Predicting Metabolic Syndrome Using Supervised Machine Learning: A Multivariate Parameter Approach †
Abstract
1. Introduction
2. Results
2.1. Demographic, Clinical, and Anthropometric Variables of the Study Subjects
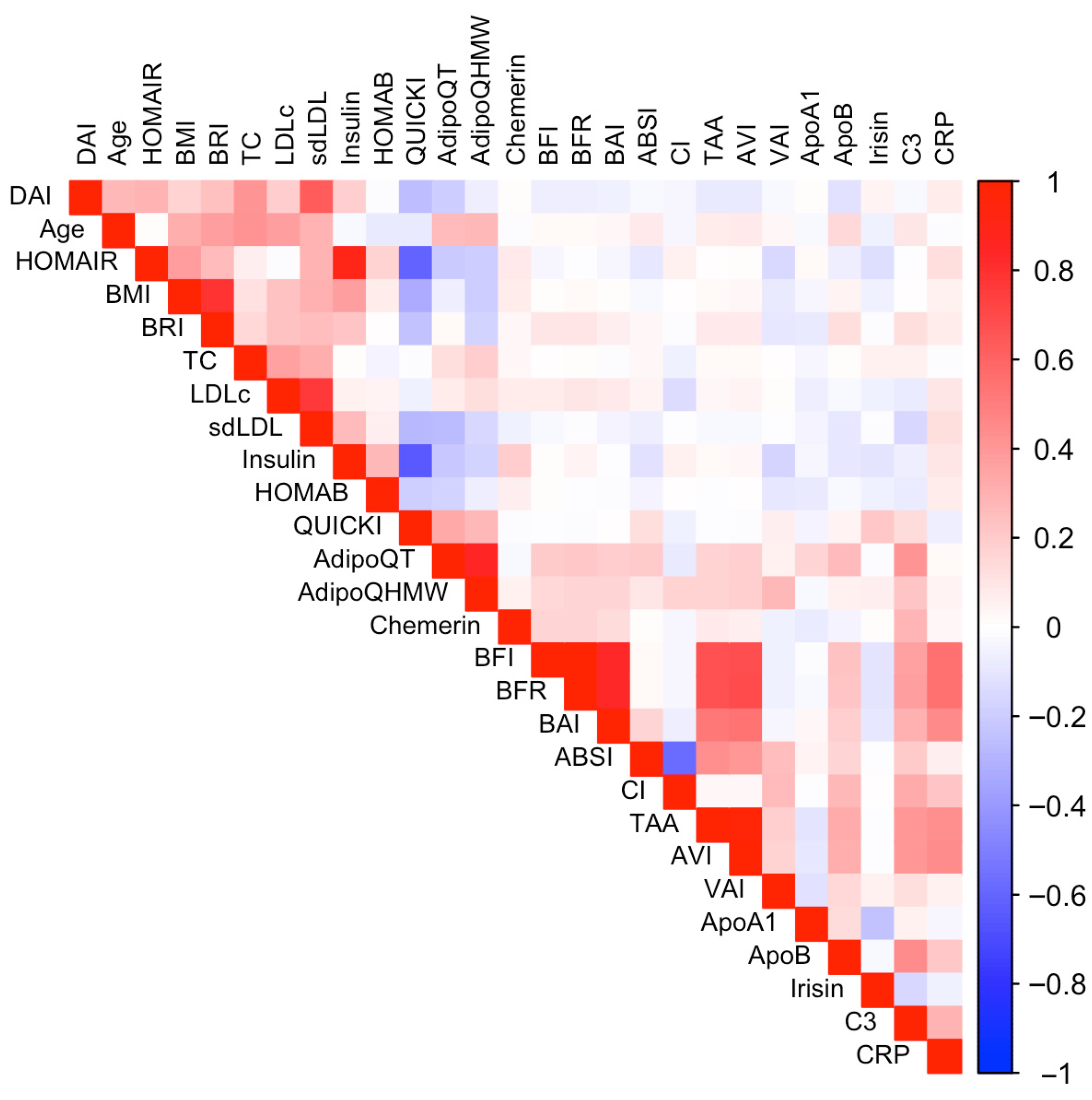
2.2. Data Pre-Processing
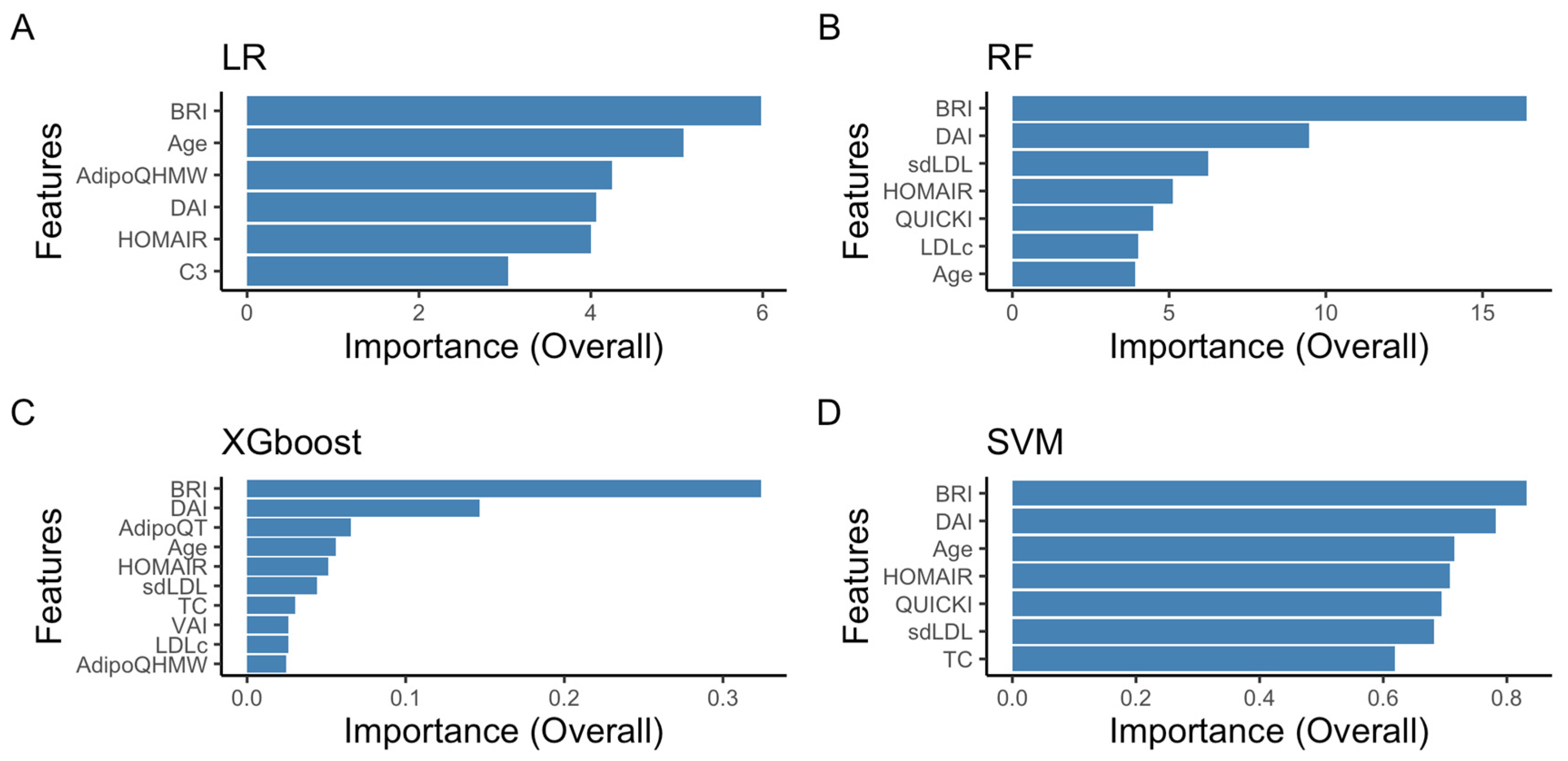
2.2.1. Recursive Feature Elimination with Cross-Validation
2.2.2. Cross-Validation: Evaluating Model Performance
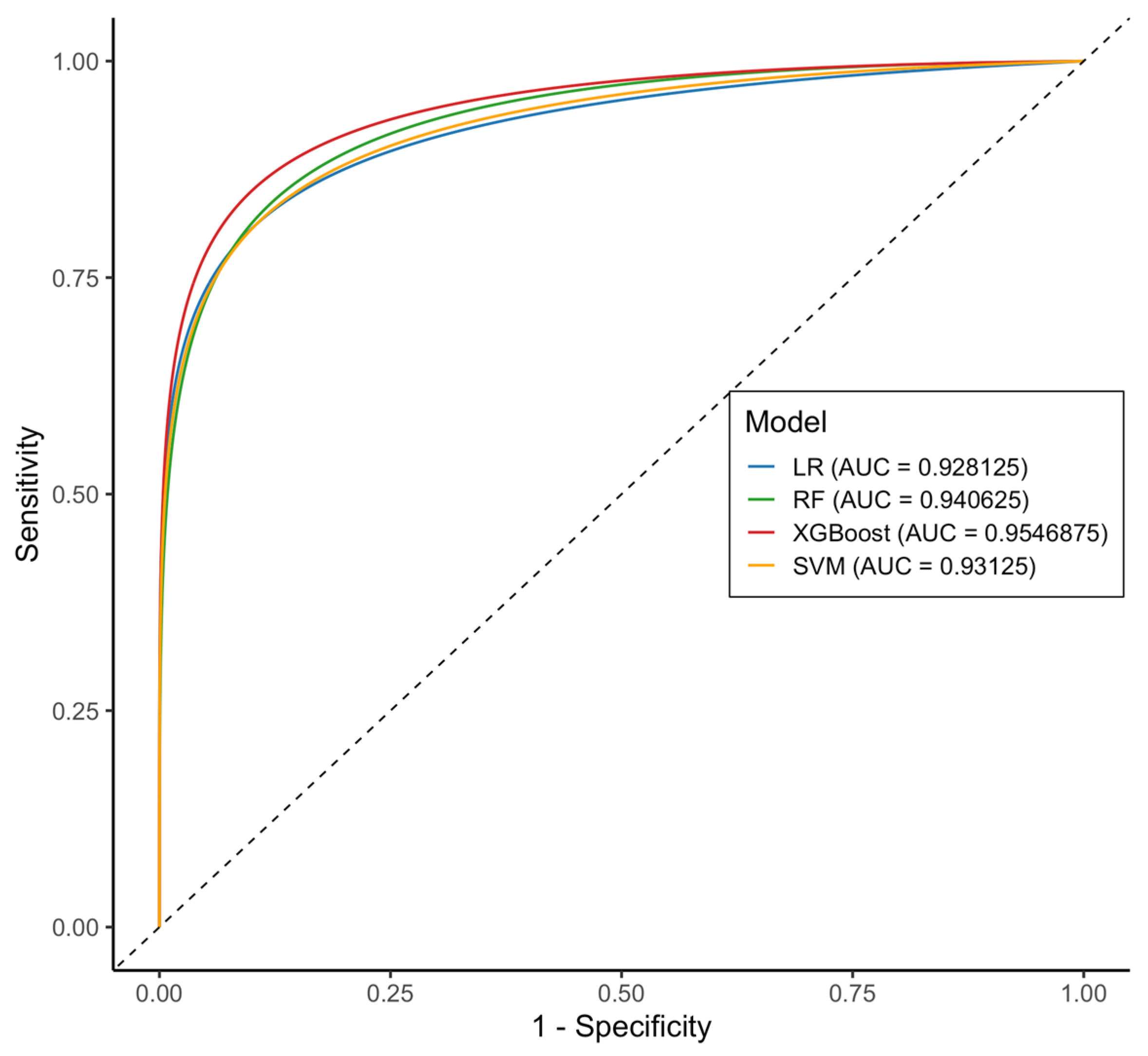
2.2.3. Test Data Performance
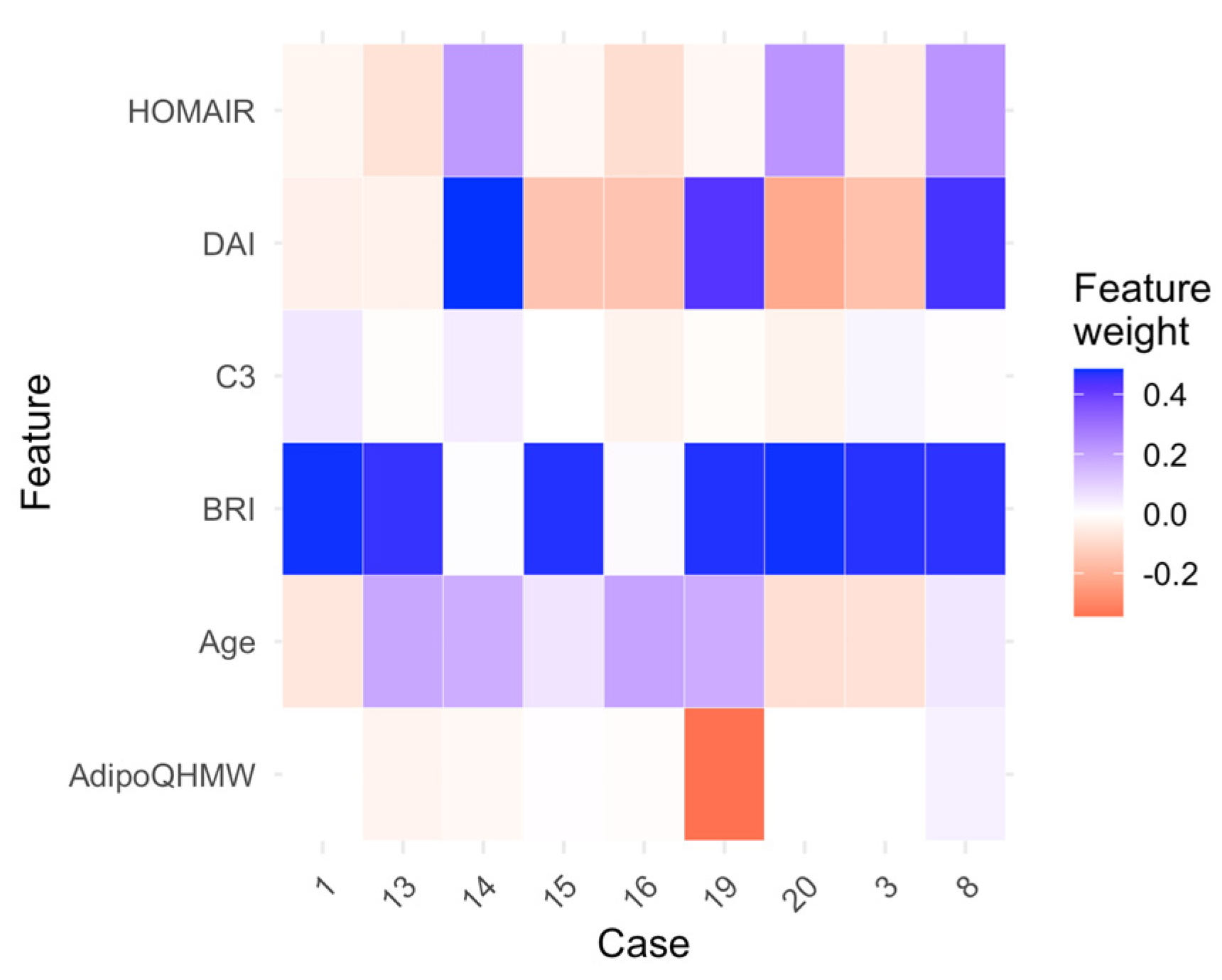
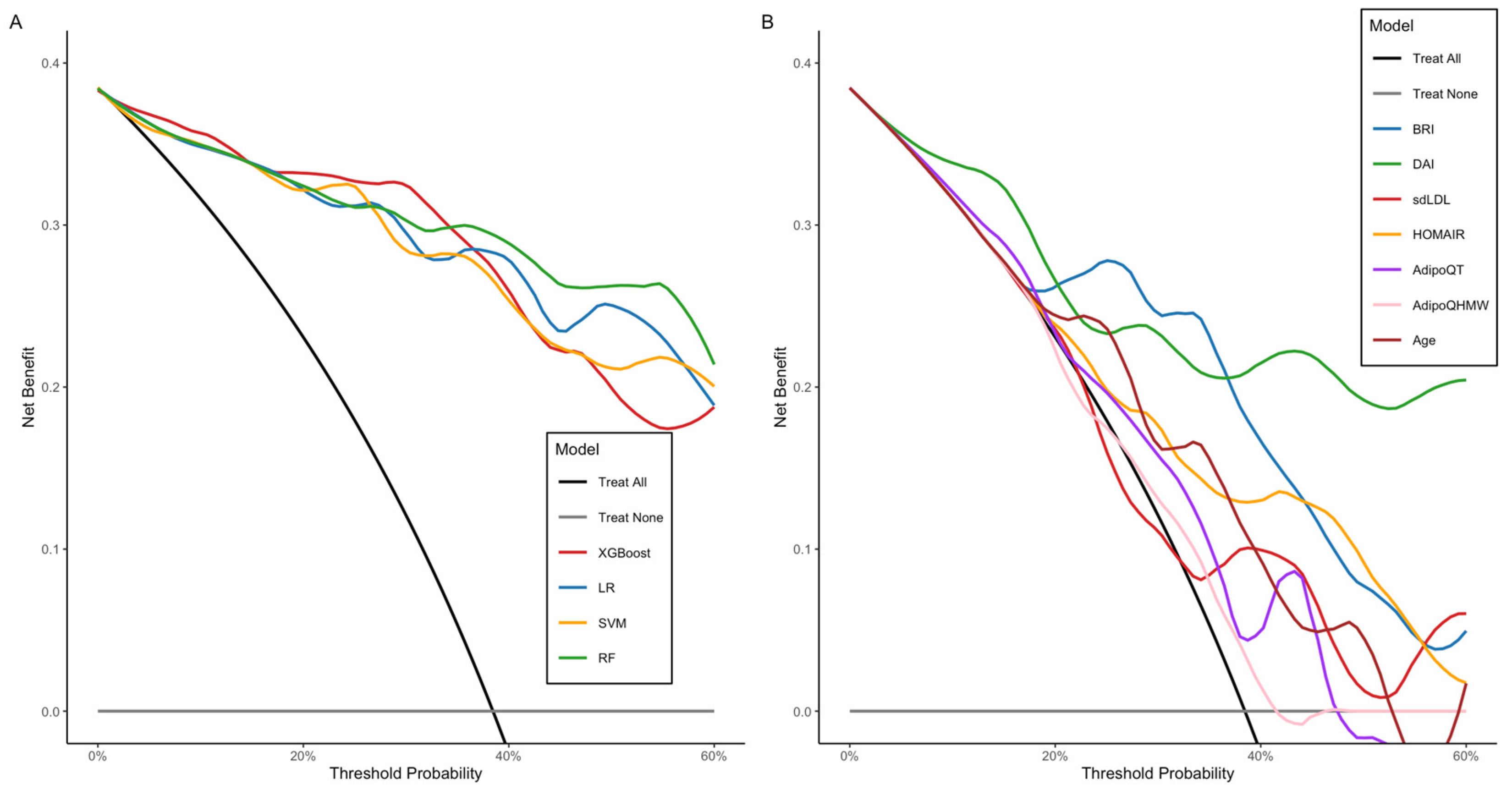
2.2.4. Random Forest and Logistic Regression as the Optimal Models: Interpretability Analysis
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Blood Sample Collection
4.3. Biochemical Parameters
4.4. Blood Pressure Measurements
4.5. Metabolic and Anthropometric Parameters
4.6. Definition of Metabolic Syndrome
4.7. Development of Predictive Models Using Supervised Machine Learning
4.7.1. Data Selection and Preprocessing
4.7.2. Cross-Validation and Feature Selection
4.7.3. Model Performance Evaluation
4.7.4. Statistics Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Model | Method (Caret) | Hyperparameter Tuning |
|---|---|---|
| Random Forest (RF) | rf | mtry = 2 |
| Support Vector Machine (SVM) | svmLinear | C = 0.01 |
| eXtreme Gradient Boosting (XGBoost) | xgbTree | nrounds = 200, eta = 0.1, max_depth = 3, gamma = 0, subsample = 0.8, colsample_bytree = 0.8, min_child_weight = 1 |
| Logistic regression (LR) | glm (binomial) | - |
Appendix B

Appendix C
References
- Lopez-Yus, M.; Horndler, C.; Borlan, S.; Bernal-Monterde, V.; Arbones-Mainar, J.M. Unraveling Adipose Tissue Dysfunction: Molecular Mechanisms, Novel Biomarkers, and Therapeutic Targets for Liver Fat Deposition. Cells 2024, 13, 380. [Google Scholar] [CrossRef]
- Goossens, G.H.; Blaak, E.E. Adipose tissue dysfunction and impaired metabolic health in human obesity: A matter of oxygen? Front. Endocrinol. 2015, 6, 55. [Google Scholar] [CrossRef]
- Hagberg, C.E.; Spalding, K.L. White adipocyte dysfunction and obesity-associated pathologies in humans. Nat. Rev. Mol. Cell Biol. 2024, 25, 270–289, Erratum in Nat. Rev. Mol. Cell Biol. 2024, 25, 333. [Google Scholar] [CrossRef]
- Bovolini, A.; Garcia, J.; Andrade, M.A.; Duarte, J.A. Metabolic Syndrome Pathophysiology and Predisposing Factors. Int. J. Sports 2021, 42, 199–214. [Google Scholar] [CrossRef]
- Swanson, K.; Wu, E.; Zhang, A.; Alizadeh, A.A.; Zou, J. From patterns to patients: Advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell 2023, 186, 1772–1791. [Google Scholar] [CrossRef] [PubMed]
- Guan, C.; Gong, A.; Zhao, Y.; Yin, C.; Geng, L.; Liu, L.; Yang, X.; Lu, J.; Xiao, B. Interpretable machine learning model for new-onset atrial fibrillation prediction in critically ill patients: A multi-center study. Crit. Care 2024, 28, 349. [Google Scholar] [CrossRef]
- Yang, H.; Yu, B.; Ou, P.; Li, X.; Lai, X.; Zhang, G.; Zhang, H. Machine learning-aided risk prediction for metabolic syndrome based on 3 years study. Sci. Rep. 2022, 12, 2248. [Google Scholar] [CrossRef]
- Shin, H.; Shim, S.; Oh, S. Machine learning-based predictive model for prevention of metabolic syndrome. PLoS ONE 2023, 18, e0286635. [Google Scholar] [CrossRef]
- Trigka, M.; Dritsas, E. Predicting the Occurrence of Metabolic Syndrome Using Machine Learning Models. Computation 2023, 11, 170. [Google Scholar] [CrossRef]
- Li, Z.; Wu, W.; Kang, H. Machine Learning-Driven Metabolic Syndrome Prediction: An International Cohort Validation Study. Healthcare 2024, 12, 2527. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Rana, S.; Pathak, P. Artificial intelligence in the management of metabolic disorders: A comprehensive review. J. Endocrinol. Investig. 2025, 48, 1525–1538. [Google Scholar] [CrossRef]
- Choubey, U.; Upadrasta, V.A.; Kaur, I.P.; Banker, H.; Kanagala, S.G.; Anamika, F.N.U.; Virmani, M.; Jain, R. From prevention to management: Exploring AI’s role in metabolic syndrome management: A comprehensive review. Egypt. J. Intern. Med. 2024, 36, 106. [Google Scholar] [CrossRef]
- Lee, M.; Park, T.; Shin, J.Y.; Park, M. A comprehensive multi-task deep learning approach for predicting metabolic syndrome with genetic, nutritional, and clinical data. Sci. Rep. 2024, 14, 17851. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Z.; Liu, C.; Sun, H.; Li, X.; Yang, Y. Integrating Artificial Intelligence in the Diagnosis and Management of Metabolic Syndrome: A Comprehensive Review. Diabetes Metab. Res. Rev. 2025, 41, e70039. [Google Scholar] [CrossRef]
- Hossain, M.F.; Hossain, S.; Akter, M.N.; Nahar, A.; Liu, B.; Faruque, M.O. Metabolic syndrome predictive modelling in Bangladesh applying machine learning approach. PLoS ONE 2024, 19, e0309869. [Google Scholar] [CrossRef] [PubMed]
- Shin, D. Prediction of metabolic syndrome using machine learning approaches based on genetic and nutritional factors: A 14-year prospective-based cohort study. BMC Med. Genom. 2024, 17, 224. [Google Scholar] [CrossRef] [PubMed]
- Goldman, O.; Ben-Assuli, O.; Ababa, S.; Rogowski, O.; Berliner, S. Predicting metabolic syndrome: Machine learning techniques for improved preventive medicine. Health Inform. J. 2025, 31. [Google Scholar] [CrossRef]
- Park, J.E.; Mun, S.; Lee, S. Metabolic Syndrome Prediction Models Using Machine Learning and Sasang Constitution Type. Evid. Based Complement. Altern. Med. 2021, 2021, 8315047. [Google Scholar] [CrossRef]
- Afzal, Z.; Schuemie, M.J.; van Blijderveen, J.C.; Sen, E.F.; Sturkenboom, M.C.; Kors, J.A. Improving sensitivity of machine learning methods for automated case identification from free-text electronic medical records. BMC Med. Inf. Decis. Mak. 2013, 13, 30. [Google Scholar] [CrossRef]
- Novianti, P.W.; Jong, V.L.; Roes, K.C.; Eijkemans, M.J. Factors affecting the accuracy of a class prediction model in gene expression data. BMC Bioinform. 2015, 16, 199. [Google Scholar] [CrossRef] [PubMed]
- Panagoulias, D.P.; Sotiropoulos, D.N.; Tsihrintzis, G.A. SVM-Based Blood Exam Classification for Predicting Defining Factors in Metabolic Syndrome Diagnosis. Electronics 2022, 11, 857. [Google Scholar] [CrossRef]
- Thomas, D.M.; Bredlau, C.; Bosy-Westphal, A.; Mueller, M.; Shen, W.; Gallagher, D.; Maeda, Y.; McDougall, A.; Peterson, C.M.; Ravussin, E.; et al. Relationships between body roundness with body fat and visceral adipose tissue emerging from a new geometrical model. Obesity 2013, 21, 2264–2271. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ma, N.; Lin, Q.; Chen, K.; Zheng, F.; Wu, J.; Dong, X.; Niu, W. Body Roundness Index and All-Cause Mortality Among US Adults. JAMA Netw. Open 2024, 7, e2415051. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, L.; Wu, Q.; Zhou, Y.; Jin, Z.; Li, Z.; Zhu, Y. Body roundness index is a superior indicator to associate with the cardio-metabolic risk: Evidence from a cross-sectional study with 17,000 Eastern-China adults. BMC Cardiovasc. Disord. 2021, 21, 97. [Google Scholar] [CrossRef] [PubMed]
- Rico-Martín, S.; Calderón-García, J.F.; Sánchez-Rey, P.; Franco-Antonio, C.; Alvarez, M.M.; Muñoz-Torrero, J.F.S. Effectiveness of body roundness index in predicting metabolic syndrome: A systematic review and meta-analysis. Obes. Rev. 2020, 21, e13023. [Google Scholar] [CrossRef]
- Reyes-Barrera, J.; Sainz-Escárrega, V.H.; Medina-Urritia, A.X.; Jorge-Galarza, E.; Osorio-Alonso, H.; Torres-Tamayo, M.; Leal-Escobar, G.; Posadas-Romero, C.; Torre-Villalvazo, I.; Juárez-Rojas, J.G. Dysfunctional adiposity index as a marker of adipose tissue morpho-functional abnormalities and metabolic disorders in apparently healthy subjects. Adipocyte 2021, 10, 142–152. [Google Scholar] [CrossRef]
- Bilgin Göçer, D.; Baş, M.; Çakır Biçer, N.; Hajhamidiasl, L. Predicting metabolic syndrome by visceral adiposity index, body roundness index, dysfunctional adiposity index, lipid accumulation product index, and body shape index in adults. Nutr. Hosp. 2022, 39, 794–802. [Google Scholar]
- Keyif, B.; Yavuzcan, A. Visceral and Dysfunctional Adiposity Indices as Predictors of Insulin Resistance and Metabolic Syndrome in Women with Polycystic Ovary Syndrome: A Cross-Sectional Study. Medicina 2025, 61, 424. [Google Scholar] [CrossRef]
- Heo, A.-S.; Lee, J.-C. Development of a Metabolic Syndrome Prediction Model Using HOMA-IR and Multivariate Factors. Appl. Sci. 2025, 15, 2985. [Google Scholar] [CrossRef]
- Shim, W.S.; Kim, H.J.; Kang, E.S.; Rhee, Y.M.; Ahn, C.W.; Lim, S.K.; Lee, H.C.; Cha, B.S. The Relationship between Metabolic Syndrome and Small Dense Low Density Lipoprotein-Cholesterol. Diabetes Metab. J. 2005, 29, 548–556. [Google Scholar]
- Zhu, X.; Chen, Y.; Zhu, M.; Hu, J. The Relationship Between Small Dense Low-Density Lipoprotein Cholesterol and Metabolic Syndrome. Diabetes Metab. Syndr. Obes. 2024, 17, 1523–1532. [Google Scholar] [CrossRef]
- Fan, J.; Liu, Y.; Yin, S.; Chen, N.; Bai, X.; Ke, Q.; Shen, J.; Xia, M. Small dense LDL cholesterol is associated with metabolic syndrome traits independently of obesity and inflammation. Nutr. Metab. 2019, 16, 7. [Google Scholar] [CrossRef]
- Matsuzawa, Y.; Funahashi, T.; Kihara, S.; Shimomura, I. Adiponectin and metabolic syndrome. Arter. Thromb. Vasc. Biol. 2004, 24, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Fisman, E.Z.; Tenenbaum, A. Adiponectin: A manifold therapeutic target for metabolic syndrome, diabetes, and coronary disease? Cardiovasc. Diabetol. 2014, 13, 103. [Google Scholar] [CrossRef] [PubMed]
- Frühbeck, G.; Catalán, V.; Rodríguez, A.; Gómez-Ambrosi, J. Adiponectin-leptin ratio: A promising index to estimate adipose tissue dysfunction. Relation with obesity-associated cardiometabolic risk. Adipocyte 2018, 7, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Frühbeck, G.; Catalán, V.; Rodríguez, A.; Ramírez, B.; Becerril, S.; Salvador, J.; Portincasa, P.; Colina, I.; Gómez-Ambrosi, J. Involvement of the leptin-adiponectin axis in inflammation and oxidative stress in the metabolic syndrome. Sci. Rep. 2017, 7, 6619. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.M.I. Therapeutic potential of adiponectin in prediabetes: Strategies, challenges, and future directions. Ther. Adv. Endocrinol. Metab. 2024, 18, 15. [Google Scholar] [CrossRef]
- Sekgala, M.D.; Sewpaul, R.; Kengne, A.P.; Mchiza, Z.; Peer, N. Clinical utility of novel anthropometric indices in identifying type 2 diabetes mellitus among South African adult females. BMC Public Health 2024, 24, 2676. [Google Scholar] [CrossRef]
- Mather, K.J.; Goldberg, R.B. Clinical use of adiponectin as a marker of metabolic dysregulation. Best. Pract. Res. Clin. Endocrinol. Metab. 2014, 28, 107–117. [Google Scholar] [CrossRef]
- Liu, Y.; Vu, V.; Sweeney, G. Examining the Potential of Developing and Implementing Use of Adiponectin-Targeted Therapeutics for Metabolic and Cardiovascular Diseases. Front. Endocrinol. 2019, 11, 842. [Google Scholar] [CrossRef]
- Byeon, H. Explainable AI using dart booster and lime algorithms for metabolic syndrome prediction in Korean adults. ICIC Express Lett. 2024, 15, 1125–1133. [Google Scholar]
- Zhang, Y.; Zhang, X.; Razbek, J.; Li, D.; Xia, W.; Bao, L.; Mao, H.; Daken, M.; Cao, M. Opening the black box: Interpretable machine learning for predictor finding of metabolic syndrome. BMC Endocr. Disord. 2022, 22, 214. [Google Scholar] [CrossRef] [PubMed]
- Gayoso-Diz, P.; Otero-González, A.; Rodriguez-Alvarez, M.X.; Gude, F.; García, F.; De Francisco, A.; Quintela, A.G. Insulin resistance (HOMA-IR) cut-off values and the metabolic syndrome in a general adult population: Effect of gender and age: EPIRCE cross-sectional study. BMC Endocr. Disord. 2013, 16, 47. [Google Scholar]
- de Luis, D.; Muñoz, M.; Izaola, O.; Gomez, J.J.L.; Rico, D.; Primo, D. Body Roundness Index (BRI) Predicts Metabolic Syndrome in Postmenopausal Women with Obesity Better than Insulin Resistance. Diabetology 2025, 6, 60. [Google Scholar] [CrossRef]
- Cahyaningsih, I.; Rokhman, M.R.; Sudikno; Postma, M.J.; van der Schans, J. Accuracy of the Modified Finnish Diabetes Risk Score (Modified FINDRISC) for detecting metabolic syndrome: Findings from the Indonesian national health survey. PLoS ONE 2025, 20, e0314824. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Romero, F.; Rodríguez-Morán, M. Abdominal volume index. An anthropometry-based index for estimation of obesity is strongly related to impaired glucose tolerance and type 2 diabetes mellitus. Arch. Med. Res. 2003, 34, 428–432. [Google Scholar] [CrossRef]
- Corona-Meraz, F.I.; Navarro-Hernández, R.E.; Ruíz-Quezada, S.L.; Madrigal-Ruíz, P.M.; Castro-Albarrán, J.; Chavarría-Ávila, E.; Guzmán-Ornelas, M.O.; Gómez-Bañuelos, E.; Petri, M.H.; Ramírez-Cedano, J.I.; et al. Inverse Relationship of the CMKLR1 Relative Expression and Chemerin Serum Levels in Obesity with Dysmetabolic Phenotype and Insulin Resistance. Mediat. Inflamm. 2016, 2016, 3085390. [Google Scholar] [CrossRef]
- Diaz-Rubio, G.I.; Corona-Meraz, F.-I.; Madrigal-Ruiz, P.-M.; Anda, J.-A.R.-D.; Gómez-Bañuelos, E.; Castro-Albarran, J.; Flores-Alvarado, L.-J.; Mercado, M.V.-D.; Pérez-Vázquez, F.d.J.; Pizano-Martínez, O.-E.; et al. CCR2/CCL2 and CMKLR1/RvE1 chemokines system levels are associated with insulin resistance in rheumatoid arthritis. PLoS ONE 2021, 16, e0246054. [Google Scholar] [CrossRef]
- Corona-Meraz, F.-I.; Mercado, M.V.-D.; Ortega, F.J.; Ruiz-Quezada, S.-L.; Guzmán-Ornelas, M.-O.; Navarro-Hernández, R.-E. Ageing influences the relationship of circulating miR-33a and miR-33b levels with insulin resistance and adiposity. Diabetes Vasc. Dis. Res. 2019, 16, 244–253. [Google Scholar] [CrossRef]
- Khairy, P.; Ouyang, D.W.; Fernandes, S.M.; Lee-Parritz, A.; Economy, K.E.; Landzberg, M.J. Pregnancy outcomes in women with congenital heart disease. Circulation 2006, 113, 517–524. [Google Scholar] [CrossRef]
- Krakauer, N.Y.; Krakauer, J.C. A new body shape index predicts mortality hazard independently of body mass index. PLoS ONE 2012, 7, e39504. [Google Scholar] [CrossRef]
- Freedman, D.S.; Thornton, J.C.; Pi-Sunyer, F.X.; Heymsfield, S.B.; Wang, J.; Pierson, R.N.; Blanck, H.M., Jr.; Gallagher, D. The body adiposity index (hip circumference ÷ height(1.5)) is not a more accurate measure of adiposity than is BMI, waist circumference, or hip circumference. Obesity 2012, 20, 2438–2444. [Google Scholar] [CrossRef] [PubMed]
- Valdez, R. A simple model-based index of abdominal adiposity. J. Clin. Epidemiol. 1991, 44, 955–956. [Google Scholar] [CrossRef] [PubMed]
- Vicente-Herrero, M.T.; Egea-Sancho, M.; Ramírez-Iñiguez-de la Torre, M.V.; López-González, Á.A. Relación predictiva de los índices de adiposidad visceral y adiposidad disfuncional con el riesgo cardiovascular en población laboral. Rev. Clínica Med. Fam. 2023, 16, 318–324. [Google Scholar] [CrossRef]
- Amato, M.C.; Giordano, C.; Galia, M.; Criscimanna, A.; Vitabile, S.; Midiri, M.; Galluzzo, A.; AlkaMeSy Study, G. Visceral Adiposity Index: A reliable indicator of visceral fat function associated with cardiometabolic risk. Diabetes Care 2010, 33, 920–922. [Google Scholar] [CrossRef]
- Grundy, S.M.; Brewer, H.B., Jr.; Cleeman, J.I.; Smith, S.C., Jr.; Lenfant, C.; American Heart Association; National Heart, Lung, and Blood Institute. Definition of metabolic syndrome: Report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Circulation 2004, 109, 433–438. [Google Scholar] [CrossRef]
- Huang, P.L. A comprehensive definition for metabolic syndrome. Dis. Model. Mech. 2009, 2, 231–237. [Google Scholar] [CrossRef]
- Saklayen, M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef]
- Awad, M.; Fraihat, S. Recursive Feature Elimination with Cross-Validation with Decision Tree: Feature Selection Method for Machine Learning-Based Intrusion Detection Systems. J. Sens. Actuator Netw. 2023, 12, 67. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer Nature: Berlin, Germany, 2013; pp. 247–518. [Google Scholar]
- Giger, M.; Csillaghy, A. Unsupervised Anomaly Detection With Variational Autoencoders Applied to Full-Disk Solar Images. Space Weather 2024, 22, 2. [Google Scholar] [CrossRef]
- Ferdowsy, F.; Rahi, K.S.A.; Jabiullah, M.I.; Habib, M.T. A machine learning approach for obesity risk prediction. Curr. Res. Behav. Sci. 2021, 2, 100053. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2009; Volume VIII, p. 213. [Google Scholar]
- Rainio, O.; Teuho, J.; Klén, R. Evaluation metrics and statistical tests for machine learning. Sci. Rep. 2024, 14, 6086, Correction in Sci. Rep. 2024, 14, 15724. [Google Scholar] [CrossRef] [PubMed]
- Shipe, M.E.; Deppen, S.A.; Farjah, F.; Grogan, E.L. Developing prediction models for clinical use using logistic regression: An overview. J. Thorac. Dis. 2019, 11, S574–S584. [Google Scholar] [CrossRef]
- Wang, B.; Liu, Y.; Sun, X.; Yin, Z.; Li, H.; Ren, Y.; Zhao, Y.; Zhang, R.; Zhang, M.; Hu, D. Prediction model and assessment of probability of incident hypertension: The Rural Chinese Cohort Study. J. Hum. Hypertens. 2021, 35, 74–84. [Google Scholar] [CrossRef]
- Huang, S.; Ma, J.-W.; Dai, H.; Luo, L. A new in-hospital mortality prediction nomogram for intensive care unit patients with acute pancreatitis. Arch. Med. Sci. 2023, 20, 61–70. [Google Scholar] [CrossRef]
- Liu, Y.; Lyu, X.; Yang, B.; Fang, Z.; Hu, D.; Shi, L.; Wu, B.; Tian, Y.; Zhang, E.; Yang, Y. Early Triage of Critically Ill Adult Patients With Mushroom Poisoning: Machine Learning Approach. JMIR Form. Res. 2023, 21, e44666. [Google Scholar] [CrossRef]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef]
- Vickers, A.J.; van Calster, B.; Steyerberg, E.W. A simple, step-by-step guide to interpreting decision curve analysis. Diagn. Progn. Res. 2019, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, M.; Singh, S.; Guestrin, C. "Why Should I Trust You?": Explaining the Predictions of Any Classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 1135–1144. [Google Scholar]
- Valdez Vega, R.I.; Noboa Velastegui, J.A.; Torres Carrillo, N.M.; Fletes Rayas, A.L.; Ruiz Quezada, S.L.; Ramos Marquez, M.E.; Alvarez Perez, I.; Navarra Hernández, R.E. Parameters to Predicting Metabolic Syndrome by Improves the Performance of the Dysfunctional Adipose Index. Presented at the European Congress of Obesity, Málaga, Spain, 11–14 May 2025. [Google Scholar]


| Groups | MetS | Non-MetS | p Value |
|---|---|---|---|
| n | 84 | 297 | |
| Sex (Men/Women) | 29:55 | 110:187 | |
| Age | 44 ± 14 | 35 ± 19 | |
| Blood Pressure (mmHg) | |||
| Systolic | 124 (13) | 108 (15) | <0.001 |
| Diastolic | 81 (14) | 71 (15) | <0.001 |
| Lipid Profile | |||
| Apo A | 129.5 (31.75) | 135.0 (50.25) | 0.207 |
| Apo B | 104 (52) | 88.5 (50.25) | 0.089 |
| HDL-C (mg/dL) | 38.6 (8.6) | 43.0 (13.3) | <0.001 |
| LDL-C (mg/dL) | 110.9 (68.7) | 101.4 (54.5) | 0.043 |
| sdLDL-C (mg/dL) | 44.1 (27.9) | 23.1 (27.82) | <0.001 |
| Total cholesterol (mg/dL) | 211 (57) | 192 (53) | <0.001 |
| Triglycerides (mg/dL) | 252 (134) | 119 (64) | <0.001 |
| Insulin Resistance Status | |||
| Fasting glucose (mg/dL) | 101.0 (22.5) | 86.0 (22.0) | <0.001 |
| Fasting insulin (μUI/mL) | 20.6 (17.5) | 12.8 (11.7) | <0.001 |
| HOMA-IR | 5.1 (3.9) | 2.5 (2.4) | <0.001 |
| HOMA-B | 15.2 (15.4) | 15.7 (14.6) | 0.008 |
| QUICKI | 0.31 (0.04) | 0.35 (0.06) | <0.001 |
| Body Adiposity Status Evaluation | |||
| ABSI | 0.080 (0.008) | 0.80 (0.009) | 0.596 |
| AVI | 17.58 (8.53) | 16.07 (17.72) | 0.531 |
| BAI | 31.25 (6.94) | 31.36 (9.32) | 0.666 |
| BFI | 8.88 (5.15) | 8.29 (5.84) | 0.334 |
| BFR | 8.88 (5.15) | 8.29 (5.73) | 0.302 |
| BMI (kg/m2) | 33.3 (6.9) | 26.7 (6.1) | <0.001 |
| BRI | 1.5 (0.9) | 0.5 (0.6) | <0.001 |
| CI | 1.25 (0.163) | 1.23 (0.168) | 0.518 |
| DAI | 2.7 (1.8) | 1.3 (0.9) | <0.001 |
| TAA | 688.00 (341) | 636.50 (284.5) | 0.533 |
| VAI | 0.20 (0.200) | 0.20 (0.198) | 0.315 |
| WC | 107.3 (15.5) | 82.8 (18.9) | <0.001 |
| Adipokines and inflammation markers | |||
| AdipoQT (ng/mL) | 4252 (4125) | 7108 (4728) | <0.001 |
| AdipoQHMW (ng/mL) | 981 (1199) | 2582 (2991) | <0.001 |
| Chemerin (ng/mL) | 129 (66) | 147 (89) | 0.191 |
| Irisin | 4560 (1994) | 4889 (2124) | 0.380 |
| CRP | 6.45 (5.40) | 5.48 (4.32) | 0.058 |
| C3 | 108.5 (47.25) | 102.0 (42) | 0.232 |
| Full Set | Reduced Set | ||||
|---|---|---|---|---|---|
| AUC-ROC | Accuracy | Size | AUC-ROC | Accuracy | |
| LR | 0.865 (±0.098) | 0.793 (±0.073) | 6 | 0.897 (±0.063) | 0.8127 (±0.069) |
| SVM | 0.861 (±0.075) | 0.778 (±0.077) | 7 | 0.882 (±0.073) | 0.7959 (±0.078) |
| XGboost | 0.881 (±0.058) | 0.851 (±0.055) | 10 | 0.882 (±0.063) | 0.854 (±0.052) |
| RF | 0.882 (±0.067) | 0.864 (±0.045) | 7 | 0.893 (±0.061) | 0.875 (±0.041) |
| Metrics | LR | RF | XGBoost | SVM |
|---|---|---|---|---|
| AUC-ROC | 0.910 (±0.048) | 0.886 (±0.068) | 0.867 (±0.063) | 0.881 (±0.068) |
| Sensitivity | 0.816 (±0.069) | 0.781 (±0.088) | 0.759 (±0.090) | 0.798 (±0.162) |
| Specificity | 0.859 (±0.146) | 0.827 (±0.155) | 0.778 (±0.179) | 0.795 (±0.098) |
| Metrics (95%, CI) | LR | RF | XGBoost | SVM |
|---|---|---|---|---|
| Accuracy | 0.86 (0.74–0.94) | 0.88 (0.76–0.96) | 0.80 (0.67–0.90) | 0.83 (0.70–0.92) |
| Sensitivity (Recall) | 0.85 (0.62–0.97) | 0.85 (0.62, 0.97) | 0.70 (0.46–0.88) | 0.75 (0.51–0.91) |
| Specificity | 0.87 (0.71–0.96) | 0.91 (0.75, 0.98) | 0.88 (0.71, 0.96) | 0.88 (0.71, 0.96) |
| PPV | 0.80 (0.58–0.95) | 0.85 (0.62, 0.97) | 0.78 (0.52, 0.94) | 0.79 (0.54–0.94) |
| NPV | 0.90 (0.74, 0.98) | 0.91 (0.75, 0.98) | 0.82 (0.65, 0.93) | 0.85 (0.68–0.95) |
| Balanced Accuracy | 0.86 (0.74, 0.94) | 0.88 (0.77, 0.96) | 0.81 (0.67, 0.90) | 0.83 (0.70–0.92) |
| AUC-ROC | 0.93 (0.86–0.99) | 0.94 (0.88–1.00) | 0.954 (0.87–1.00) | 0.931 (0.86–0.99) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdez Vega, R.I.; Noboa-Velástegui, J.A.; Fletes-Rayas, A.L.; Álvarez, I.; Ramos-Marquez, M.E.; Ruíz-Quezada, S.L.; Torres-Carrillo, N.M.; Navarro-Hernández, R.E. Predicting Metabolic Syndrome Using Supervised Machine Learning: A Multivariate Parameter Approach. Int. J. Mol. Sci. 2025, 26, 9897. https://doi.org/10.3390/ijms26209897
Valdez Vega RI, Noboa-Velástegui JA, Fletes-Rayas AL, Álvarez I, Ramos-Marquez ME, Ruíz-Quezada SL, Torres-Carrillo NM, Navarro-Hernández RE. Predicting Metabolic Syndrome Using Supervised Machine Learning: A Multivariate Parameter Approach. International Journal of Molecular Sciences. 2025; 26(20):9897. https://doi.org/10.3390/ijms26209897
Chicago/Turabian StyleValdez Vega, Rodolfo Iván, Jacqueline Alejandra Noboa-Velástegui, Ana Lilia Fletes-Rayas, Iñaki Álvarez, Martha Eloisa Ramos-Marquez, Sandra Luz Ruíz-Quezada, Nora Magdalena Torres-Carrillo, and Rosa Elena Navarro-Hernández. 2025. "Predicting Metabolic Syndrome Using Supervised Machine Learning: A Multivariate Parameter Approach" International Journal of Molecular Sciences 26, no. 20: 9897. https://doi.org/10.3390/ijms26209897
APA StyleValdez Vega, R. I., Noboa-Velástegui, J. A., Fletes-Rayas, A. L., Álvarez, I., Ramos-Marquez, M. E., Ruíz-Quezada, S. L., Torres-Carrillo, N. M., & Navarro-Hernández, R. E. (2025). Predicting Metabolic Syndrome Using Supervised Machine Learning: A Multivariate Parameter Approach. International Journal of Molecular Sciences, 26(20), 9897. https://doi.org/10.3390/ijms26209897






