Accumulation of Lymphoid Progenitors with Defective B Cell Differentiation and of Putative Natural Killer Progenitors in Aging Human Bone Marrow
Abstract
1. Introduction
2. Results
2.1. Single Cell Transcriptome Studies and the Developmental Trajectory of Human HSPC
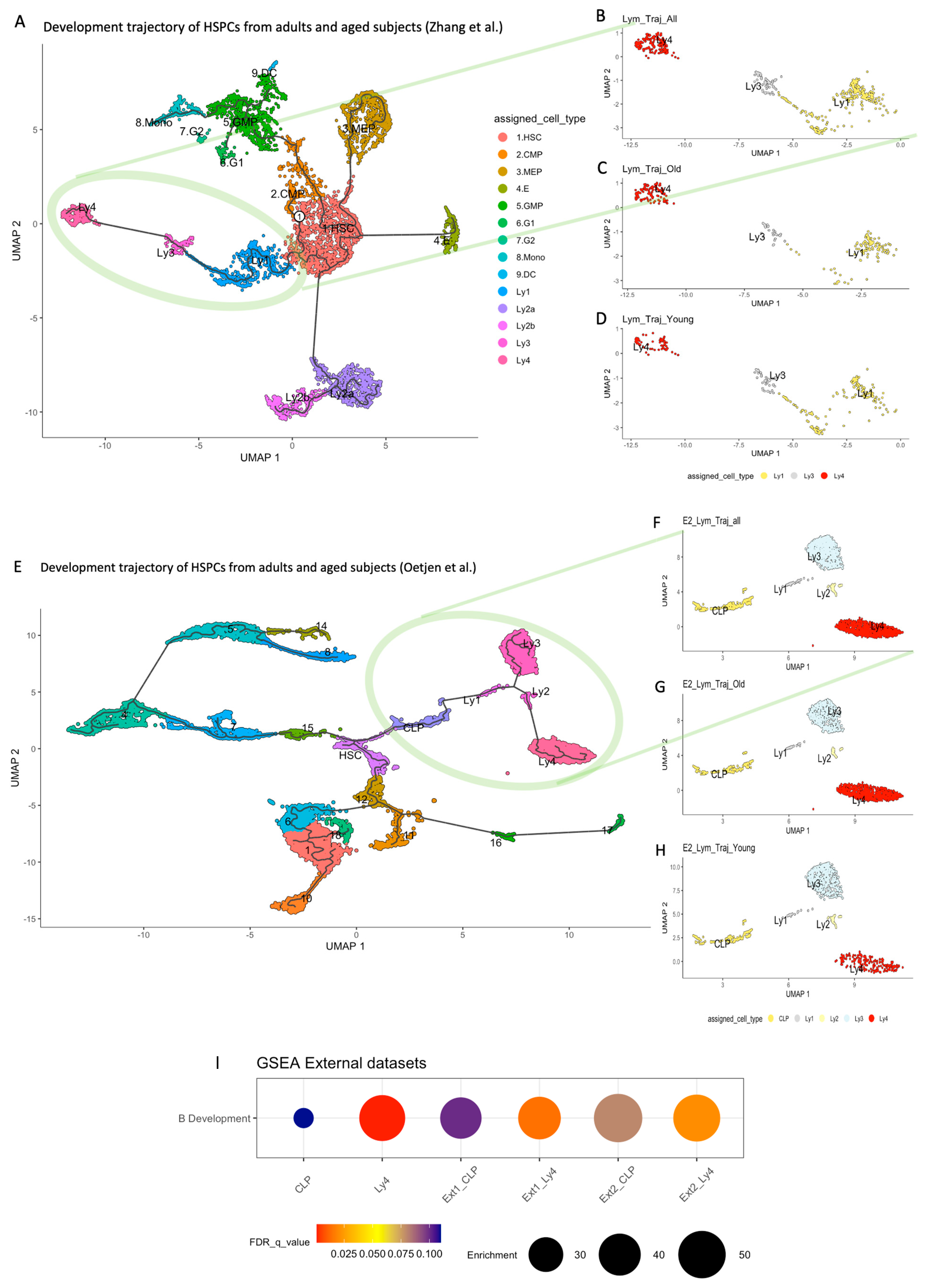
2.2. Developmental Trajectory of Lymphocytic Clusters upon Aging
2.3. Differences Between Old and Young in Each of the Clusters in the Lymphoid Compartment
2.4. GSEA of the Lymphoid Clusters
2.5. Accumulation of Memory-like NK Progenitors in Ly4 Compartment
2.6. Accumulation of Ly4 Analogous Population in the Lymphoid Progenitors in Other Human Bone Marrow Datasets
3. Discussion
4. Materials and Methods
4.1. Human Specimen
4.2. Single-Cell RNA Sequencing of HSPCs
4.3. Data Processing
4.4. Toolkit for Analyzing Single-Cell Transcriptome Data
- Gene sets used for annotation of cell clusters:
4.5. Cell Cycle Annotation
4.6. Gene Set Enrichment Analysis (GSEA)
4.7. Generating the Gene Set “B Cell Development”
4.8. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernandez Maestre, I.; Harris, A.S.; Amor, C. Aging and immunity: The age-old tango. Genes Dev. 2025, 39, 948–974. [Google Scholar] [CrossRef]
- de Mol, J.; Kuiper, J.; Tsiantoulas, D.; Foks, A.C. The Dynamics of B Cell Aging in Health and Disease. Front. Immunol. 2021, 12, 733566. [Google Scholar] [CrossRef]
- Andersson, R.; Mejia-Ramirez, E.; Florian, M.C. Haematopoietic ageing in health and lifespan. Nat. Cell Biol. 2025, 27, 1398–1410. [Google Scholar] [CrossRef]
- Li, H.; Cote, P.; Kuoch, M.; Ezike, J.; Frenis, K.; Afanassiev, A.; Greenstreet, L.; Tanaka-Yano, M.; Tarantino, G.; Zhang, S.; et al. The dynamics of hematopoiesis over the human lifespan. Nat. Methods 2025, 22, 422–434. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Sanjuan-Pla, A.; Thongjuea, S.; Carrelha, J.; Giustacchini, A.; Gambardella, A.; Macaulay, I.; Mancini, E.; Luis, T.C.; Mead, A.; et al. Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun. 2016, 7, 11075. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Song, B.; Carlino, M.J.; Li, G.; Ferchen, K.; Chen, M.; Thompson, E.N.; Kain, B.N.; Schnell, D.; Thakkar, K.; et al. An immunophenotype-coupled transcriptomic atlas of human hematopoietic progenitors. Nat. Immunol. 2024, 25, 703–715. [Google Scholar] [CrossRef]
- Hennrich, M.L.; Romanov, N.; Horn, P.; Jaeger, S.; Eckstein, V.; Steeples, V.; Ye, F.; Ding, X.; Poisa-Beiro, L.; Lai, M.C.; et al. Cell-specific proteome analyses of human bone marrow reveal molecular features of age-dependent functional decline. Nat. Commun. 2018, 9, 4004. [Google Scholar] [CrossRef]
- van Deursen, J.M. The role of senescent cells in ageing. Nature 2014, 509, 439–446. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; LeBrasseur, N.K.; Childs, B.G.; van de Sluis, B.; Kirkland, J.L.; van Deursen, J.M. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Tchkonia, T.; Zhu, Y.; van Deursen, J.; Campisi, J.; Kirkland, J.L. Cellular senescence and the senescent secretory phenotype: Therapeutic opportunities. J. Clin. Investig. 2013, 123, 966–972. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Childs, B.G.; Durik, M.; Wijers, M.E.; Sieben, C.J.; Zhong, J.; Saltness, R.A.; Jeganathan, K.B.; Verzosa, G.C.; Pezeshki, A.; et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature 2016, 530, 184–189. [Google Scholar] [CrossRef]
- Vaziri, H.; Dragowska, W.; Allsopp, R.C.; Thomas, T.E.; Harley, C.B.; Lansdorp, P.M. Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc. Natl. Acad. Sci. USA 1994, 91, 9857–9860. [Google Scholar] [CrossRef]
- Fali, T.; Fabre-Mersseman, V.; Yamamoto, T.; Bayard, C.; Papagno, L.; Fastenackels, S.; Zoorab, R.; Koup, R.A.; Boddaert, J.; Sauce, D.; et al. Elderly human hematopoietic progenitor cells express cellular senescence markers and are more susceptible to pyroptosis. JCI Insight 2018, 3, e95319. [Google Scholar] [CrossRef]
- Casella, G.; Munk, R.; Kim, K.M.; Piao, Y.; De, S.; Abdelmohsen, K.; Gorospe, M. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019, 47, 7294–7305. [Google Scholar] [CrossRef]
- Munoz-Espin, D.; Canamero, M.; Maraver, A.; Gomez-Lopez, G.; Contreras, J.; Murillo-Cuesta, S.; Rodriguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed cell senescence during mammalian embryonic development. Cell 2013, 155, 1104–1118. [Google Scholar] [CrossRef]
- Sperka, T.; Wang, J.; Rudolph, K.L. DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 2012, 13, 579–590. [Google Scholar] [CrossRef]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Kowalczyk, M.S.; Tirosh, I.; Heckl, D.; Rao, T.N.; Dixit, A.; Haas, B.J.; Schneider, R.K.; Wagers, A.J.; Ebert, B.L.; Regev, A. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res. 2015, 25, 1860–1872. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, K.; Chandra, T.; Kiselev, V.; Flores-Santa Cruz, D.; Macaulay, I.C.; Park, H.J.; Li, J.; Kent, D.G.; Kumar, R.; Pask, D.C.; et al. Proliferation Drives Aging-Related Functional Decline in a Subpopulation of the Hematopoietic Stem Cell Compartment. Cell Rep. 2017, 19, 1503–1511. [Google Scholar] [CrossRef] [PubMed]
- Flach, J.; Bakker, S.T.; Mohrin, M.; Conroy, P.C.; Pietras, E.M.; Reynaud, D.; Alvarez, S.; Diolaiti, M.E.; Ugarte, F.; Forsberg, E.C.; et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature 2014, 512, 198–202. [Google Scholar] [CrossRef]
- Beerman, I.; Bock, C.; Garrison, B.S.; Smith, Z.D.; Gu, H.; Meissner, A.; Rossi, D.J. Proliferation-dependent alterations of the DNA methylation landscape underlie hematopoietic stem cell aging. Cell Stem Cell 2013, 12, 413–425. [Google Scholar] [CrossRef]
- Sun, D.; Luo, M.; Jeong, M.; Rodriguez, B.; Xia, Z.; Hannah, R.; Wang, H.; Le, T.; Faull, K.F.; Chen, R.; et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 2014, 14, 673–688. [Google Scholar] [CrossRef]
- Mann, M.; Mehta, A.; de Boer, C.G.; Kowalczyk, M.S.; Lee, K.; Haldeman, P.; Rogel, N.; Knecht, A.R.; Farouq, D.; Regev, A.; et al. Heterogeneous Responses of Hematopoietic Stem Cells to Inflammatory Stimuli Are Altered with Age. Cell Rep. 2018, 25, 2992–3005.e5. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Thoma, J.; Landry, J.; Sauer, S.; Yamamoto, A.; Eckstein, V.; Romanov, N.; Raffel, S.; Hoffmann, G.F.; Bork, P.; et al. Glycogen accumulation, central carbon metabolism, and aging of hematopoietic stem and progenitor cells. Sci. Rep. 2020, 10, 11597. [Google Scholar] [CrossRef] [PubMed]
- Poisa-Beiro, L.; Landry, J.J.M.; Raffel, S.; Tanaka, M.; Zaugg, J.; Gavin, A.C.; Ho, A.D. Glucose Metabolism and Aging of Hematopoietic Stem and Progenitor Cells. Int. J. Mol. Sci. 2022, 23, 3028. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Wang, Y.; Shao, L.; Laberge, R.M.; Demaria, M.; Campisi, J.; Janakiraman, K.; Sharpless, N.E.; Ding, S.; Feng, W.; et al. Clearance of senescent cells by ABT263 rejuvenates aged hematopoietic stem cells in mice. Nat. Med. 2016, 22, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Chaib, S.; Tchkonia, T.; Kirkland, J.L. Cellular senescence and senolytics: The path to the clinic. Nat. Med. 2022, 28, 1556–1568. [Google Scholar] [CrossRef]
- Poisa-Beiro, L.; Landry, J.J.M.; Yan, B.; Kardorff, M.; Eckstein, V.; Villacorta, L.; Krammer, P.H.; Zaugg, J.; Gavin, A.C.; Benes, V.; et al. A Senescent Cluster in Aged Human Hematopoietic Stem Cell Compartment as Target for Senotherapy. Int. J. Mol. Sci. 2025, 26, 787. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Haghverdi, L.; Lun, A.T.L.; Morgan, M.D.; Marioni, J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018, 36, 421–427. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J.; et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Nestorowa, S.; Hamey, F.K.; Pijuan Sala, B.; Diamanti, E.; Shepherd, M.; Laurenti, E.; Wilson, N.K.; Kent, D.G.; Gottgens, B. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood 2016, 128, e20–e31. [Google Scholar] [CrossRef]
- Pellin, D.; Loperfido, M.; Baricordi, C.; Wolock, S.L.; Montepeloso, A.; Weinberg, O.K.; Biffi, A.; Klein, A.M.; Biasco, L. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat. Commun. 2019, 10, 2395. [Google Scholar] [CrossRef] [PubMed]
- Pliner, H.A.; Shendure, J.; Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods 2019, 16, 983–986. [Google Scholar] [CrossRef]
- Xie, X.; Liu, M.; Zhang, Y.; Wang, B.; Zhu, C.; Wang, C.; Li, Q.; Huo, Y.; Guo, J.; Xu, C.; et al. Single-cell transcriptomic landscape of human blood cells. Natl. Sci. Rev. 2021, 8, nwaa180. [Google Scholar] [CrossRef]
- Massey, F.J. The Kolmogorov-Smirnov Test for Goodness of Fit. J. Am. Stat. Assoc. 1951, 46, 68–78. [Google Scholar] [CrossRef]
- Haddad, R.; Guardiola, P.; Izac, B.; Thibault, C.; Radich, J.; Delezoide, A.L.; Baillou, C.; Lemoine, F.M.; Gluckman, J.C.; Pflumio, F.; et al. Molecular characterization of early human T/NK and B-lymphoid progenitor cells in umbilical cord blood. Blood 2004, 104, 3918–3926. [Google Scholar] [CrossRef]
- Lee, R.D.; Munro, S.A.; Knutson, T.P.; LaRue, R.S.; Heltemes-Harris, L.M.; Farrar, M.A. Single-cell analysis identifies dynamic gene expression networks that govern B cell development and transformation. Nat. Commun. 2021, 12, 6843. [Google Scholar] [CrossRef]
- Stewart, A.; Ng, J.C.; Wallis, G.; Tsioligka, V.; Fraternali, F.; Dunn-Walters, D.K. Single-Cell Transcriptomic Analyses Define Distinct Peripheral B Cell Subsets and Discrete Development Pathways. Front. Immunol. 2021, 12, 602539. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.; Tergaonkar, V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 2022, 43, 210–229. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Pico, A.R.; Kelder, T.; van Iersel, M.P.; Hanspers, K.; Conklin, B.R.; Evelo, C. WikiPathways: Pathway editing for the people. PLoS Biol. 2008, 6, e184. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Wang, X. Molecular Regulation of NK Cell Maturation. Front. Immunol. 2020, 11, 1945. [Google Scholar] [CrossRef]
- Renoux, V.M.; Zriwil, A.; Peitzsch, C.; Michaelsson, J.; Friberg, D.; Soneji, S.; Sitnicka, E. Identification of a Human Natural Killer Cell Lineage-Restricted Progenitor in Fetal and Adult Tissues. Immunity 2015, 43, 394–407. [Google Scholar] [CrossRef]
- Crinier, A.; Milpied, P.; Escaliere, B.; Piperoglou, C.; Galluso, J.; Balsamo, A.; Spinelli, L.; Cervera-Marzal, I.; Ebbo, M.; Girard-Madoux, M.; et al. High-Dimensional Single-Cell Analysis Identifies Organ-Specific Signatures and Conserved NK Cell Subsets in Humans and Mice. Immunity 2018, 49, 971–986.e5. [Google Scholar] [CrossRef]
- Wang, D.; Malarkannan, S. Transcriptional Regulation of Natural Killer Cell Development and Functions. Cancers 2020, 12, 1591. [Google Scholar] [CrossRef]
- Yang, C.; Siebert, J.R.; Burns, R.; Gerbec, Z.J.; Bonacci, B.; Rymaszewski, A.; Rau, M.; Riese, M.J.; Rao, S.; Carlson, K.S.; et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single-cell transcriptome. Nat. Commun. 2019, 10, 3931. [Google Scholar] [CrossRef]
- Crinier, A.; Dumas, P.Y.; Escaliere, B.; Piperoglou, C.; Gil, L.; Villacreces, A.; Vely, F.; Ivanovic, Z.; Milpied, P.; Narni-Mancinelli, E.; et al. Single-cell profiling reveals the trajectories of natural killer cell differentiation in bone marrow and a stress signature induced by acute myeloid leukemia. Cell. Mol. Immunol. 2021, 18, 1290–1304. [Google Scholar] [CrossRef]
- Guo, C.; Wu, M.; Huang, B.; Zhao, R.; Jin, L.; Fu, B.; Wang, P.; Wang, D.; Zheng, M.; Fang, J.; et al. Single-cell transcriptomics reveal a unique memory-like NK cell subset that accumulates with ageing and correlates with disease severity in COVID-19. Genome Med. 2022, 14, 46. [Google Scholar] [CrossRef]
- Oetjen, K.A.; Lindblad, K.E.; Goswami, M.; Gui, G.; Dagur, P.K.; Lai, C.; Dillon, L.W.; McCoy, J.P.; Hourigan, C.S. Human bone marrow assessment by single-cell RNA sequencing, mass cytometry, and flow cytometry. JCI Insight 2018, 3, e124928. [Google Scholar] [CrossRef]
- Zhang, Y.; Xie, X.; Huang, Y.; Liu, M.; Li, Q.; Luo, J.; He, Y.; Yin, X.; Ma, S.; Cao, W.; et al. Temporal molecular program of human hematopoietic stem and progenitor cells after birth. Dev. Cell 2022, 57, 2745–2760.e6. [Google Scholar] [CrossRef] [PubMed]
- de Haan, G.; Lazare, S.S. Aging of hematopoietic stem cells. Blood 2018, 131, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Orkin, S.H.; Zon, L.I. Hematopoiesis: An evolving paradigm for stem cell biology. Cell 2008, 132, 631–644. [Google Scholar] [CrossRef] [PubMed]
- Svendsen, F.A.; Yang, D.; Kim, K.; Lazare, S.; Skinder, N.; Zwart, E.; Mura-Meszaros, A.; Ausema, A.; von Eyss, B.; de Haan, G.; et al. A comprehensive transcriptome signature of murine hematopoietic stem cell aging. Blood 2021, 138, 439–451. [Google Scholar] [CrossRef]
- Young, K.; Borikar, S.; Bell, R.; Kuffler, L.; Philip, V.; Trowbridge, J.J. Progressive alterations in multipotent hematopoietic progenitors underlie lymphoid cell loss in aging. J. Exp. Med. 2016, 213, 2259–2267. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Teo, Y.V.; Hinthorn, S.J.; Webb, A.E.; Neretti, N. Single-cell transcriptomics of peripheral blood in the aging mouse. Aging 2023, 15, 6–20. [Google Scholar] [CrossRef]
- Martyshkina, Y.S.; Tereshchenko, V.P.; Bogdanova, D.A.; Rybtsov, S.A. Reliable Hallmarks and Biomarkers of Senescent Lymphocytes. Int. J. Mol. Sci. 2023, 24, 15653. [Google Scholar] [CrossRef] [PubMed]
- Gergues, M.; Bari, R.; Koppisetti, S.; Gosiewska, A.; Kang, L.; Hariri, R.J. Senescence, NK cells, and cancer: Navigating the crossroads of aging and disease. Front. Immunol. 2025, 16, 1565278. [Google Scholar] [CrossRef] [PubMed]
- Ben-Porath, I.; Weinberg, R.A. The signals and pathways activating cellular senescence. Int. J. Biochem. Cell Biol. 2005, 37, 961–976. [Google Scholar] [CrossRef] [PubMed]
- d’Adda di Fagagna, F. Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef]
- Ou, H.L.; Schumacher, B. DNA damage responses and p53 in the aging process. Blood 2018, 131, 488–495. [Google Scholar] [CrossRef]
- Hewitt, G.; Jurk, D.; Marques, F.D.; Correia-Melo, C.; Hardy, T.; Gackowska, A.; Anderson, R.; Taschuk, M.; Mann, J.; Passos, J.F. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 2012, 3, 708. [Google Scholar] [CrossRef]
- Picelli, S.; Bjorklund, A.K.; Faridani, O.R.; Sagasser, S.; Winberg, G.; Sandberg, R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 2013, 10, 1096–1098. [Google Scholar] [CrossRef]
- Hennig, B.P.; Velten, L.; Racke, I.; Tu, C.S.; Thoms, M.; Rybin, V.; Besir, H.; Remans, K.; Steinmetz, L.M. Large-Scale Low-Cost NGS Library Preparation Using a Robust Tn5 Purification and Tagmentation Protocol. G3 Genes Genomes Genet. 2018, 8, 79–89. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Street, K.; Risso, D.; Fletcher, R.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H., 2nd; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
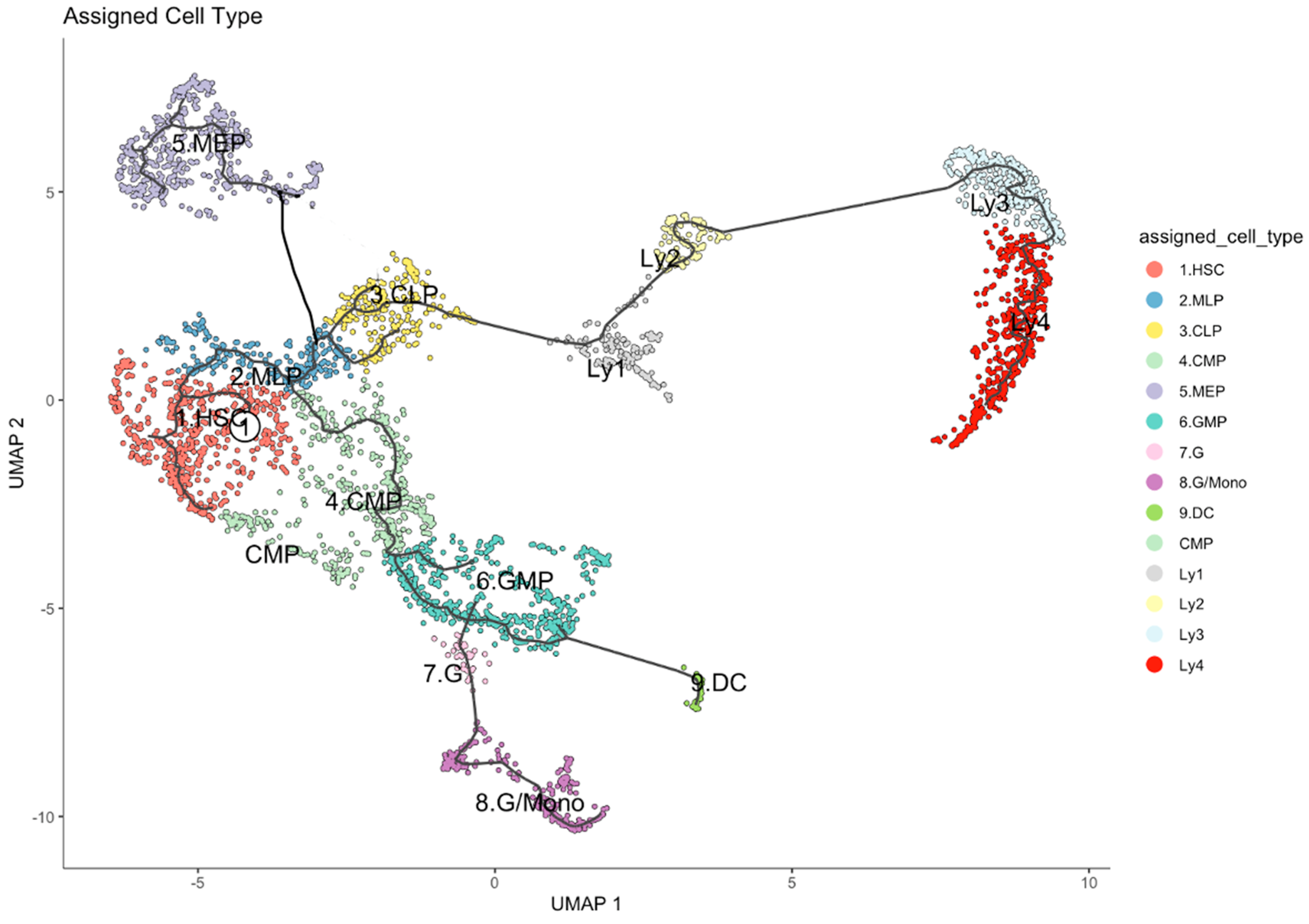
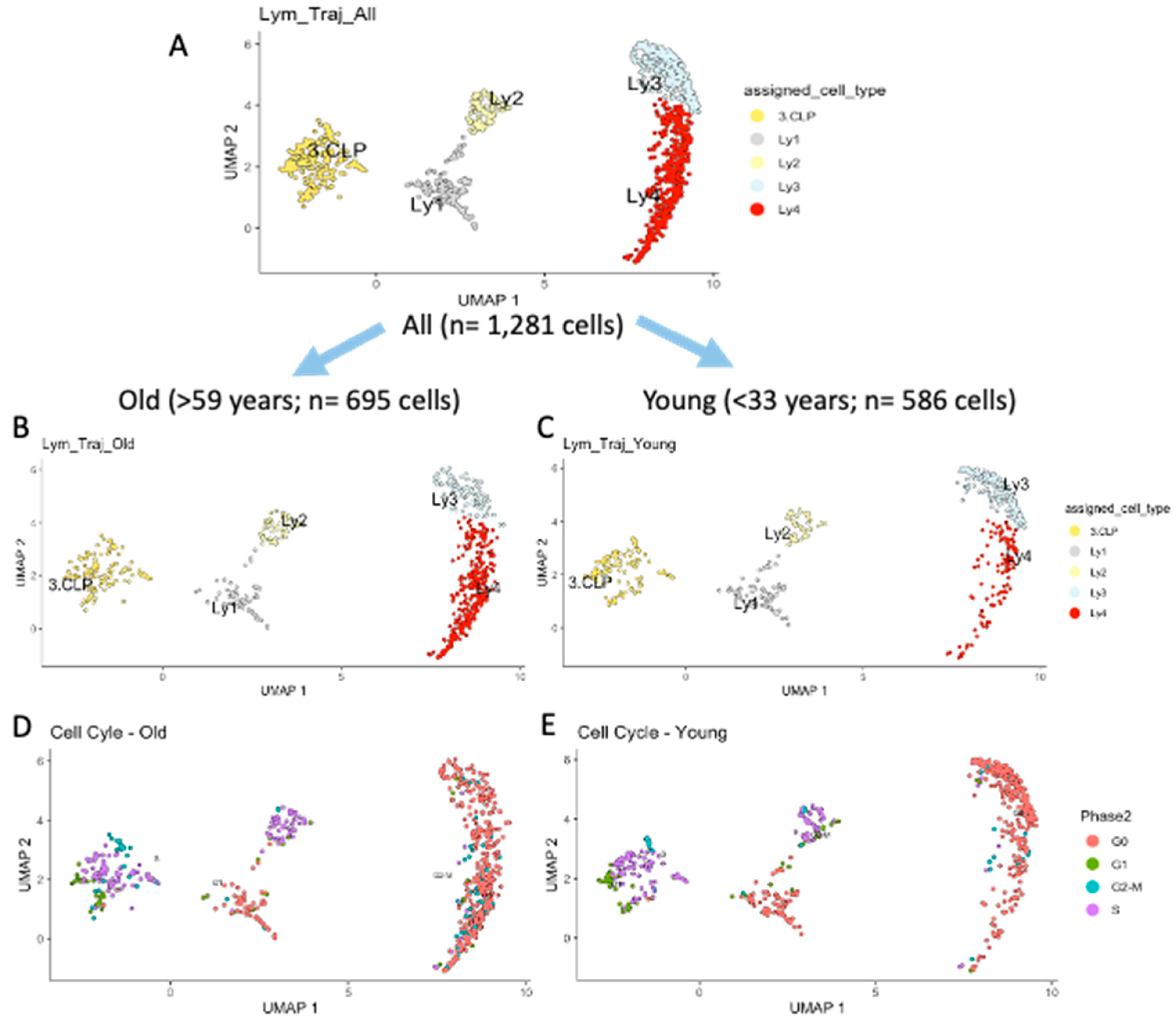


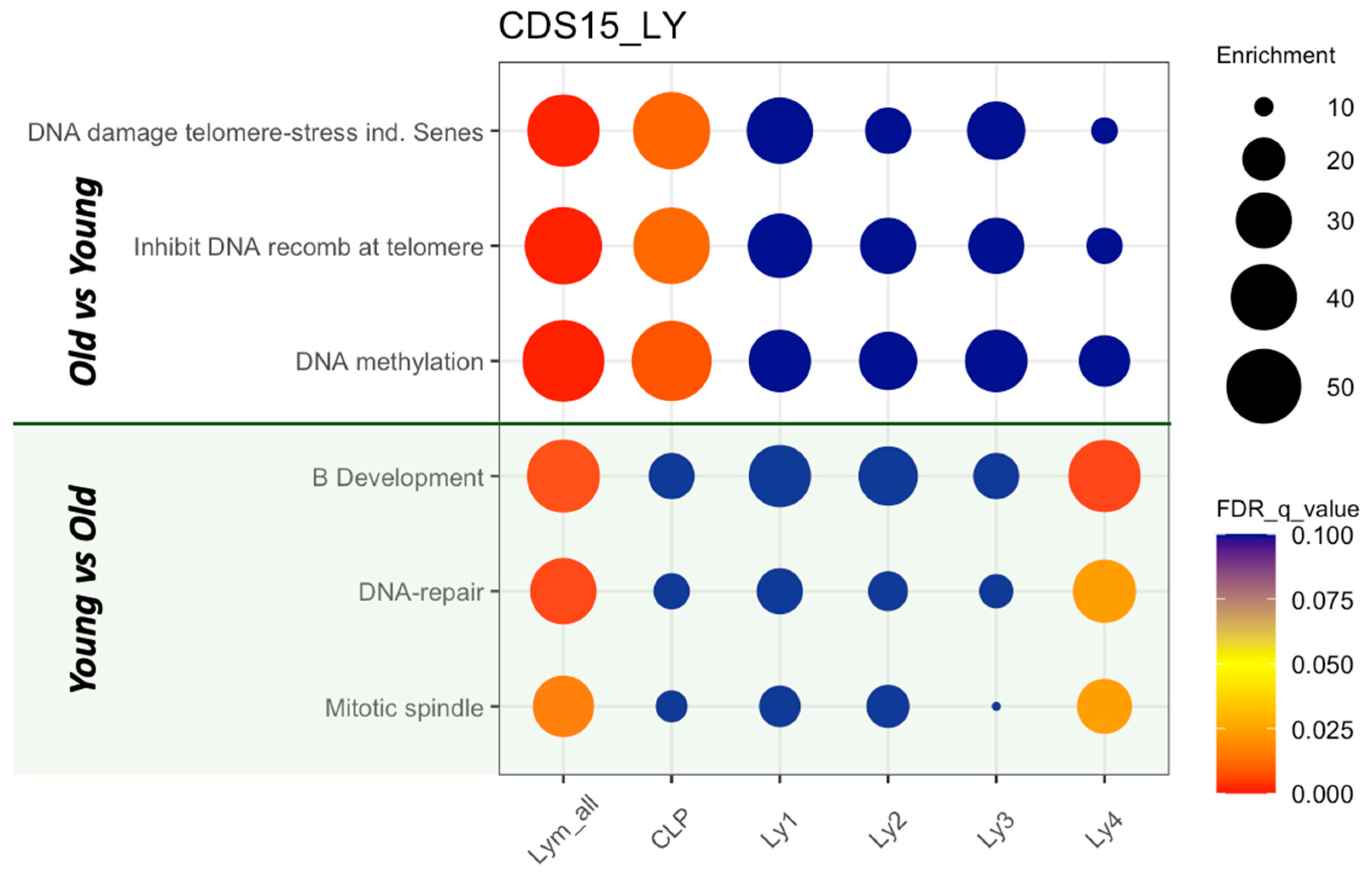
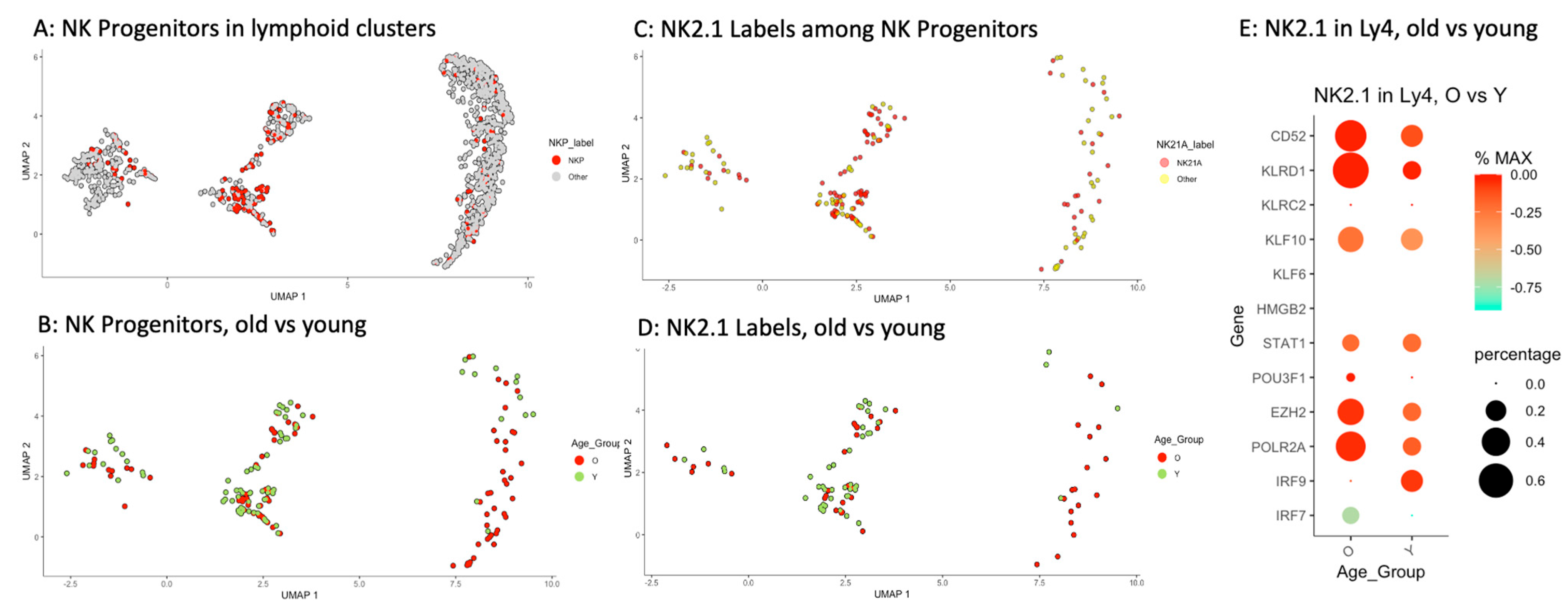
| Lymphoid Clusters | Re-Defined Lymphoid Clusters | All n = 1281 Cells % | Old n = 695 Cells % | Young n = 586 Cells % | p Value |
|---|---|---|---|---|---|
| CLP | CLP | 20.2 | 18.1 | 22.7 | n.s. |
| Ly1 | PreProB | 10.9 | 9.5 | 13.1 | n.s. |
| Ly2 | ProB | 10.1 | 9.2 | 11.3 | n.s. |
| Ly3 | PreB1 | 22.8 | 13.8 | 33.4 | *** |
| CLP+Ly1+ Ly2+Ly3 | Early Progenitors | 64.0 | 50.1 | 80.5 | *** |
| Ly4 | PreB2 | 36.0 | 49.9 | 19.5 | *** |
| Old | Young | p Values | ||||||
|---|---|---|---|---|---|---|---|---|
| Total n | NKP | NK2.1 | Total n | NKP | NK2.1 | NKP | NK2.1 | |
| CLP | 126 | 12 | 6 | 133 | 14 | 4 | n.s. | n.s. |
| Ly1 | 62 | 25 | 14 | 77 | 42 | 27 | * | * |
| Ly2 | 64 | 12 | 10 | 66 | 16 | 11 | n.s. | n.s. |
| Ly3 | 96 | 5 | 2 | 196 | 10 | 3 | n.s. | n.s. |
| Ly4 | 347 | 33 | 15 | 114 | 3 | 1 | *** | *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poisa-Beiro, L.; Landry, J.J.M.; Cherdintsev, A.; Kardorff, M.; Eckstein, V.; Villacorta, L.; Zaugg, J.; Gavin, A.-C.; Benes, V.; Raffel, S.; et al. Accumulation of Lymphoid Progenitors with Defective B Cell Differentiation and of Putative Natural Killer Progenitors in Aging Human Bone Marrow. Int. J. Mol. Sci. 2025, 26, 10467. https://doi.org/10.3390/ijms262110467
Poisa-Beiro L, Landry JJM, Cherdintsev A, Kardorff M, Eckstein V, Villacorta L, Zaugg J, Gavin A-C, Benes V, Raffel S, et al. Accumulation of Lymphoid Progenitors with Defective B Cell Differentiation and of Putative Natural Killer Progenitors in Aging Human Bone Marrow. International Journal of Molecular Sciences. 2025; 26(21):10467. https://doi.org/10.3390/ijms262110467
Chicago/Turabian StylePoisa-Beiro, Laura, Jonathan J. M. Landry, Aleksandr Cherdintsev, Michael Kardorff, Volker Eckstein, Laura Villacorta, Judith Zaugg, Anne-Claude Gavin, Vladimir Benes, Simon Raffel, and et al. 2025. "Accumulation of Lymphoid Progenitors with Defective B Cell Differentiation and of Putative Natural Killer Progenitors in Aging Human Bone Marrow" International Journal of Molecular Sciences 26, no. 21: 10467. https://doi.org/10.3390/ijms262110467
APA StylePoisa-Beiro, L., Landry, J. J. M., Cherdintsev, A., Kardorff, M., Eckstein, V., Villacorta, L., Zaugg, J., Gavin, A.-C., Benes, V., Raffel, S., & Ho, A. D. (2025). Accumulation of Lymphoid Progenitors with Defective B Cell Differentiation and of Putative Natural Killer Progenitors in Aging Human Bone Marrow. International Journal of Molecular Sciences, 26(21), 10467. https://doi.org/10.3390/ijms262110467







