Assessment of Microbiome-Based Pathogen Detection Using Illumina Short-Read and Nanopore Long-Read Sequencing in 144 Patients Undergoing Bronchoalveolar Lavage in a University Hospital in Germany
Abstract
1. Introduction
2. Results
2.1. Culture-Based Diagnostics
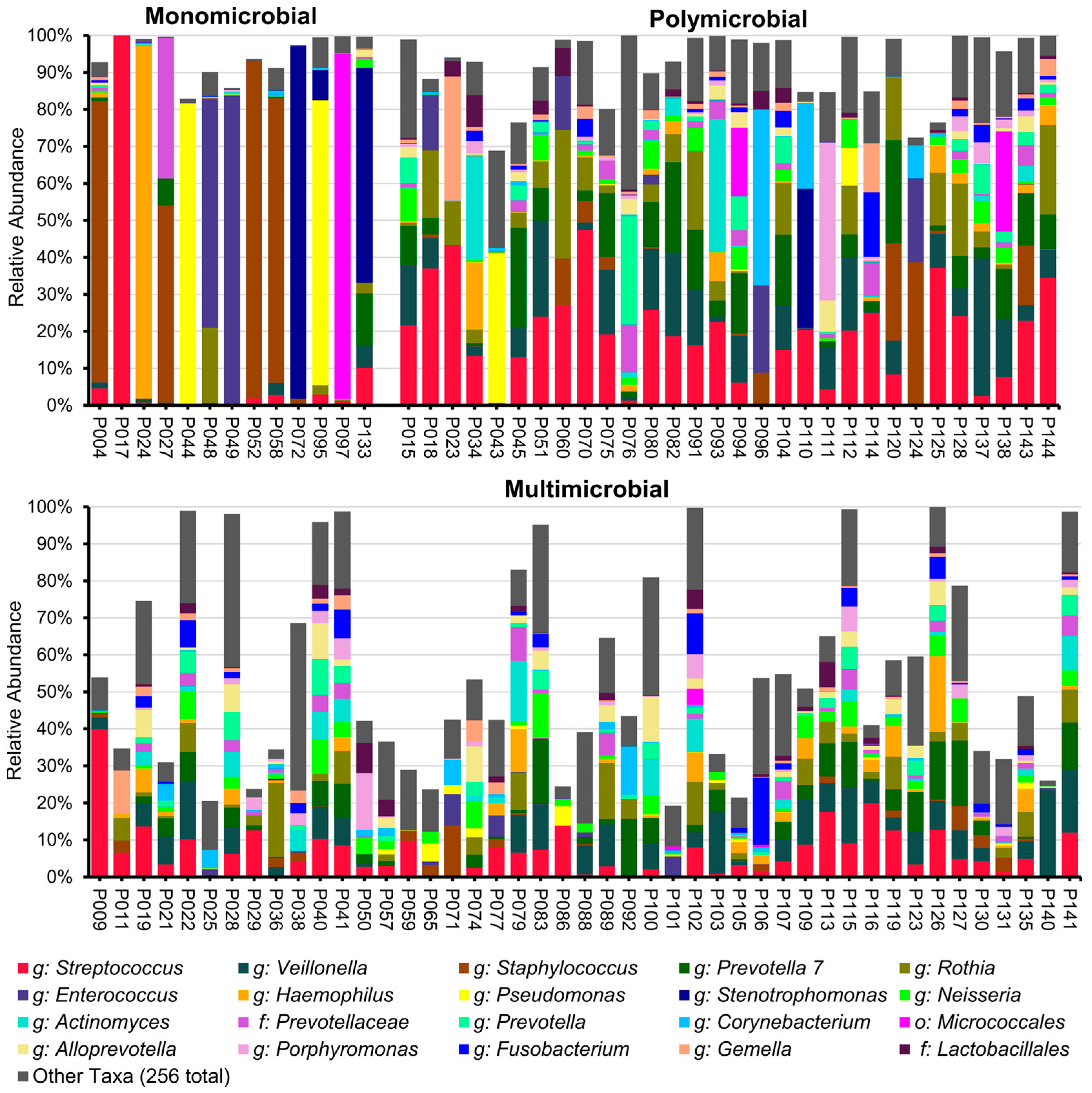
2.2. Results of Illumina 16S rRNA Gene V4 Region Sequencing
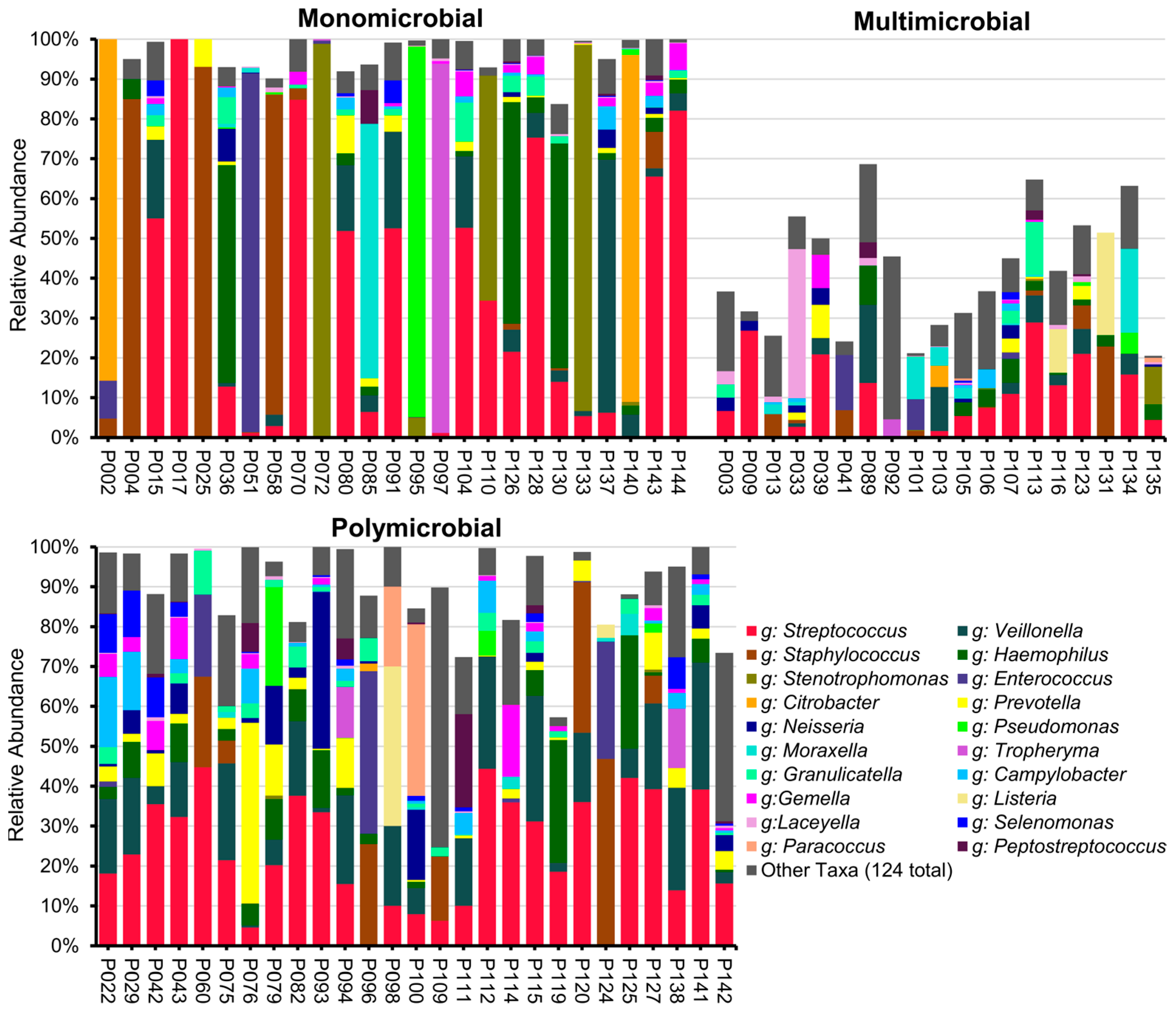
2.3. Results of ONT 16S rRNA Gene Full-Length Sequencing
2.4. DNA Yield of BALF Samples
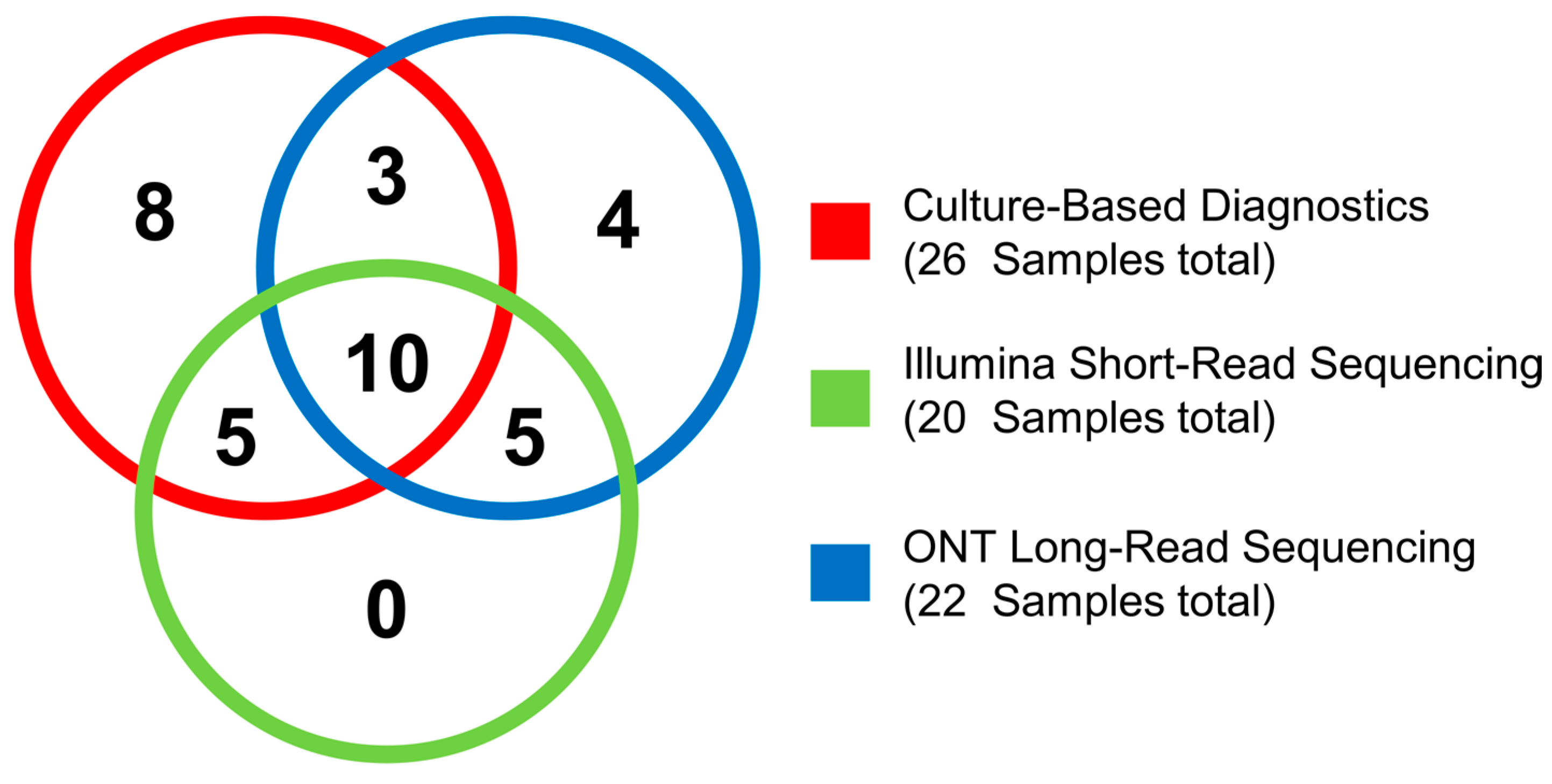
2.5. Validation of Culture-Based Diagnostics with NGS
2.6. Expanding Bacterial Pathogen Detection by NGS
3. Discussion
4. Materials and Methods
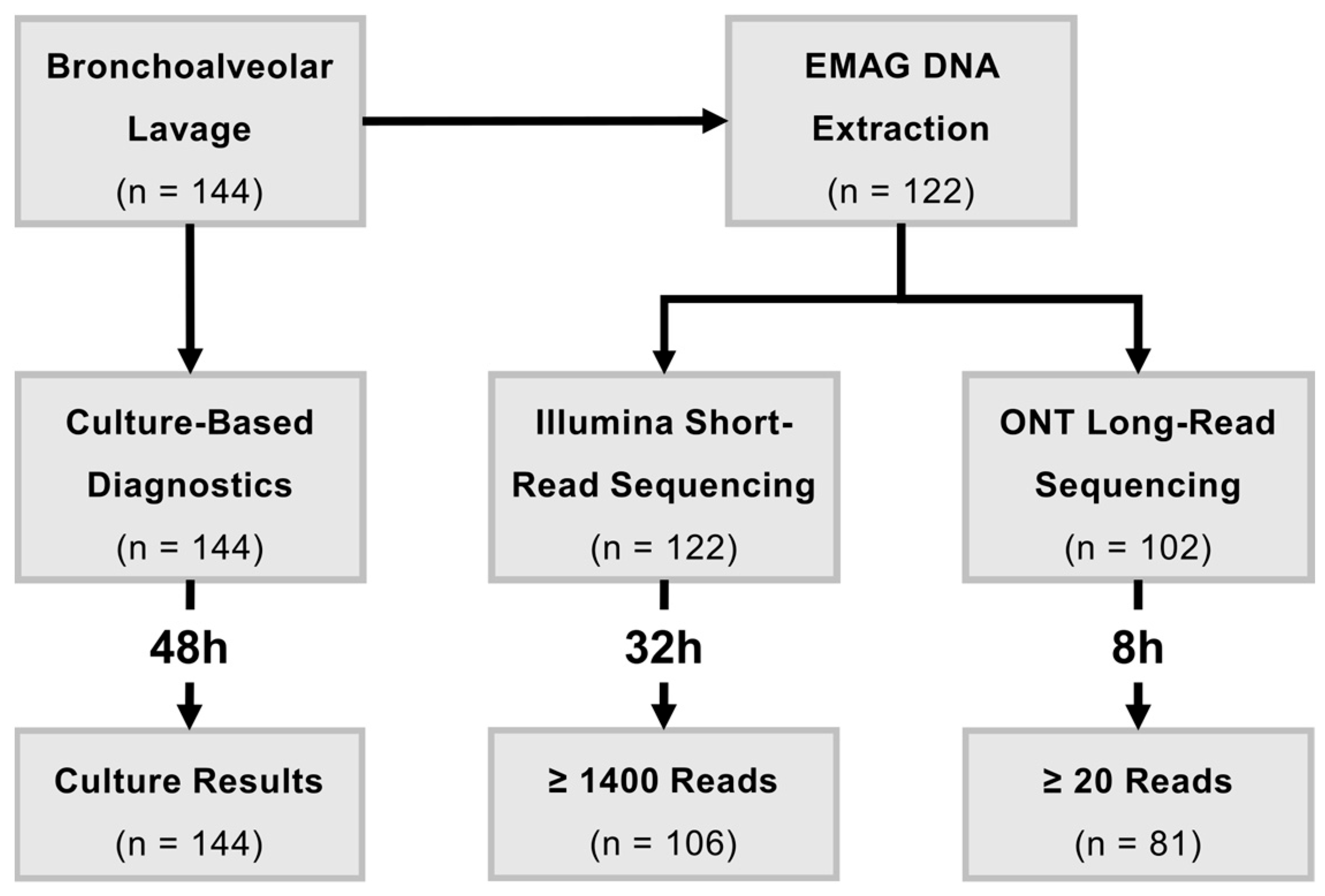
4.1. Sample Acquisition and Culture-Based Diagnostics
4.2. DNA Extraction
4.3. Control of Contaminations
4.4. Short-Read Sequencing with Illumina
4.5. Bioinformatic Analysis of Illumina Short-Read Sequencing Data
4.6. Long-Read Sequencing with Oxford Nanopore Technologies
4.7. Bioinformatic Analysis of ONT Long-Read Sequencing Data
4.8. Comparitive Microbiome Analysis
4.9. Statistic Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BALF | Bronchoalveolar lavage fluid |
| IQR | Interquartile range |
| LRTI | Lower respiratory tract infection |
| NGS | Next-generation sequencing |
| ONT | Oxford Nanopore Technologies |
| OTU | Operational taxonomic unit |
References
- Marsland, B.J.; Trompette, A.; Gollwitzer, E.S. The Gut–Lung Axis in Respiratory Disease. Ann. Am. Thorac. Soc. 2015, 12, S150–S156. [Google Scholar] [CrossRef]
- Natalini, J.G.; Singh, S.; Segal, L.N. The Dynamic Lung Microbiome in Health and Disease. Nat. Rev. Microbiol. 2023, 21, 222–235. [Google Scholar] [CrossRef]
- Beck, J.M.; Young, V.B.; Huffnagle, G.B. The Microbiome of the Lung. Transl. Res. 2012, 160, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Fromentin, M.; Ricard, J.-D.; Roux, D. Respiratory Microbiome in Mechanically Ventilated Patients: A Narrative Review. Intensive Care Med. 2021, 47, 292–306. [Google Scholar] [CrossRef] [PubMed]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The Respiratory Tract Microbiome and Lung Inflammation: A Two-Way Street. Mucosal Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Mendez, R.; Banerjee, S.; Bhattacharya, S.K.; Banerjee, S. Lung Inflammation and Disease: A Perspective on Microbial Homeostasis and Metabolism. IUBMB Life 2019, 71, 152–165. [Google Scholar] [CrossRef]
- Moffatt, M.F.; Cookson, W.O. The Lung Microbiome in Health and Disease. Clin. Med. 2017, 17, 525–529. [Google Scholar] [CrossRef]
- Yagi, K.; Huffnagle, G.B.; Lukacs, N.W.; Asai, N. The Lung Microbiome during Health and Disease. Int. J. Mol. Sci. 2021, 22, 10872. [Google Scholar] [CrossRef]
- Martin-Loeches, I.; Dickson, R.; Torres, A.; Hanberger, H.; Lipman, J.; Antonelli, M.; de Pascale, G.; Bozza, F.; Vincent, J.L.; Murthy, S.; et al. The Importance of Airway and Lung Microbiome in the Critically Ill. Crit. Care 2020, 24, 537. [Google Scholar] [CrossRef]
- Wu, B.G.; Segal, L.N. The Lung Microbiome and Its Role in Pneumonia. Clin. Chest Med. 2018, 39, 677–689. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef] [PubMed]
- Molyneaux, P.L.; Mallia, P.; Cox, M.J.; Footitt, J.; Willis-Owen, S.A.G.; Homola, D.; Trujillo-Torralbo, M.-B.; Elkin, S.; Kon, O.M.; Cookson, W.O.C.; et al. Outgrowth of the Bacterial Airway Microbiome after Rhinovirus Exacerbation of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2013, 188, 1224. [Google Scholar] [CrossRef] [PubMed]
- Man, W.H.; de Steenhuijsen Piters, W.A.A.; Bogaert, D. The Microbiota of the Respiratory Tract: Gatekeeper to Respiratory Health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- GBD 2016 Lower Respiratory Infections Collaborators Estimates of the Global, Regional, and National Morbidity, Mortality, and Aetiologies of Lower Respiratory Infections in 195 Countries, 1990–2016: A Systematic Analysis for the Global Burden of Disease Study 2016. Lancet Infect. Dis. 2018, 18, 1191–1210. [CrossRef]
- Bender, R.G.; Sirota, S.B.; Swetschinski, L.R.; Dominguez, R.-M.V.; Novotney, A.; Wool, E.E.; Ikuta, K.S.; Vongpradith, A.; Rogowski, E.L.B.; Doxey, M.; et al. Global, Regional, and National Incidence and Mortality Burden of Non-COVID-19 Lower Respiratory Infections and Aetiologies, 1990–2021: A Systematic Analysis from the Global Burden of Disease Study 2021. Lancet Infect. Dis. 2024, 24, 974–1002. [Google Scholar] [CrossRef]
- Kalantar, K.L.; Langelier, C.R. Host-Microbe Metagenomics: A Lens To Refocus Our Perspective on Infectious and Inflammatory Diseases. mSystems 2021, 6, e0040421. [Google Scholar] [CrossRef]
- Shoar, S.; Musher, D.M. Etiology of Community-Acquired Pneumonia in Adults: A Systematic Review. Pneumonia 2020, 12, 11. [Google Scholar] [CrossRef]
- Gu, W.; Deng, X.; Lee, M.; Sucu, Y.D.; Arevalo, S.; Stryke, D.; Federman, S.; Gopez, A.; Reyes, K.; Zorn, K.; et al. Rapid Pathogen Detection by Metagenomic Next-Generation Sequencing of Infected Body Fluids. Nat. Med. 2021, 27, 115–124. [Google Scholar] [CrossRef]
- Zhu, N.; Zhou, D.; Yuan, R.; Ruzetuoheti, Y.; Li, J.; Zhang, X.; Li, S. Identification and Comparison of Chlamydia Psittaci, Legionella and Mycoplasma Pneumonia Infection. Clin. Respir. J. 2023, 17, 384–393. [Google Scholar] [CrossRef]
- Laboratory Diagnosis of Infectious Diseases. Available online: https://accessmedicine.mhmedical.com/content.aspx?sectionid=265477006&bookid=3095 (accessed on 10 September 2025).
- Bador, J.; Nicolas, B.; Chapuis, A.; Varin, V.; Dullier-Taillefumier, N.; de Curraize, C.; Amoureux, L.; Putot, A.; Neuwirth, C. 16S rRNA PCR on Clinical Specimens: Impact on Diagnosis and Therapeutic Management. Med. Mal. Infect. 2020, 50, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Charalampous, T.; Kay, G.L.; Richardson, H.; Aydin, A.; Baldan, R.; Jeanes, C.; Rae, D.; Grundy, S.; Turner, D.J.; Wain, J.; et al. Nanopore Metagenomics Enables Rapid Clinical Diagnosis of Bacterial Lower Respiratory Infection. Nat. Biotechnol. 2019, 37, 783–792. [Google Scholar] [CrossRef]
- Kitsios, G.D. Translating Lung Microbiome Profiles into the Next-Generation Diagnostic Gold Standard for Pneumonia: A Clinical Investigator’s Perspective. mSystems 2018, 3, e00153-17. [Google Scholar] [CrossRef]
- Mahnic, A.; Breznik, V.; Bombek Ihan, M.; Rupnik, M. Comparison Between Cultivation and Sequencing Based Approaches for Microbiota Analysis in Swabs and Biopsies of Chronic Wounds. Front. Med. 2021, 8, 607255. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Prescott, H.C.; Martinez, F.J.; Curtis, J.L.; Lama, V.N.; Huffnagle, G.B. Analysis of Culture-Dependent versus Culture-Independent Techniques for Identification of Bacteria in Clinically Obtained Bronchoalveolar Lavage Fluid. J. Clin. Microbiol. 2014, 52, 3605–3613. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, J.; Zhang, L.; Shen, Z.; Xiao, Y.; Zhang, G.; Chen, M.; Chen, F.; Liu, L.; Wang, Y.; et al. Highly Diverse Sputum Microbiota Correlates with the Disease Severity in Patients with Community-Acquired Pneumonia: A Longitudinal Cohort Study. Respir. Res. 2024, 25, 223. [Google Scholar] [CrossRef] [PubMed]
- Yoo, I.Y.; Kang, O.K.; Lee, M.K.; Kim, Y.J.; Cho, S.Y.; Huh, K.; Kang, C.I.; Chung, D.R.; Peck, K.R.; Huh, H.J.; et al. Comparison of 16S Ribosomal RNA Targeted Sequencing and Culture for Bacterial Identification in Normally Sterile Body Fluid Samples: Report of a 10-Year Clinical Laboratory Review. Ann. Lab. Med. 2020, 40, 63–67. [Google Scholar] [CrossRef]
- Zachariah, P.; Ryan, C.; Nadimpalli, S.; Coscia, G.; Kolb, M.; Smith, H.; Foca, M.; Saiman, L.; Planet, P.J. Culture-Independent Analysis of Pediatric Bronchoalveolar Lavage Specimens. Ann. Am. Thorac. Soc. 2018, 15, 1047–1056. [Google Scholar] [CrossRef]
- Dickson, R.P.; Schultz, M.J.; van der Poll, T.; Schouten, L.R.; Falkowski, N.R.; Luth, J.E.; Sjoding, M.W.; Brown, C.A.; Chanderraj, R.; Huffnagle, G.B.; et al. Lung Microbiota Predict Clinical Outcomes in Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2020, 201, 555–563. [Google Scholar] [CrossRef]
- Li, Y.; Sun, B.; Tang, X.; Liu, Y.-L.; He, H.-Y.; Li, X.-Y.; Wang, R.; Guo, F.; Tong, Z.-H. Application of Metagenomic Next-Generation Sequencing for Bronchoalveolar Lavage Diagnostics in Critically Ill Patients. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 369–374. [Google Scholar] [CrossRef]
- Carney, S.M.; Clemente, J.C.; Cox, M.J.; Dickson, R.P.; Huang, Y.J.; Kitsios, G.D.; Kloepfer, K.M.; Leung, J.M.; LeVan, T.D.; Molyneaux, P.L.; et al. Methods in Lung Microbiome Research. Am. J. Respir. Cell Mol. Biol. 2020, 62, 283–299. [Google Scholar] [CrossRef]
- Wang, J.; Han, Y.; Feng, J. Metagenomic Next-Generation Sequencing for Mixed Pulmonary Infection Diagnosis. BMC Pulm. Med. 2019, 19, 252. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Duan, Z.; Zeng, W.; Xie, S.; Xie, M.; Fu, H.; Ye, Q.; Xu, T.; Xie, L. Clinical Metagenomics Assessments Improve Diagnosis and Outcomes in Community-Acquired Pneumonia. BMC Infect. Dis. 2021, 21, 352. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Falkowski, N.R.; Huffnagle, G.B.; Curtis, J.L. Bacterial Topography of the Healthy Human Lower Respiratory Tract. mBio 2017, 8, e02287-16. [Google Scholar] [CrossRef] [PubMed]
- Wypych, T.P.; Wickramasinghe, L.C.; Marsland, B.J. The Influence of the Microbiome on Respiratory Health. Nat. Immunol. 2019, 20, 1279–1290. [Google Scholar] [CrossRef]
- Gill, C.; van de Wijgert, J.H.H.M.; Blow, F.; Darby, A.C. Evaluation of Lysis Methods for the Extraction of Bacterial DNA for Analysis of the Vaginal Microbiota. PLoS ONE 2016, 11, e0163148. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best Practices for Analysing Microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Allaband, C.; McDonald, D.; Vázquez-Baeza, Y.; Minich, J.J.; Tripathi, A.; Brenner, D.A.; Loomba, R.; Smarr, L.; Sandborn, W.J.; Schnabl, B.; et al. Microbiome 101: Studying, Analyzing, and Interpreting Gut Microbiome Data for Clinicians. Clin. Gastroenterol. Hepatol. 2019, 17, 218–230. [Google Scholar] [CrossRef]
- Drengenes, C.; Eagan, T.M.L.; Haaland, I.; Wiker, H.G.; Nielsen, R. Exploring Protocol Bias in Airway Microbiome Studies: One versus Two PCR Steps and 16S rRNA Gene Region V3 V4 versus V4. BMC Genom. 2021, 22, 3. [Google Scholar] [CrossRef]
- Lao, H.-Y.; Wong, L.L.-Y.; Hui, Y.; Ng, T.T.-L.; Chan, C.T.-M.; Lo, H.W.-H.; Yau, M.C.-Y.; Leung, E.C.-M.; Wong, R.C.-W.; Ho, A.Y.-M.; et al. The Clinical Utility of Nanopore 16S rRNA Gene Sequencing for Direct Bacterial Identification in Normally Sterile Body Fluids. Front. Microbiol. 2024, 14, 1324494. [Google Scholar] [CrossRef]
- Lai, L.M.; Zhu, X.Y.; Zhao, R.; Chen, Q.; Liu, J.J.; Liu, Y.; Yuan, L. Tropheryma Whipplei Detected by Metagenomic Next-Generation Sequencing in Bronchoalveolar Lavage Fluid. Diagn. Microbiol. Infect. Dis. 2024, 109, 116374, Erratum in Diagn. Microbiol. Infect. Dis. 2024, 110, 116393. [Google Scholar] [CrossRef]
- Morgand, M.; Leclercq, A.; Maury, M.M.; Bracq-Dieye, H.; Thouvenot, P.; Vales, G.; Lecuit, M.; Charlier, C. Listeria Monocytogenes-Associated Respiratory Infections: A Study of 38 Consecutive Cases. Clin. Microbiol. Infect. 2018, 24, 1339.e1–1339.e5. [Google Scholar] [CrossRef]
- Chen, Y.; Feng, W.; Ye, K.; Guo, L.; Xia, H.; Guan, Y.; Chai, L.; Shi, W.; Zhai, C.; Wang, J.; et al. Application of Metagenomic Next-Generation Sequencing in the Diagnosis of Pulmonary Infectious Pathogens From Bronchoalveolar Lavage Samples. Front. Cell. Infect. Microbiol. 2021, 11, 541092. [Google Scholar] [CrossRef]
- Langelier, C.; Zinter, M.S.; Kalantar, K.; Yanik, G.A.; Christenson, S.; O’Donovan, B.; White, C.; Wilson, M.; Sapru, A.; Dvorak, C.C.; et al. Metagenomic Sequencing Detects Respiratory Pathogens in Hematopoietic Cellular Transplant Patients. Am. J. Respir. Crit. Care Med. 2018, 197, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Chapman, R.; Jones, L.; D’Angelo, A.; Suliman, A.; Anwar, M.; Bagby, S. Nanopore-Based Metagenomic Sequencing in Respiratory Tract Infection: A Developing Diagnostic Platform. Lung 2023, 201, 171–179. [Google Scholar] [CrossRef]
- Ma, L.; Zhu, C.; Yan, T.; Hu, Y.; Zhou, J.; Li, Y.; Du, F.; Zhou, J. Illumina and Nanopore Sequencing in Culture-Negative Samples from Suspected Lower Respiratory Tract Infection Patients. Front. Cell. Infect. Microbiol. 2024, 14, 1230650. [Google Scholar] [CrossRef]
- Ye, J.; Huang, K.; Xu, Y.; Chen, N.; Tu, Y.; Huang, J.; Shao, L.; Kong, W.; Zhao, D.; Xie, Y. Clinical Application of Nanopore-Targeted Sequencing Technology in Bronchoalveolar Lavage Fluid from Patients with Pulmonary Infections. Microbiol. Spectr. 2024, 12, e00026-24. [Google Scholar] [CrossRef]
- Sun, G.; Liu, W.; Zheng, Q.; Shan, Q.; Hou, H. Ratio of Procalcitonin/Simpson’s Dominance Index Predicted the Short-Term Prognosis of Patients with Severe Bacterial Pneumonia. Front. Cell. Infect. Microbiol. 2023, 13, 1175747. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Cobas, A.E.; Ginevra, C.; Rusniok, C.; Jarraud, S.; Buchrieser, C. The Respiratory Tract Microbiome, the Pathogen Load, and Clinical Interventions Define Severity of Bacterial Pneumonia. Cell Rep. Med. 2023, 4, 101167. [Google Scholar] [CrossRef]
- Abdill, R.J.; Adamowicz, E.M.; Blekhman, R. Public Human Microbiome Data Are Dominated by Highly Developed Countries. PLoS Biol. 2022, 20, e3001536. [Google Scholar] [CrossRef]
- Gaulke, C.A.; Sharpton, T.J. The Influence of Ethnicity and Geography on Human Gut Microbiome Composition. Nat. Med. 2018, 24, 1495–1496. [Google Scholar] [CrossRef]
- Dabrowski, A.N.; Shrivastav, A.; Conrad, C.; Komma, K.; Weigel, M.; Dietert, K.; Gruber, A.D.; Bertrams, W.; Wilhelm, J.; Schmeck, B.; et al. Peptidoglycan Recognition Protein 4 Limits Bacterial Clearance and Inflammation in Lungs by Control of the Gut Microbiota. Front. Immunol. 2019, 10, 2106. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- NCBI RefSeq Targeted Loci Project. Available online: https://www.ncbi.nlm.nih.gov/refseq/targetedloci/ (accessed on 29 February 2024).
- Available online: https://github.com/OpenGene/fastplong (accessed on 30 April 2025).
- Curry, K.D.; Wang, Q.; Nute, M.G.; Tyshaieva, A.; Reeves, E.; Soriano, S.; Wu, Q.; Graeber, E.; Finzer, P.; Mendling, W.; et al. Emu: Species-Level Microbial Community Profiling of Full-Length 16S rRNA Oxford Nanopore Sequencing Data. Nat. Methods 2022, 19, 845–853. [Google Scholar] [CrossRef] [PubMed]
- Roller, B.R.K.; Stoddard, S.F.; Schmidt, T.M. Exploiting rRNA Operon Copy Number to Investigate Bacterial Reproductive Strategies. Nat. Microbiol. 2016, 1, 16160. [Google Scholar] [CrossRef] [PubMed]
- Gujju, V.R.; Akram, B.; Shibib, D.R.; McGhee, M.A.; Drevets, D.A. Bordetella Bronchiseptica Infections in Patients with HIV/AIDS: A Case Report and Review of the Literature. Medicine 2021, 100, e28244. [Google Scholar] [CrossRef]
- Yao, Y.; Falgenhauer, L.; Falgenhauer, J.; Hauri, A.M.; Heinmüller, P.; Domann, E.; Chakraborty, T.; Imirzalioglu, C. Carbapenem-Resistant Citrobacter Spp. as an Emerging Concern in the Hospital-Setting: Results From a Genome-Based Regional Surveillance Study. Front. Cell. Infect. Microbiol. 2021, 11, 744431. [Google Scholar] [CrossRef]
- Dalhoff, K.; Abele-Horn, M.; Andreas, S.; Deja, M.; Ewig, S.; Gastmeier, P.; Gatermann, S.; Gerlach, H.; Grabein, B.; Heußel, C.; et al. S3-Leitlinie zur Epidemiologie, Diagnostik und Therapie erwachsener Patienten mit nosokomialer Pneumonie–Update 2017. Available online: https://register.awmf.org/assets/guidelines/020-013l_S3_Nosokomiale_Pneumonie_Erwachsener_2017-11.pdf (accessed on 27 January 2024).
- Ewig, S.; Kolditz, M.; Pletz, M.; Altiner, A.; Albrich, W.; Droemann, D.; Flick, H.; Gatermann, S.; Krüger, S.; Nehls, W.; et al. S3-Leitlinie zur Behandlung von Erwachsenen Patienten mit Ambulant Erworbener Pneumonie–Update 2021. Available online: https://register.awmf.org/assets/guidelines/020-020l_S3_Behandlung-von-erwachsenen-Patienten-mit-ambulant-erworbener-Pneumonie__2021-05.pdf (accessed on 27 January 2024).
- Koufakis, T.; Chatzopoulou, M.; Margaritis, A.; Tsiakalou, M.; Gabranis, I. Pneumonia by Listeria Monocytogenes: A Common Infection by an Uncommon Pathogen. Case Rep. Infect. Dis. 2015, 2015, 627073. [Google Scholar] [CrossRef] [PubMed]
- Viswanath, L.S.; Gunalan, A.; Jamir, I.; S, B.; S, A.; K, A.; Biswas, R. Prevotella oris: A Lesser Known Etiological Agent of Pleural Effusion. Anaerobe 2022, 78, 102644. [Google Scholar] [CrossRef]
- Ryan, M.P.; Pembroke, J.T.; Adley, C.C. Differentiating the Growing Nosocomial Infectious Threats Ralstonia Pickettii and Ralstonia Insidiosa. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 1245–1247. [Google Scholar] [CrossRef]
- Furuta, A.; Brokaw, A.; Manuel, G.; Dacanay, M.; Marcell, L.; Seepersaud, R.; Rajagopal, L.; Adams Waldorf, K. Bacterial and Host Determinants of Group B Streptococcal Infection of the Neonate and Infant. Front. Microbiol. 2022, 13, 820365. [Google Scholar] [CrossRef]
- Garriss, G.; Nannapaneni, P.; Simões, A.S.; Browall, S.; Subramanian, K.; Sá-Leão, R.; Goossens, H.; de Lencastre, H.; Henriques-Normark, B. Genomic Characterization of the Emerging Pathogen Streptococcus pseudopneumoniae. mBio 2019, 10, e01286-19. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Liu, R.; Qiu, J.; Wei, C.; Pan, D.; Xiang, T.; Cheng, N. Pulmonary Infection Caused by Tropheryma Whipplei: A Case Report and Review of the Literature. J. Med. Case Rep. 2024, 18, 613. [Google Scholar] [CrossRef] [PubMed]

| Culture-Based Diagnostics | Corresponding Genera Detected by Illumina Short-Read Sequencing | Corresponding Species Detected by ONT Long-Read Sequencing | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | No. of Reports | >20% | ≥10% <20% | ≥3% <10% | <3% | Absent | >20% | ≥10% <20% | ≥3% <10% | <3% | Absent |
| A. odontolyticus | 1 | - | - | - | 1 | - | - | - | - | - | - |
| B. bronchiseptica | 1 | - | - | 1 | - | - | - | - | 1 | - | - |
| C. koseri | 2 | - | - | - | - | 2 | 2 | - | - | - | - |
| E. faecalis | 2 | 1 | - | - | - | 1 | 1 | - | - | - | 1 |
| E. faecium | 4 | 2 | - | 1 | 1 | - | - | 1 | - | - | 2 |
| E. coli | 2 | - | - | - | - | - | - | - | - | - | - |
| H. influenzae | 1 | 1 | - | - | - | - | - | - | - | - | - |
| H. parahaemolyticus | 1 | 1 | - | - | - | - | 1 | - | - | - | - |
| L. reuteri | 1 | - | 1 | - | - | - | - | - | - | - | - |
| L. salivarius | 1 | - | - | - | - | 1 | - | - | - | - | 1 |
| Saproph. Neisseria | 12 | - | - | 6 | 3 | 3 | - | 1 | 1 | 2 | 5 |
| N. flava | 2 | 1 | 1 | - | - | - | - | - | - | - | 2 |
| P. melaninogenica | 5 | - | 1 | 3 | 1 | - | - | - | - | 3 | 1 |
| P. aeruginosa | 5 | 3 | - | - | 1 | 1 | 1 | - | - | - | 3 |
| Rothia sp. | 3 | 1 | - | 2 | - | - | - | - | - | - | 2 |
| R. mucilaginosa | 5 | 1 | 3 | 1 | - | - | - | - | - | 2 | 2 |
| S. marcescens | 1 | - | - | - | - | 1 | - | - | - | - | 1 |
| Coagulase-neg. Staphylococcus | 9 | 2 | - | 1 | 2 | 4 | 2 | - | 1 | 3 | 2 |
| S. aureus | 11 | 3 | 1 | - | 4 | 3 | 1 | - | 1 | 1 | 5 |
| S. epidermidis | 4 | - | 1 | 1 | 2 | - | 1 | - | - | 1 | 1 |
| S. maltophilia | 3 | 3 | - | - | - | - | 2 | - | - | - | 1 |
| α-haemolytic Streptococcus | 35 | 10 | 11 | 9 | 5 | - | 16 | 3 | 3 | 2 | 2 |
| S. mitis | 3 | - | 1 | 1 | 1 | - | - | - | - | - | 3 |
| S. parasanguinis | 1 | - | - | 1 | - | - | - | - | - | - | 1 |
| S. pneumoniae | 1 | 1 | - | - | - | - | 1 | - | - | - | - |
| S. salivarius | 1 | - | - | 1 | - | - | - | - | 1 | - | - |
| V. parvula | 1 | - | 1 | - | - | - | - | - | - | 1 | - |
| Sum | 118 | 30 | 21 | 28 | 21 | 16 | 28 | 5 | 7 | 16 | 35 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bitter, M.; Weigel, M.; Mengel, J.P.; Ott, B.; Windhorst, A.C.; Tello, K.; Imirzalioglu, C.; Hain, T. Assessment of Microbiome-Based Pathogen Detection Using Illumina Short-Read and Nanopore Long-Read Sequencing in 144 Patients Undergoing Bronchoalveolar Lavage in a University Hospital in Germany. Int. J. Mol. Sci. 2025, 26, 9841. https://doi.org/10.3390/ijms26209841
Bitter M, Weigel M, Mengel JP, Ott B, Windhorst AC, Tello K, Imirzalioglu C, Hain T. Assessment of Microbiome-Based Pathogen Detection Using Illumina Short-Read and Nanopore Long-Read Sequencing in 144 Patients Undergoing Bronchoalveolar Lavage in a University Hospital in Germany. International Journal of Molecular Sciences. 2025; 26(20):9841. https://doi.org/10.3390/ijms26209841
Chicago/Turabian StyleBitter, Merle, Markus Weigel, Jan Philipp Mengel, Benjamin Ott, Anita C. Windhorst, Khodr Tello, Can Imirzalioglu, and Torsten Hain. 2025. "Assessment of Microbiome-Based Pathogen Detection Using Illumina Short-Read and Nanopore Long-Read Sequencing in 144 Patients Undergoing Bronchoalveolar Lavage in a University Hospital in Germany" International Journal of Molecular Sciences 26, no. 20: 9841. https://doi.org/10.3390/ijms26209841
APA StyleBitter, M., Weigel, M., Mengel, J. P., Ott, B., Windhorst, A. C., Tello, K., Imirzalioglu, C., & Hain, T. (2025). Assessment of Microbiome-Based Pathogen Detection Using Illumina Short-Read and Nanopore Long-Read Sequencing in 144 Patients Undergoing Bronchoalveolar Lavage in a University Hospital in Germany. International Journal of Molecular Sciences, 26(20), 9841. https://doi.org/10.3390/ijms26209841






