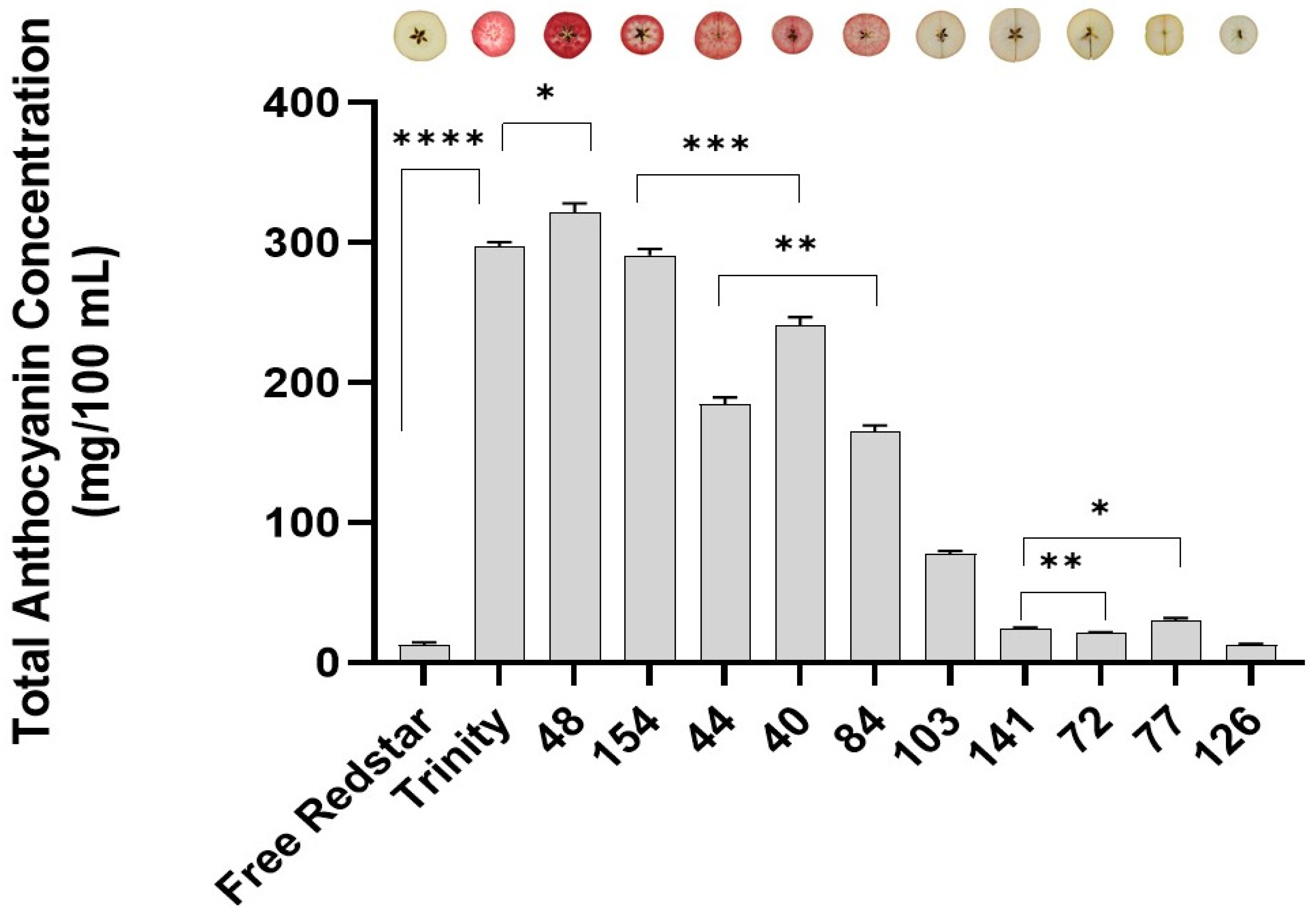

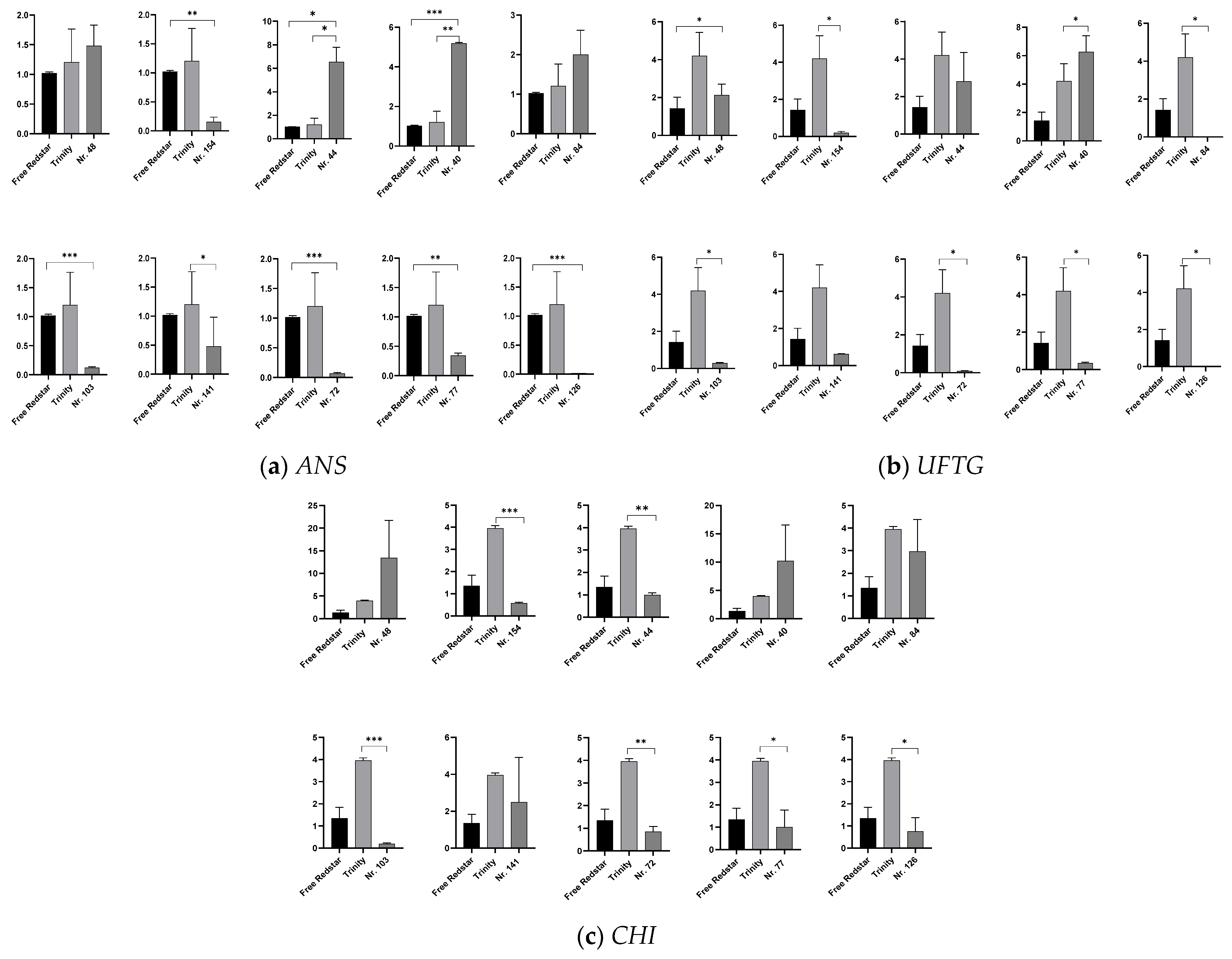
| Cultivar | Average Anthocyanin Concentration | SE Difference |
|---|---|---|
| Free Redstar | 9.482 | - |
| Trinity | 297.472 | 3.078 |
| 48 | 321.403 | 6.681 |
| 154 | 290.723 | 4.779 |
| 44 | 184.556 | 5070 |
| 40 | 241.094 | 5.647 |
| 84 | 164.864 | 4643 |
| 103 | 77.586 | 2197 |
| 141 | 24.403 | 0.9361 |
| 72 | 20.797 | 1173 |
| 77 | 29.968 | 1.801 |
| 126 | 12.489 | 1.081 |
| Free Redstar | Trinity | 48 | 154 | 44 | 40 | 84 | 103 | 141 | 72 | 77 | 126 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Free Redstar | **** | **** | **** | **** | **** | **** | **** | **** | **** | **** | * | |
| Trinity | * | ns | **** | **** | **** | **** | **** | **** | **** | **** | ||
| 48 | *** | **** | **** | **** | **** | **** | **** | **** | **** | |||
| 154 | **** | *** | **** | **** | **** | **** | **** | **** | ||||
| 44 | **** | ** | **** | **** | **** | **** | **** | |||||
| 40 | **** | **** | **** | **** | **** | **** | ||||||
| 84 | **** | **** | **** | **** | **** | |||||||
| 103 | **** | **** | **** | **** | ||||||||
| 141 | ** | * | **** | |||||||||
| 72 | ** | *** | ||||||||||
| 77 | **** |
| Gen ID | Gene Acronym Used in This Study | Gene Function | Oligo Sequence | |
|---|---|---|---|---|
| 3′ | 5′ | |||
| LOC103400025 | N3Dioxy | naringenin 3-dioxygenase | ggcttcatcgtgtccagtca | gcctgctgctgtttgagttc |
| LOC103402727 | LAR1 | leucoanthocyanidin reductase | gatgtggacagggctgatcc | agccatcgaagcactcatcc |
| LOC103403397 | CYP865B1 | cytochrome P450 86B1, flavoprotein reductase activity | tagcagcctcttttgcgtca | atccgcaaactcgtccactt |
| LOC103422716 | Cyt450 | cytochrome P450 oxydase activity | tagtggaggaattggcaggg | tggctctccaggacgtctta |
| LOC103428452 | CER1 | very-long-chain aldehyde decarbonylase CER1-like | gacacttacctggggctacg | catctggcgattcctcctcc |
| LOC103437875 | F3Mo | flavonoid 3′-monooxygenase | gttccccatcactctctggc | tcgaacctcttgtgcagctt |
| LOC103445140 | AlcFAred | alcohol-forming fatty acyl-CoA reductase | agttatcatccgcccatccg | tgtacagctctaccatgcgc |
| LOC103418919 | CYP86A4 | cytochrome P450, fatty acid hydrolase activity | tcaagttactcaggccgctg | gagcaaccatcactcaccca |
| LOC103423436 | TropRed | tropinone reductase | cctaaccctattcggccacc | ggagtacgctagaaaccgct |
| LOC103404168 | PalmTransf | omega-hydroxypalmitate O-feruloyl transferase | cctcgaccaaaacattgcgg | atggaggagctgtcaatggc |
| Structural genes Kondo et al. [67] | ANS | anthocyanin synthase | caatttggcctcaaacacct | gagcttcaacaccaagtgct |
| UFGT | UDP:flavonoid 3-O-glycosyltransferase | tccctttcactagccatgcaag | gtggaggatggagtttttacc | |
| CHI | chalcone isomerase | attatctctgctgggtca | gggaggagatggtcgaagga | |
| Ref. [95] | ACTIN | actin protein | gactgtgaaactgcgaatggctca | catgaatcatcagagcaacgggca |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keller-Przybyłkowicz, S.E.; Oskiera, M.; Walencik, A.; Lewandowski, M. Molecular Interaction of Genes Related to Anthocyanin, Lipid and Wax Biosynthesis in Apple Red-Fleshed Fruits. Int. J. Mol. Sci. 2025, 26, 10987. https://doi.org/10.3390/ijms262210987
Keller-Przybyłkowicz SE, Oskiera M, Walencik A, Lewandowski M. Molecular Interaction of Genes Related to Anthocyanin, Lipid and Wax Biosynthesis in Apple Red-Fleshed Fruits. International Journal of Molecular Sciences. 2025; 26(22):10987. https://doi.org/10.3390/ijms262210987
Chicago/Turabian StyleKeller-Przybyłkowicz, Sylwia Elżbieta, Michał Oskiera, Agnieszka Walencik, and Mariusz Lewandowski. 2025. "Molecular Interaction of Genes Related to Anthocyanin, Lipid and Wax Biosynthesis in Apple Red-Fleshed Fruits" International Journal of Molecular Sciences 26, no. 22: 10987. https://doi.org/10.3390/ijms262210987
APA StyleKeller-Przybyłkowicz, S. E., Oskiera, M., Walencik, A., & Lewandowski, M. (2025). Molecular Interaction of Genes Related to Anthocyanin, Lipid and Wax Biosynthesis in Apple Red-Fleshed Fruits. International Journal of Molecular Sciences, 26(22), 10987. https://doi.org/10.3390/ijms262210987





