Genome-Wide Identification of the LsaPHR1 Gene Family and Preliminary Functional Validation of LsaPHR1.1 in Phosphorus Tolerance in Lactuca sativa
Abstract
1. Introduction
2. Results
2.1. Identification of the PHR1 Genes in the Lettuce Genome
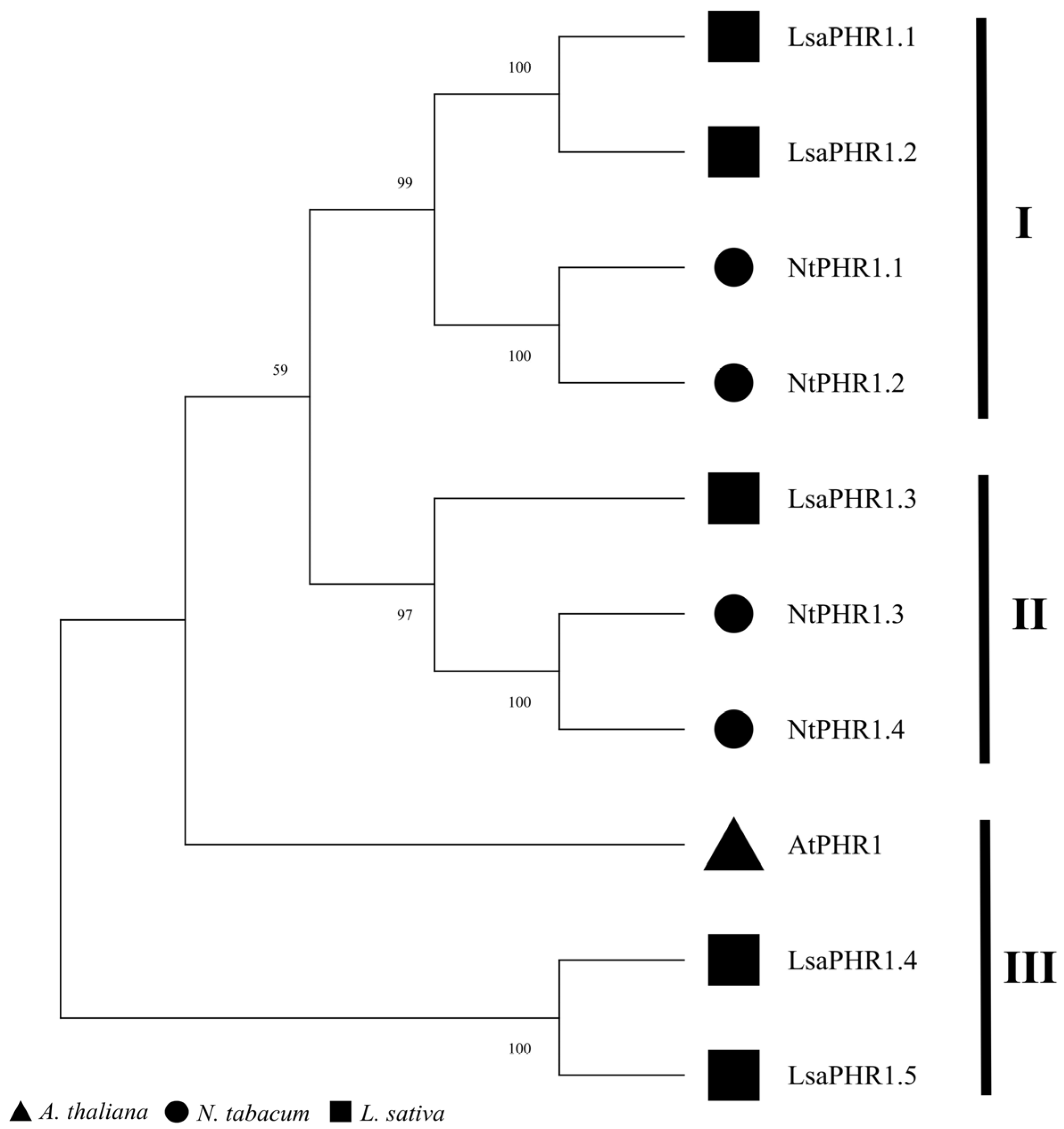
2.2. Phylogenetic Analysis of the LsaPHR1 Gene Family
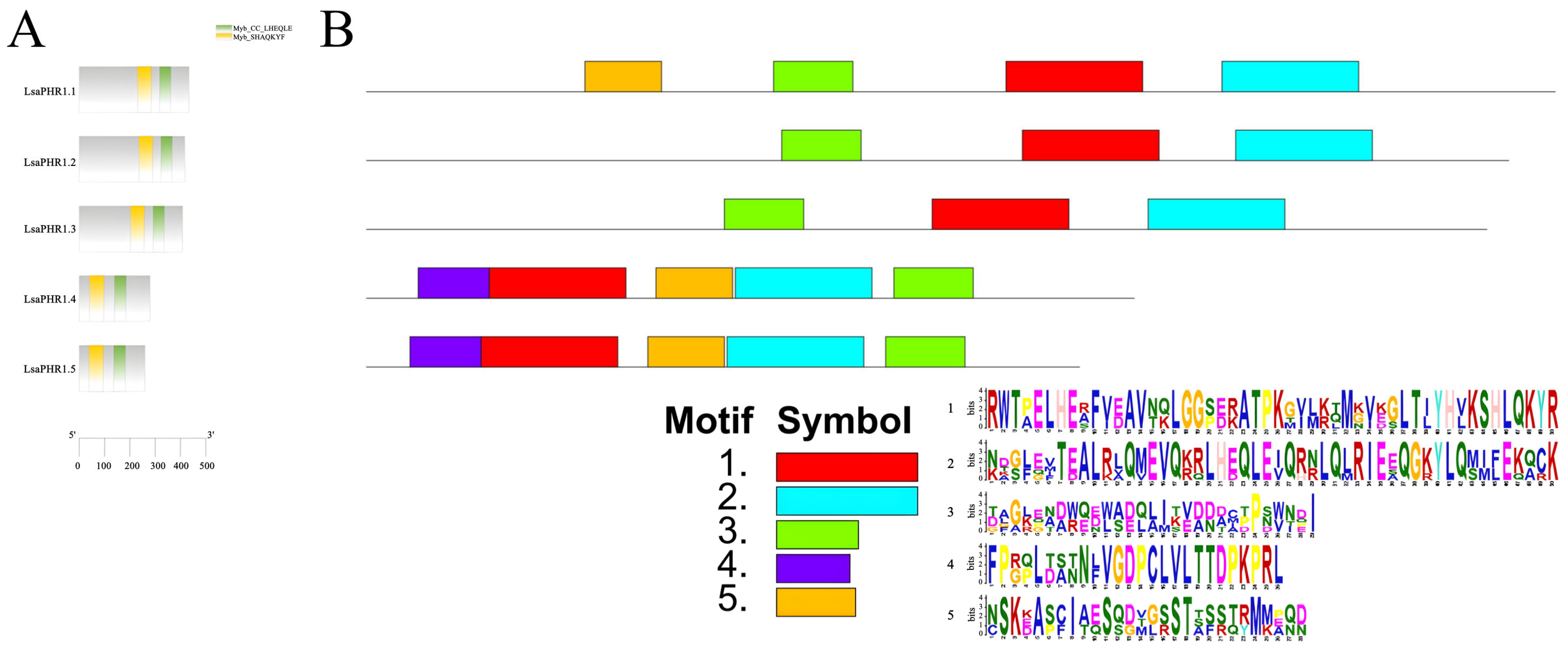
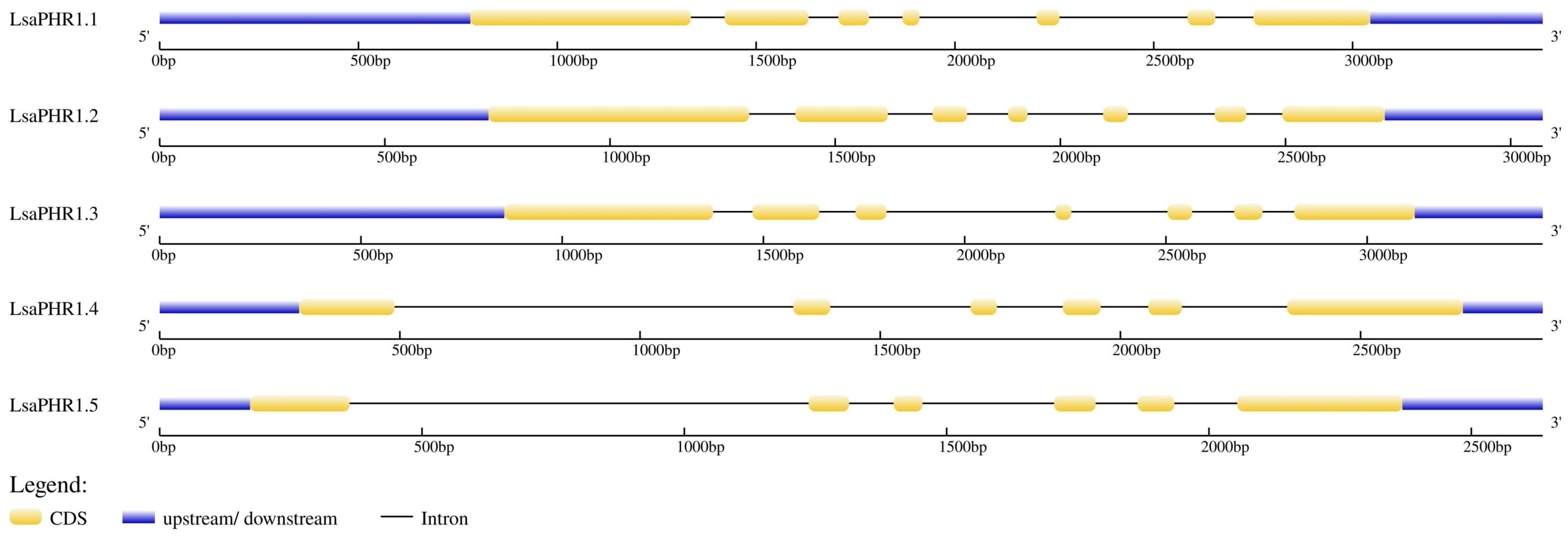
2.3. Conservation and Structural Analysis of LsaPHR1s
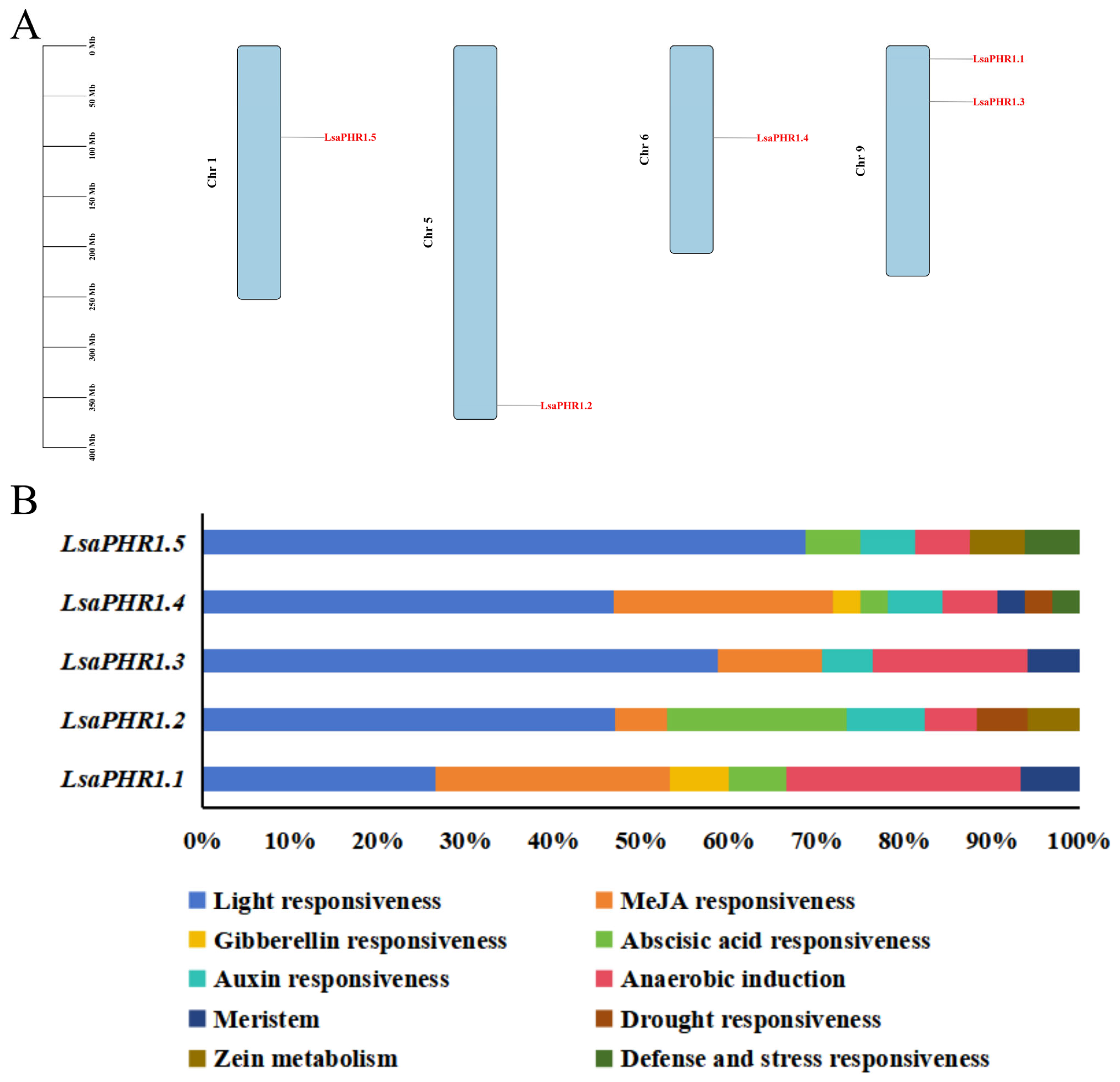
2.4. Chromosomal Distribution and Cis-Acting Elements of LsaPHR1s
2.5. Expression Patterns of LsaPHR1s Under Phosphate, Phytohormone, and Dark Treatments
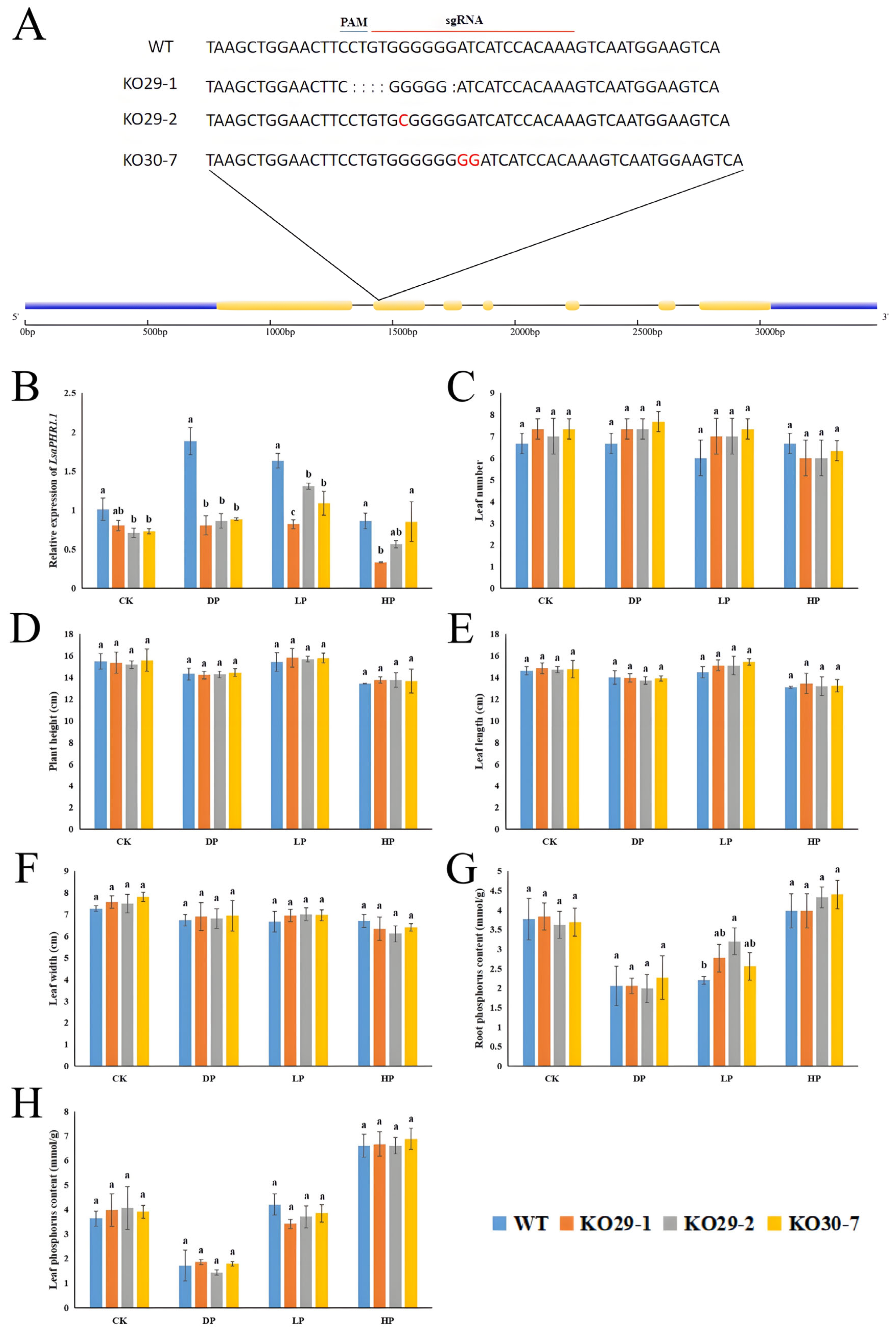
2.6. Editing LsaPHR1.1 Has Little Effect on Lettuce Morphology and Phosphorus Content
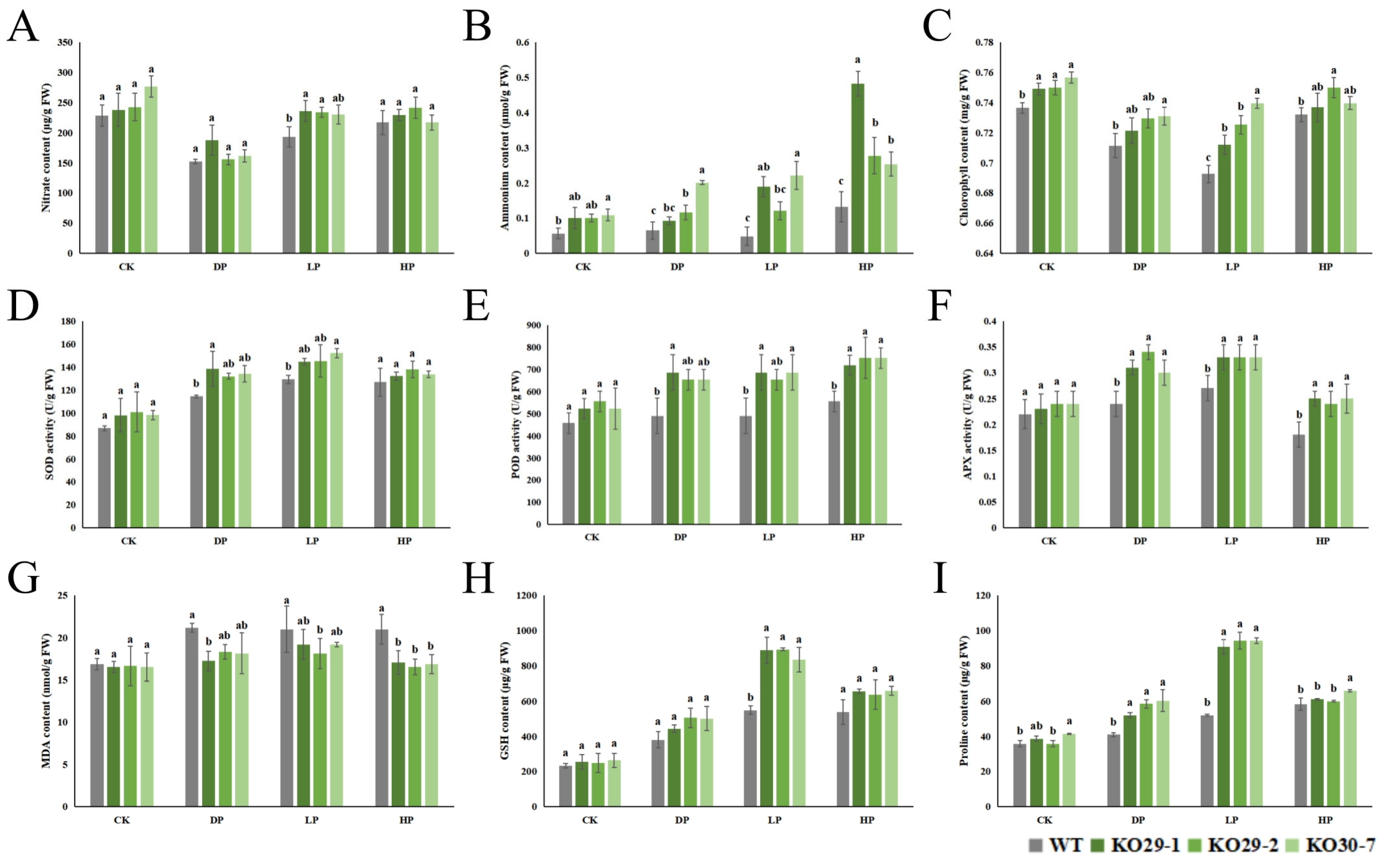
2.7. Lsaphr1.1 Improves Nitrogen Level, Antioxidant Capacity, and Photosynthesis, Improving Phosphorus Stress Tolerance in Lettuce
3. Discussion
4. Methods and Materials
4.1. Plant Materials
4.2. Identification and Characterization of the LsaPHR1 Gene Family
4.3. Multiple Sequence Alignment and Phylogenetic Tree Construction
4.4. Conservation and Structural Analysis of LsaPHR1
4.5. Chromosomal Location and Cis-Acting Element Analysis
4.6. Expression Analysis by qRT-PCR
4.7. The Genetic Transformation of LsaPHR1.1 and the Experimental Treatments of Their Offsprings
4.8. Phenotypic Analysis and Determination of Physiological Indices
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krouk, G.; Kiba, T. Nitrogen and Phosphorus interactions in plants: From agronomic to physiological and molecular insights. Curr. Opin. Plant Biol. 2020, 57, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Wu, W. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 2021, 63, 34–52. [Google Scholar] [CrossRef] [PubMed]
- Holford, I.C.R. Soil phosphorus: Its measurement, and its uptake by plants. Soil Res. 1997, 35, 227. [Google Scholar] [CrossRef]
- Subhashini, D.V. Effect of NPK Fertilizers and Co-inoculation with Phosphate-Solubilizing Arbuscular Mycorrhizal Fungus and Potassium-Mobilizing Bacteria on Growth, Yield, Nutrient Acquisition, and Quality of Tobacco (Nicotiana tabacum L.). Commun. Soil Sci. Plant Anal. 2016, 47, 328–337. [Google Scholar] [CrossRef]
- Guo, M.; Ruan, W.; Li, C.; Huang, F.; Zeng, M.; Liu, Y.; Yu, Y.; Ding, X.; Wu, Y.; Wu, Z.; et al. Integrative Comparison of the Role of the PHOSPHATE RESPONSE1 Subfamily in Phosphate Signaling and Homeostasis in Rice. Plant Physiol. 2015, 168, 1762–1776. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.; Jiang, H.; Jiang, C.; Du, Y.; Gong, C.; Wang, W.; Zhu, S.; Han, G.; Cheng, B. Systematic Identification, Evolution and Expression Analysis of the Zea mays PHT1 Gene Family Reveals Several New Members Involved in Root Colonization by Arbuscular Mycorrhizal Fungi. Int. J. Mol. Sci. 2016, 17, 930. [Google Scholar] [CrossRef]
- Wu, P.; Shou, H.; Xu, G.; Lian, X. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 2013, 16, 205–212. [Google Scholar] [CrossRef]
- Martin, A.C.; del Pozo, J.C.; Iglesias, J.; Rubio, V.; Solano, R.; de la Pena, A.; Leyva, A.; Paz-Ares, J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000, 24, 559–567. [Google Scholar] [CrossRef]
- Li, R.; An, J.-P.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. Overexpression of MdPHR1 Enhanced Tolerance to Phosphorus Deficiency by Increasing MdPAP10 Transcription in Apple (Malus × Domestica). J. Plant Growth Regul. 2020, 40, 1753–1763. [Google Scholar] [CrossRef]
- Chen, N.; Tong, S.; Yang, J.; Qin, J.; Wang, W.; Chen, K.; Shi, W.; Li, J.; Liu, J.; Jiang, Y. PtoWRKY40 interacts with PtoPHR1-LIKE3 while regulating the phosphate starvation response in poplar. Plant Physiol. 2022, 190, 2688–2705. [Google Scholar] [CrossRef]
- Zhu, J.; Lau, K.; Puschmann, R.; Harmel, R.K.; Zhang, Y.; Pries, V.; Gaugler, P.; Broger, L.; Dutta, A.K.; Jessen, H.J.; et al. Two bifunctional inositol pyrophosphate kinases/phosphatases control plant phosphate homeostasis. eLife 2019, 8, e43582. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Hu, Q.; Xiao, X.; Yao, D.; Ge, S.; Ye, J.; Li, H.; Cai, R.; Liu, R.; Meng, F.; et al. Mechanism of phosphate sensing and signaling revealed by rice SPX1-PHR2 complex structure. Nat. Commun. 2021, 12, 7040. [Google Scholar] [CrossRef] [PubMed]
- Guan, Z.; Zhang, Q.; Zhang, Z.; Zuo, J.; Chen, J.; Liu, R.; Savarin, J.; Broger, L.; Cheng, P.; Wang, Q.; et al. Mechanistic insights into the regulation of plant phosphate homeostasis by the rice SPX2–PHR2 complex. Nat. Commun. 2022, 13, 1581. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Hu, J.; Ai, Z.; Pang, L.; Li, Y.; Zhu, M. Light exposure during storage preserving soluble sugar and l-ascorbic acid content of minimally processed romaine lettuce (Lactuca sativa L.var. longifolia). Food Chem. 2013, 136, 273–278. [Google Scholar] [CrossRef]
- Ninkuu, V.; Zhou, Y.; Liu, H.; Sun, S.; Liu, Z.; Liu, Y.; Yang, J.; Hu, M.; Guan, L.; Sun, X. Regulation of Nitrogen Metabolism by COE2 under Low Sulfur Stress in Arabidopsis. Plant Sci. 2024, 346, 112137. [Google Scholar] [CrossRef]
- Xing, G.; Jin, M.; Yue, P.; Ren, C.; Hao, J.; Zhao, Y.; Zhao, X.; Sun, Z.; Hou, S. Role of SiPHR1 in the Response to Low Phosphate in Foxtail Millet via Comparative Transcriptomic and Co-Expression Network Analyses. Int. J. Mol. Sci. 2023, 24, 12786. [Google Scholar] [CrossRef]
- Raghothama, K.G.; Karthikeyan, A.S. Phosphate Acquisition. Plant Soil 2005, 274, 37–49. [Google Scholar] [CrossRef]
- Veneklaas, E.J.; Lambers, H.; Bragg, J.; Finnegan, P.M.; Lovelock, C.E.; Plaxton, W.C.; Price, C.A.; Scheible, W.; Shane, M.W.; White, P.J.; et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012, 195, 306–320. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Miao, J.; Guo, J.; Shi, Z.; He, M.; Chen, Y.; Zhao, X.; Li, B.; Han, F.; et al. A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann. Bot. 2013, 111, 1139–1153. [Google Scholar] [CrossRef]
- Valdés-López, O.; Arenas Huertero, C.; Ramírez, M.; Girard, L.; Sánchez, F.; Vance, C.P.; Luis Reyes, J.; Hernández, G. Essential role of MYB transcription factor: PvPHR1 and microRNA: PvmiR399 in phosphorus-deficiency signalling in common bean roots. Plant Cell Environ. 2008, 31, 1834–1843. [Google Scholar] [CrossRef]
- Ren, F.; Guo, Q.-Q.; Chang, L.-L.; Chen, L.; Zhao, C.-Z.; Zhong, H.; Li, X.-B. Brassica napus PHR1 Gene Encoding a MYB-Like Protein Functions in Response to Phosphate Starvation. PLoS ONE 2012, 7, e44005. [Google Scholar] [CrossRef]
- Xue, Y.; Xiao, B.-X.; Zhu, S.; Mo, X.; Liang, C.; Tian, J.; Liao, H. GmPHR25, a GmPHR member up-regulated by phosphate starvation, controls phosphate homeostasis in soybean. J. Exp. Bot. 2017, 68, 4951–4967. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Sun, F.; Wang, M.; Su, L.; Li, R.; Caetano-Anollés, G. Genome-wide analysis of the MYB-CC gene family of maize. Genetica 2018, 147, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lang, Y.; Liu, Z. Basic Helix-Loop-Helix (bHLH) transcription factor family in Yellow horn (Xanthoceras sorbifolia Bunge): Genome-wide characterization, chromosome location, phylogeny, structures and expression patterns. Int. J. Biol. Macromol. 2020, 160, 711–723. [Google Scholar] [CrossRef] [PubMed]
- Lei, M.; Zhu, C.; Liu, Y.; Karthikeyan, A.S.; Bressan, R.A.; Raghothama, K.G.; Liu, D. Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol. 2010, 189, 1084–1095. [Google Scholar] [CrossRef]
- Karthikeyan, A.S.; Varadarajan, D.K.; Jain, A.; Held, M.A.; Carpita, N.C.; Raghothama, K.G. Phosphate starvation responses are mediated by sugar signaling in Arabidopsis. Planta 2006, 225, 907–918. [Google Scholar] [CrossRef]
- Jiang, C.; Gao, X.; Liao, L.; Harberd, N.P.; Fu, X. Phosphate Starvation Root Architecture and Anthocyanin Accumulation Responses Are Modulated by the Gibberellin-DELLA Signaling Pathway in Arabidopsis. Plant Physiol. 2007, 145, 1460–1470. [Google Scholar] [CrossRef]
- Klecker, M.; Gasch, P.; Peisker, H.; Dörmann, P.; Schlicke, H.; Grimm, B.; Mustroph, A. A Shoot-Specific Hypoxic Response of Arabidopsis Sheds Light on the Role of the Phosphate-Responsive Transcription Factor PHOSPHATE STARVATION RESPONSE1. Plant Physiol. 2014, 165, 774–790. [Google Scholar] [CrossRef]
- Liu, Y.; Xie, Y.; Wang, H.; Ma, X.; Yao, W.; Wang, H. Light and Ethylene Coordinately Regulate the Phosphate Starvation Response through Transcriptional Regulation of PHOSPHATE STARVATION RESPONSE1. Plant Cell 2017, 29, 2269–2284. [Google Scholar] [CrossRef]
- Vidal, E.A.; Alvarez, J.M.; Araus, V.; Riveras, E.; Brooks, M.D.; Krouk, G.; Ruffel, S.; Lejay, L.; Crawford, N.M.; Coruzzi, G.M.; et al. Nitrate in 2020: Thirty Years from Transport to Signaling Networks. Plant Cell 2020, 32, 2094–2119. [Google Scholar] [CrossRef]
- Guo, F.Q.; Young, J.; Crawford, N.M. The Nitrate Transporter AtNRT1.1 (CHL1) Functions in Stomatal Opening and Contributes to Drought Susceptibility in Arabidopsis. Plant Cell 2002, 15, 107–117. [Google Scholar] [CrossRef]
- Martin, T.; Oswald, O.; Graham, I.A. Arabidopsis Seedling Growth, Storage Lipid Mobilization, and Photosynthetic Gene Expression Are Regulated by Carbon:Nitrogen Availability. Plant Physiol. 2002, 128, 472–481. [Google Scholar] [CrossRef][Green Version]
- Smith, F.W.; Jackson, W.A. Nitrogen Enhancement of Phosphate Transport in Roots of Zea mays L. Plant Physiol. 1987, 84, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- de Magalhães, J.V.; Alves, V.M.C.; de Novais, R.F.; Mosquim, P.R.; Magalhães, J.R.; Filho, A.F.C.B.; Hubert, D.M. Nitrate uptake by corn under increasing periods of phosphorus starvation. J. Plant Nutr. 1998, 21, 1753–1763. [Google Scholar] [CrossRef]
- Tian, W.H.; Ye, J.Y.; Cui, M.Q.; Chang, J.B.; Liu, Y.; Li, G.X.; Wu, Y.R.; Xu, J.M.; Harberd, N.P.; Mao, C.Z.; et al. A transcription factor STOP1-centered pathway coordinates ammonium and phosphate acquisition in Arabidopsis. Mol. Plant 2021, 14, 1554–1568. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Kiba, T.; Yanagisawa, S. Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of SPX genes. Plant J. 2019, 102, 448–466. [Google Scholar] [CrossRef]
- Mittler, R.; Zandalinas, S.I.; Fichman, Y.; Van Breusegem, F. Reactive oxygen species signalling in plant stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 663–679. [Google Scholar] [CrossRef]
- Choudhury, F.K.; Rivero, R.M.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant J. 2016, 90, 856–867. [Google Scholar] [CrossRef]
- Shin, R.; Berg, R.H.; Schachtman, D.P. Reactive Oxygen Species and Root Hairs in Arabidopsis Root Response to Nitrogen, Phosphorus and Potassium Deficiency. Plant Cell Physiol. 2005, 46, 1350–1357. [Google Scholar] [CrossRef]
- Xu, H.X.; Weng, X.Y.; Yang, Y. Effect of phosphorus deficiency on the photosynthetic characteristics of rice plants. Russ. J. Plant Physiol. 2007, 54, 741–748. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; I Hurwitz, D.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2019, 48, D265–D268. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc 2.0: An improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010, 2, 1090–1103. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef]
- Pan, W.; Liu, X.; Li, D.; Zhang, H. Establishment of an Efficient Genome Editing System in Lettuce Without Sacrificing Specificity. Front. Plant Sci. 2022, 13, 930592. [Google Scholar] [CrossRef]
- Arnon, D.L. Copper enzymes in isolated chloroplasts, polyphenol oxidase in Beta vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Molecular Weight (Da) | No. of Amino Acids | pI | Instability Index | Grand Average of Hydropathicity | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| LsaPHR1.1 | LOC111881312 | 48,138.67 | 435 | 5.55 | 66.66 | −0.789 | Nucleus |
| LsaPHR1.2 | LOC111895520 | 45,858.76 | 418 | 5.51 | 61.23 | −0.759 | Nucleus |
| LsaPHR1.3 | LOC111903878 | 45,775.19 | 410 | 5.30 | 47.47 | −0.786 | Nucleus |
| LsaPHR1.4 | LOC111895914 | 30,769.89 | 281 | 6.10 | 53.52 | −0.41 | Nucleus |
| LsaPHR1.5 | LOC111915887 | 28,849.99 | 261 | 6.60 | 52.03 | −0.458 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qian, Y.; Liu, X.; Wang, B.; Li, D.; Wu, Z.; Tong, J. Genome-Wide Identification of the LsaPHR1 Gene Family and Preliminary Functional Validation of LsaPHR1.1 in Phosphorus Tolerance in Lactuca sativa. Int. J. Mol. Sci. 2025, 26, 10466. https://doi.org/10.3390/ijms262110466
Qian Y, Liu X, Wang B, Li D, Wu Z, Tong J. Genome-Wide Identification of the LsaPHR1 Gene Family and Preliminary Functional Validation of LsaPHR1.1 in Phosphorus Tolerance in Lactuca sativa. International Journal of Molecular Sciences. 2025; 26(21):10466. https://doi.org/10.3390/ijms262110466
Chicago/Turabian StyleQian, Yuxuan, Xue Liu, Baoju Wang, Dayong Li, Zhanhui Wu, and Jing Tong. 2025. "Genome-Wide Identification of the LsaPHR1 Gene Family and Preliminary Functional Validation of LsaPHR1.1 in Phosphorus Tolerance in Lactuca sativa" International Journal of Molecular Sciences 26, no. 21: 10466. https://doi.org/10.3390/ijms262110466
APA StyleQian, Y., Liu, X., Wang, B., Li, D., Wu, Z., & Tong, J. (2025). Genome-Wide Identification of the LsaPHR1 Gene Family and Preliminary Functional Validation of LsaPHR1.1 in Phosphorus Tolerance in Lactuca sativa. International Journal of Molecular Sciences, 26(21), 10466. https://doi.org/10.3390/ijms262110466




