Mitochondrial Gene Regulation and Pain Susceptibility: A Multi-Omics Causal Inference Study
Abstract
1. Introduction
2. Results
2.1. Integrated Multi-Omics SMR Identifies Candidate Mitochondrial Genes Associated with Pain Phenotypes
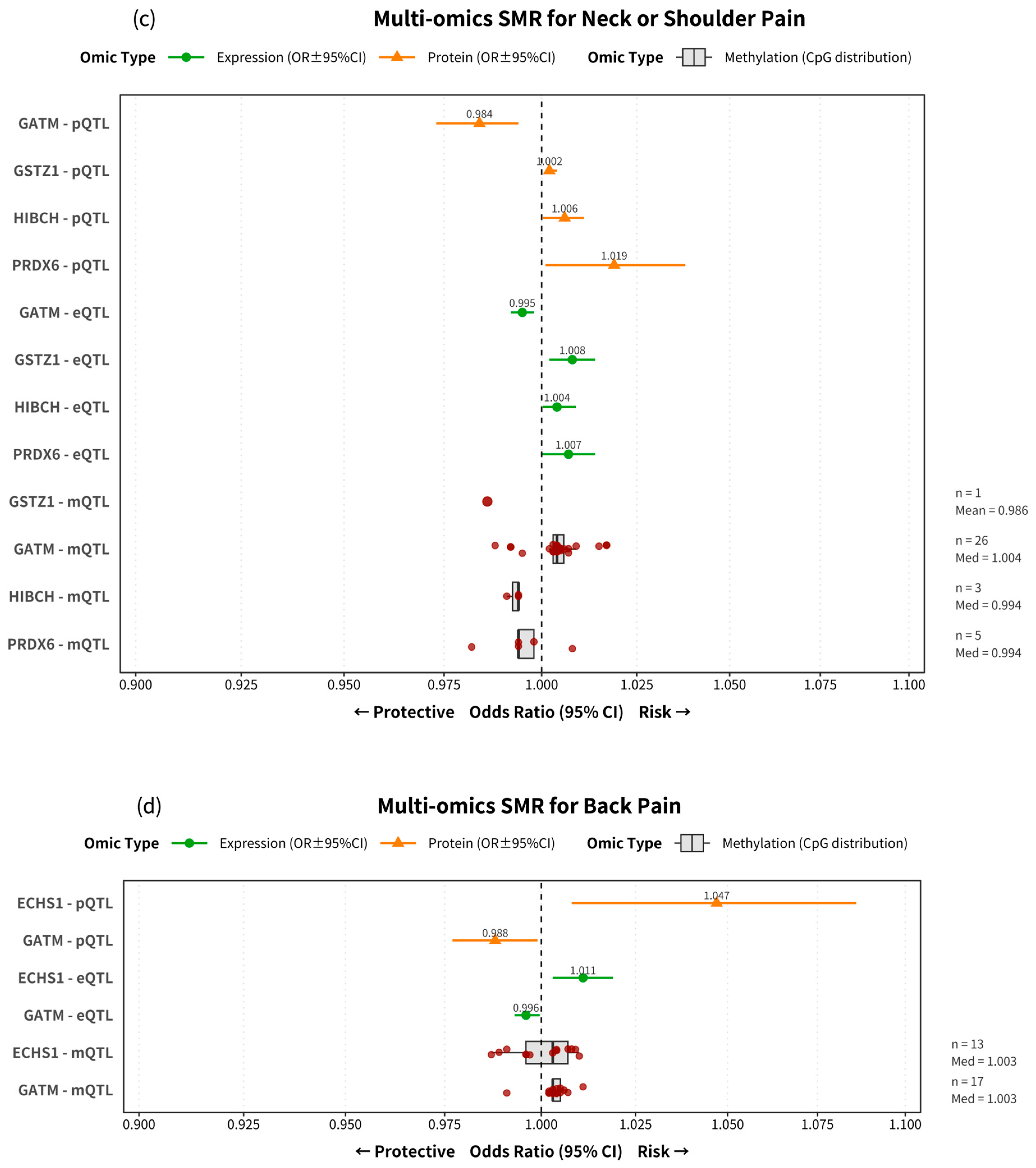
2.2. Effect Sizes and Directionality Reveal Diverse and Complex Roles of Mitochondrial Genes
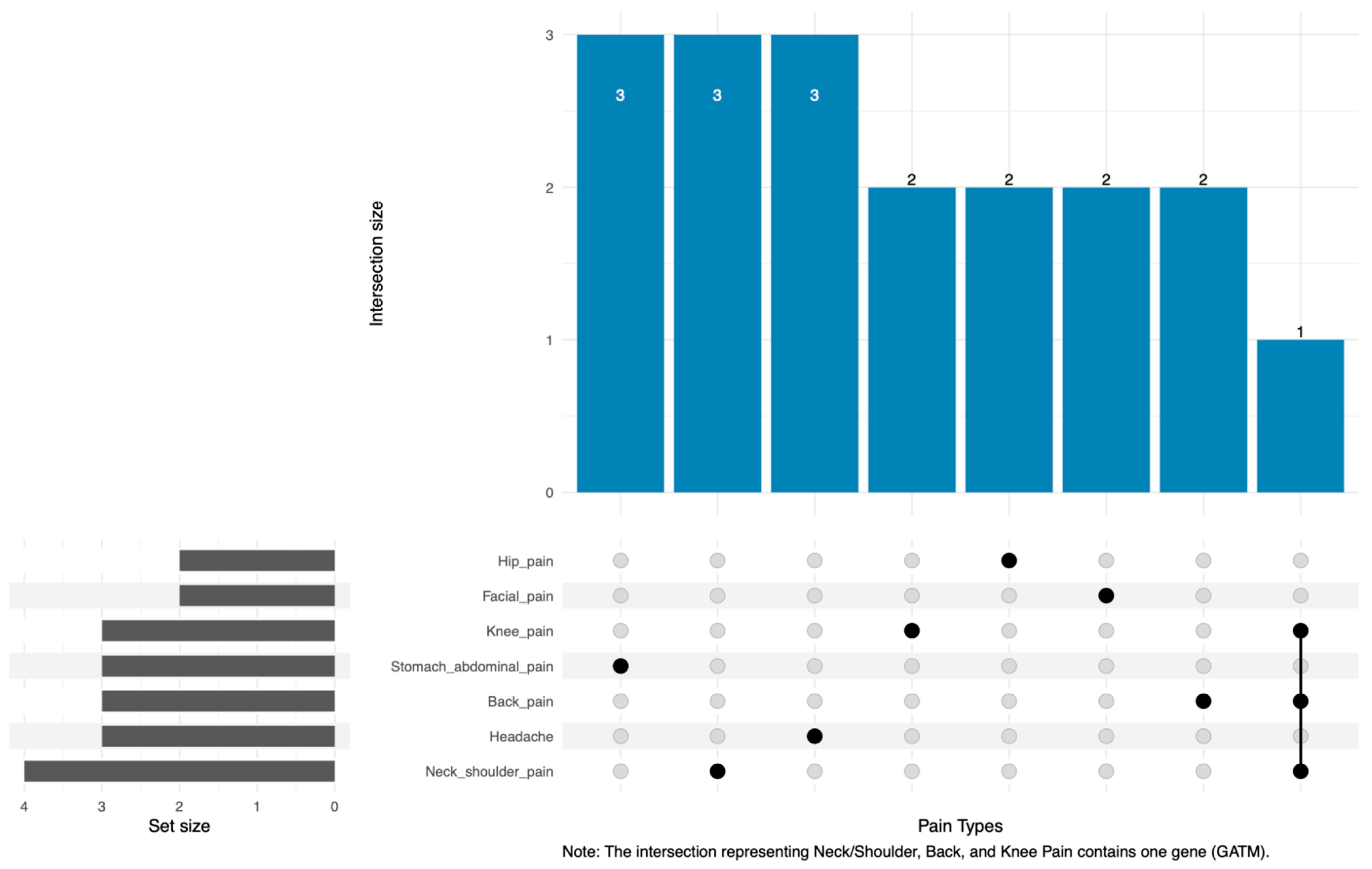
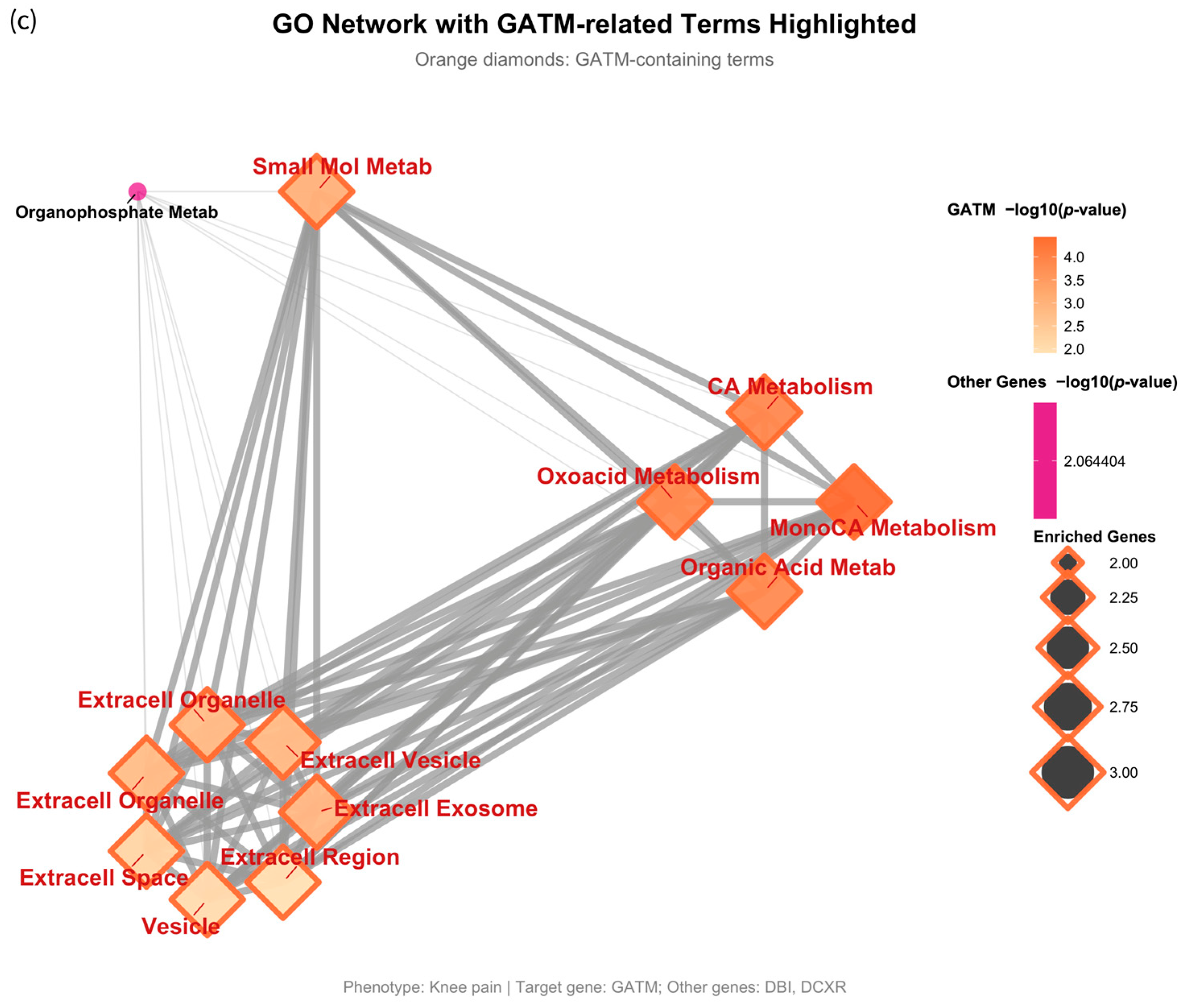
2.3. Shared Gene Patterns and Functional Insights from Enrichment Analysis
3. Discussion
4. Materials and Methods
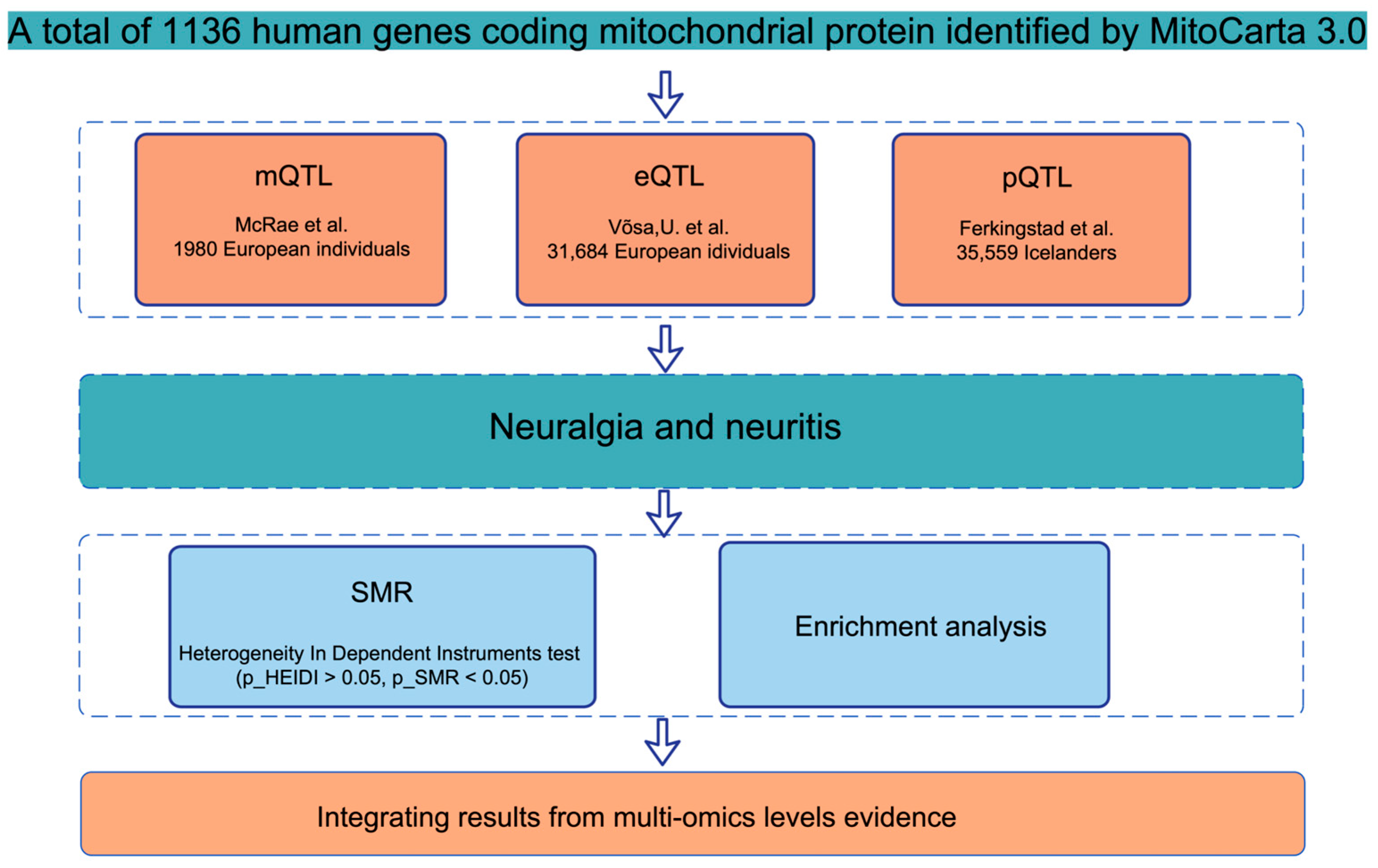
4.1. Comprehensive Study Design and Data Integration
4.2. Acquisition and Processing of mQTL, eQTL, and pQTL Data
4.2.1. DNA Methylation Quantitative Trait Loci (mQTL)
4.2.2. Gene Expression Quantitative Trait Loci (eQTL)
4.2.3. Protein Quantitative Trait Loci (pQTL)
4.3. Acquisition of Outcome GWAS Data and Quality Control of Instrumental Variables
- Cis-region Selection: SNPs located within ±1000 kb of the target gene were considered.
- Significance Threshold: A significance level of p < 5 × 10−8 was required for m/eQTL analyses, while a threshold of p < 1.8 × 10−9 was set for pQTL analyses.
- LD Pruning: SNPs exhibiting strong linkage disequilibrium (r2 > 0.9, based on the 1000 Genomes EUR dataset) were excluded.
- Weak Instrument Filtering: SNPs with an F-statistic of less than 10 were removed from consideration.
- Allele Frequency Concordance: SNPs demonstrating frequency discrepancies greater than 0.2 between the LD reference, QTL, and GWAS datasets were filtered out.
- Harmonization: A meticulous alignment of effect alleles and sizes between the exposure and outcome datasets was performed to ensure consistent estimation in the SMR analysis.
4.4. Statistical Analysis and Causal Inference Through Summary-Data-Based Mendelian Randomization (SMR)
4.5. Colocalization Analysis for Evaluating Shared Genetic Signals Between Traits
4.6. Enrichment Analysis
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| mtDNA | Mitochondrial DNA |
| SMR | Summary-data-based Mendelian randomization |
| GWAS | Genome-wide association study |
| mQTL | DNA methylation quantitative trait loci |
| eQTL | Gene expression quantitative trait loci |
| pQTL | Protein quantitative trait loci |
| LD | Linkage disequilibrium |
| SNP | Single nucleotide polymorphism |
| FDR | False discovery rate |
| HEIDI | Heterogeneity in dependent instruments |
| GO | Gene ontology |
| BP | Biological process |
| MF | Molecular function |
| CC | Cellular component |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ETFA | Electron transfer flavoprotein subunit alpha |
| GRHPR | Glyoxylate reductase/hydroxypyruvate reductase |
| MMAB | Metabolism of cobalamin associated B |
| FASN | Fatty acid synthase |
| SPHK2 | Sphingosine kinase 2 |
| GATM | Glycine amidinotransferase |
| GSTZ1 | Glutathione S-transferase zeta 1 |
| HIBCH | 3-Hydroxyisobutyryl-CoA hydrolase |
| PRDX6 | Peroxiredoxin 6 |
| ACSF2 | Acyl-CoA synthetase family member 2 |
| ECHS1 | Enoyl-CoA hydratase, short chain 1 |
| NME4 | NME/NM23 nucleoside diphosphate kinase 4 |
| RMDN1 | Regulator of microtubule dynamics 1 |
| QDPR | Quinoid dihydropteridine reductase |
| FAHD1 | Fumarylacetoacetate hydrolase domain containing 1 |
| MCL1 | MCL1 apoptosis regulator |
| DBI | Diazepam binding inhibitor |
| DCXR | Dicarbonyl and L-xylulose reductase |
References
- Awad-Igbaria, Y.; Ben-Menashe, A.; Sakas, R.; Edelman, D.; Fishboom, T.; Shamir, A.; Soustiel, J.F.; Palzur, E. Novel Insight into TRPV1-Induced Mitochondrial Dysfunction in Neuropathic Pain. Brain J. Neurol. 2025, 148, 2563–2578. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, N.; Papadopoulos, V. Role of Mitochondrial Dysfunction in Neuropathy. Int. J. Mol. Sci. 2025, 26, 3195. [Google Scholar] [CrossRef]
- Zajączkowska, R.; Kocot-Kępska, M.; Leppert, W.; Wrzosek, A.; Mika, J.; Wordliczek, J. Mechanisms of Chemotherapy-Induced Peripheral Neuropathy. Int. J. Mol. Sci. 2019, 20, 1451. [Google Scholar] [CrossRef]
- Taschler, B.; Smith, S.M.; Nichols, T.E. Causal Inference on Neuroimaging Data with Mendelian Randomisation. Neuroimage 2022, 258, 119385. [Google Scholar] [CrossRef]
- Fang, Q.; Fan, H.; Li, Q.; Zhang, M.; Zhou, Z.; Du, J.; Huang, J. Multi-Omic Insight Into the Molecular Networks in the Pathogenesis of Coronary Artery Disease. J. Am. Heart Assoc. 2025, 14, e037203. [Google Scholar] [CrossRef]
- Wang, M.; Xu, S. Statistical Power in Genome-Wide Association Studies and Quantitative Trait Locus Mapping. Heredity 2019, 123, 287–306. [Google Scholar] [CrossRef]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An Updated Mitochondrial Proteome Now with Sub-Organelle Localization and Pathway Annotations. Nucleic Acids Res. 2021, 49, D1541–D1547. [Google Scholar] [CrossRef]
- McRae, A.F.; Marioni, R.E.; Shah, S.; Yang, J.; Powell, J.E.; Harris, S.E.; Gibson, J.; Henders, A.K.; Bowdler, L.; Painter, J.N.; et al. Identification of 55,000 Replicated DNA Methylation QTL. Sci. Rep. 2018, 8, 17605. [Google Scholar] [CrossRef]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-Scale Cis- and Trans-eQTL Analyses Identify Thousands of Genetic Loci and Polygenic Scores That Regulate Blood Gene Expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-Scale Integration of the Plasma Proteome with Genetics and Disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef] [PubMed]
- Pinti, M.V.; Fink, G.K.; Hathaway, Q.A.; Durr, A.J.; Kunovac, A.; Hollander, J.M. Mitochondrial Dysfunction in Type 2 Diabetes Mellitus: An Organ-Based Analysis. Am. J. Physiol.-Endocrinol. Metab. 2019, 316, E268–E285. [Google Scholar] [CrossRef]
- Haodong, Z.; Baoping, C.; Jiongjiong, C.; Jia, C.; Hui, L.; Yu, W.; Bocheng, D. Dissecting the Genetic Etiology of Intestinal Obstruction: Mendelian Randomization Identifies Potential Therapeutic Targets. Preprint MedRxiv. 2025. [Google Scholar] [CrossRef]
- Zuber, V.; Grinberg, N.F.; Gill, D.; Manipur, I.; Slob, E.A.W.; Patel, A.; Wallace, C.; Burgess, S. Combining Evidence from Mendelian Randomization and Colocalization: Review and Comparison of Approaches. Am. J. Hum. Genet. 2022, 109, 767–782. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Liu, J.Z.; Zhang, W.; Hauberg, M.; Shi, H.; Boocock, J.; Pickrell, J.; E Jaffe, A.; The CommonMind Consortium; Pasaniuc, B.; et al. CommonMind Consortium; Pasaniuc, B.; et al. A Bayesian Framework for Multiple Trait Colocalization from Summary Association Statistics. Bioinforma. Oxf. Engl. 2018, 34, 2538–2545. [Google Scholar] [CrossRef]
- Hemani, G.; Bowden, J.; Davey Smith, G. Evaluating the Potential Role of Pleiotropy in Mendelian Randomization Studies. Hum. Mol. Genet. 2018, 27, R195–R208. [Google Scholar] [CrossRef]
- Cao, X.; Sun, H.; Feng, R.; Mazumder, R.; Najar, C.F.B.A.; Li, Y.I.; de Jager, P.L.; Bennett, D.; Consortium, T.A.D.F.G.; Dey, K.K.; et al. Integrative Multi-Omics QTL Colocalization Maps Regulatory Architecture in Aging Human Brain. Preprint MedRxiv. 2025. [Google Scholar] [CrossRef]
- Wallace, C. A More Accurate Method for Colocalisation Analysis Allowing for Multiple Causal Variants. PLoS Genet. 2021, 17, e1009440. [Google Scholar] [CrossRef]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Wallimann, T.; Tokarska-Schlattner, M.; Schlattner, U. The Creatine Kinase System and Pleiotropic Effects of Creatine. Amino Acids 2011, 40, 1271–1296. [Google Scholar] [CrossRef]
- Kreider, R.B.; Stout, J.R. Creatine in Health and Disease. Nutrients 2021, 13, 447. [Google Scholar] [CrossRef]
- Amiri, E.; Sheikholeslami-Vatani, D. The Role of Resistance Training and Creatine Supplementation on Oxidative Stress, Antioxidant Defense, Muscle Strength, and Quality of Life in Older Adults. Front. Public Health 2023, 11, 1062832. [Google Scholar] [CrossRef]
- Munoz, H.I.; Gonzales, E.B.; Sumien, N. Effects of Creatine Supplementation on Nociception in Young Male and Female Mice. Pharmacol. Rep. PR 2018, 70, 316–321. [Google Scholar] [CrossRef]
- Meftahi, G.H.; Hatef, B.; Pirzad Jahromi, G. Creatine Activity as a Neuromodulator in the Central Nervous System. Arch. Razi Inst. 2023, 78, 1169–1175. [Google Scholar] [CrossRef]
- Wu, K.; Shieh, J.-S.; Qin, L.; Guo, J.J. Mitochondrial Mechanisms in the Pathogenesis of Chronic Inflammatory Musculoskeletal Disorders. Cell Biosci. 2024, 14, 76. [Google Scholar] [CrossRef]
- Morciano, G.; Giorgi, C.; Balestra, D.; Marchi, S.; Perrone, D.; Pinotti, M.; Pinton, P. Mcl-1 Involvement in Mitochondrial Dynamics Is Associated with Apoptotic Cell Death. Mol. Biol. Cell 2016, 27, 20–34. [Google Scholar] [CrossRef]
- Sancho, M.; Leiva, D.; Lucendo, E.; Orzáez, M. Understanding MCL1: From Cellular Function and Regulation to Pharmacological Inhibition. Febs J. 2022, 289, 6209–6234. [Google Scholar] [CrossRef]
- Wajner, M.; Coelho, J.C. Neurological Dysfunction in Methylmalonic Acidaemia Is Probably Related to the Inhibitory Effect of Methylmalonate on Brain Energy Production. J. Inherit. Metab. Dis. 1997, 20, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Ni, Y.; Li, X.; Zhuang, X.; Liu, Y.; Liu, X.; Chen, S. Urinary Methylmalonic Acid as an Indicator of Early Vitamin B12 Deficiency and Its Role in Polyneuropathy in Type 2 Diabetes. J. Diabetes Res. 2014, 2014, 921616. [Google Scholar] [CrossRef] [PubMed]
- Togha, M.; Jahromi, S.R.; Ghorbani, Z.; Martami, F.; Seifishahpar, M.; Seifishahpar, M. Serum Vitamin B12 and Methylmalonic Acid Status in Migraineurs: A Case-Control Study. Headache 2019, 59, 1492–1503. [Google Scholar] [CrossRef]
- García-González, A.; Gaxiola-Robles, R.; Zenteno-Savín, T. Oxidative Stress in Patients with Rheumatoid Arthritis. Rev. Investig. Clin. Organo Hosp. Enfermedades Nutr. 2015, 67, 46–53. [Google Scholar] [CrossRef]
- Sui, B.-D.; Xu, T.-Q.; Liu, J.-W.; Wei, W.; Zheng, C.-X.; Guo, B.-L.; Wang, Y.-Y.; Yang, Y.-L. Understanding the Role of Mitochondria in the Pathogenesis of Chronic Pain. Postgrad. Med. J. 2013, 89, 709–714. [Google Scholar] [CrossRef] [PubMed]
- Yorns, W.R.; Hardison, H.H. Mitochondrial Dysfunction in Migraine. Semin. Pediatr. Neurol. 2013, 20, 188–193. [Google Scholar] [CrossRef]
- Cramer, S.D.; Ferree, P.M.; Lin, K.; Milliner, D.S.; Holmes, R.P. The Gene Encoding Hydroxypyruvate Reductase (GRHPR) Is Mutated in Patients with Primary Hyperoxaluria Type II. Hum. Mol. Genet. 1999, 8, 2063–2069. [Google Scholar] [CrossRef]
- Salles, J.; Sargueil, F.; Knoll-Gellida, A.; Witters, L.A.; Shy, M.; Jiang, H.; Cassagne, C.; Garbay, B. Fatty Acid Synthase Expression during Peripheral Nervous System Myelination. Mol. Brain Res. 2002, 101, 52–58. [Google Scholar] [CrossRef]
- Squillace, S.; Spiegel, S.; Salvemini, D. Targeting the Sphingosine-1-Phosphate Axis for Developing Non-Narcotic Pain Therapeutics. Trends Pharmacol. Sci. 2020, 41, 851–867. [Google Scholar] [CrossRef]
- Carrasco, C.; Naziroǧlu, M.; Rodríguez, A.B.; Pariente, J.A. Neuropathic Pain: Delving into the Oxidative Origin and the Possible Implication of Transient Receptor Potential Channels. Front. Physiol. 2018, 9, 95. [Google Scholar] [CrossRef]
- Ferdinandusse, S.; Waterham, H.R.; Heales, S.J.R.; Brown, G.K.; Hargreaves, I.P.; Taanman, J.-W.; Gunny, R.; Abulhoul, L.; Wanders, R.J.A.; Clayton, P.T.; et al. HIBCH Mutations Can Cause Leigh-like Disease with Combined Deficiency of Multiple Mitochondrial Respiratory Chain Enzymes and Pyruvate Dehydrogenase. Orphanet J. Rare Dis. 2013, 8, 188. [Google Scholar] [CrossRef]
- Asuni, A.A.; Guridi, M.; Sanchez, S.; Sadowski, M.J. Antioxidant Peroxiredoxin 6 Protein Rescues Toxicity Due to Oxidative Stress and Cellular Hypoxia in Vitro, and Attenuates Prion-Related Pathology in Vivo. Neurochem. Int. 2015, 90, 152–165. [Google Scholar] [CrossRef]
- Wagner, K.; Lee, K.S.S.; Yang, J.; Hammock, B.D. Epoxy Fatty Acids Mediate Analgesia in Murine Diabetic Neuropathy. Eur. J. Pain Lond. Engl. 2017, 21, 456–465. [Google Scholar] [CrossRef]
- Wang, B.; Qin, Y.; Bao, Y.; Chen, S.; Zheng, J.; Lin, S.; Zheng, K.; Duan, S. Deficiency in the Conserved ECHS1 Gene Causes Leigh Syndrome by Impairing Mitochondrial Respiration Efficiency and Suppressing ADRB2-PKA Signaling. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2025, 1871, 167930. [Google Scholar] [CrossRef]
- Lacombe, M.-L.; Tokarska-Schlattner, M.; Boissan, M.; Schlattner, U. The Mitochondrial Nucleoside Diphosphate Kinase (NDPK-D/NME4), a Moonlighting Protein for Cell Homeostasis. Lab. Invest. 2018, 98, 582–588. [Google Scholar] [CrossRef]
- Oishi, K.; Okano, H.; Sawa, H. RMD-1, a Novel Microtubule-Associated Protein, Functions in Chromosome Segregation in Caenorhabditis Elegans. J. Cell Biol. 2007, 179, 1149–1162. [Google Scholar] [CrossRef]
- Latremoliere, A.; Costigan, M. GCH1, BH4 and Pain. Curr. Pharm. Biotechnol. 2011, 12, 1728–1741. [Google Scholar] [CrossRef]
- Lim, T.K.Y.; Rone, M.B.; Lee, S.; Antel, J.P.; Zhang, J. Mitochondrial and Bioenergetic Dysfunction in Trauma-Induced Painful Peripheral Neuropathy. Mol. Pain 2015, 11, 58. [Google Scholar] [CrossRef]
- Enna, S.J.; McCarson, K.E. The Role of GABA in the Mediation and Perception of Pain. Adv. Pharmacol. San Diego Calif. 2006, 54, 1–27. [Google Scholar] [CrossRef]
- Wada, R.; Yagihashi, S. Role of Advanced Glycation End Products and Their Receptors in Development of Diabetic Neuropathy. Ann. N. Y. Acad. Sci. 2005, 1043, 598–604. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Davey Smith, G. Reading Mendelian Randomisation Studies: A Guide, Glossary, and Checklist for Clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef]
- Chepelev, I.; Harley, I.T.W.; Harley, J.B. Modeling of Horizontal Pleiotropy Identifies Possible Causal Gene Expression in Systemic Lupus Erythematosus. Front. Lupus 2023, 1, 1234578. [Google Scholar] [CrossRef]
- Relton, C.L.; Smith, G.D. Mendelian Randomization: Applications and Limitations in Epigenetic Studies. Epigenomics 2015, 7, 1239–1243. [Google Scholar] [CrossRef]
- Arvanitis, M.; Tayeb, K.; Strober, B.J.; Battle, A. Redefining Tissue Specificity of Genetic Regulation of Gene Expression in the Presence of Allelic Heterogeneity. Am. J. Hum. Genet. 2022, 109, 223–239. [Google Scholar] [CrossRef]
- Sonawane, A.R.; Platig, J.; Fagny, M.; Chen, C.-Y.; Paulson, J.N.; Lopes-Ramos, C.M.; DeMeo, D.L.; Quackenbush, J.; Glass, K.; Kuijjer, M.L. Understanding Tissue-Specific Gene Regulation. Cell Rep. 2017, 21, 1077–1088. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomisation (STROBE-MR): Explanation and Elaboration. BMJ 2021, n2233. [Google Scholar] [CrossRef] [PubMed]
- Elsworth, B.; Lyon, M.; Alexander, T.; Liu, Y.; Matthews, P.; Hallett, J.; Bates, P.; Palmer, T.; Haberland, V.; Smith, G.D.; et al. The MRC IEU OpenGWAS Data Infrastructure. Preprint bioRxiv 2020. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of Summary Data from GWAS and eQTL Studies Predicts Complex Trait Gene Targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef]
- Sun, Z.; Yun, Z.; Lin, J.; Sun, X.; Wang, Q.; Duan, J.; Li, C.; Zhang, X.; Xu, S.; Wang, Z.; et al. Comprehensive Mendelian Randomization Analysis of Plasma Proteomics to Identify New Therapeutic Targets for the Treatment of Coronary Heart Disease and Myocardial Infarction. J. Transl. Med. 2024, 22, 404. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- GTEx Consortium Genetic Effects on Gene Expression across Human Tissues. Nature 2017, 550, 204–213. [CrossRef] [PubMed]
- Morrow, J.D.; Glass, K.; Cho, M.H.; Hersh, C.P.; Pinto-Plata, V.; Celli, B.; Marchetti, N.; Criner, G.; Bueno, R.; Washko, G.; et al. Human Lung DNA Methylation Quantitative Trait Loci Colocalize with Chronic Obstructive Pulmonary Disease Genome-Wide Association Loci. Am. J. Respir. Crit. Care Med. 2018, 197, 1275–1284. [Google Scholar] [CrossRef]
- Yoshiji, S.; Butler-Laporte, G.; Lu, T.; Willett, J.D.S.; Su, C.-Y.; Nakanishi, T.; Morrison, D.R.; Chen, Y.; Liang, K.; Hultström, M.; et al. Proteome-Wide Mendelian Randomization Implicates Nephronectin as an Actionable Mediator of the Effect of Obesity on COVID-19 Severity. Nat. Metab. 2023, 5, 248–264. [Google Scholar] [CrossRef]



| Neuralgia Phenotype | Significant mQTL Associations (CpG Sites) 1 | Significant mQTL Associations (Unique Genes) 1 | Significant eQTL Associations (Unique Genes) 1 | Significant pQTL Associations (Unique Genes) 1 | Integrated Genes (Significant Across m/e/pQTL) 2 | Evidence of Strong Colocalization 3 |
|---|---|---|---|---|---|---|
| Headache | 1797 | 368 | 82 | 10 | ETFA, GRHPR, MMAB | No |
| Facial pain | 646 | 243 | 45 | 8 | FASN, SPHK2 | No |
| Neck or shoulder pain | 1275 | 361 | 75 | 10 | GATM, GSTZ1, HIBCH, PRDX6 | No |
| Back pain | 1340 | 373 | 78 | 8 | ACSF2, ECHS1, GATM | No |
| Stomach or abdominal pain | 993 | 318 | 63 | 11 | NME4, RMDN1, QDPR | No |
| Hip pain | 1014 | 317 | 67 | 7 | FAHD1, MCL1 | YES (MCL1 mQTL) |
| Knee pain | 1421 | 394 | 93 | 9 | DBI, DCXR, GATM | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.-C. Mitochondrial Gene Regulation and Pain Susceptibility: A Multi-Omics Causal Inference Study. Int. J. Mol. Sci. 2025, 26, 8690. https://doi.org/10.3390/ijms26178690
Liu C-C. Mitochondrial Gene Regulation and Pain Susceptibility: A Multi-Omics Causal Inference Study. International Journal of Molecular Sciences. 2025; 26(17):8690. https://doi.org/10.3390/ijms26178690
Chicago/Turabian StyleLiu, Chien-Cheng. 2025. "Mitochondrial Gene Regulation and Pain Susceptibility: A Multi-Omics Causal Inference Study" International Journal of Molecular Sciences 26, no. 17: 8690. https://doi.org/10.3390/ijms26178690
APA StyleLiu, C.-C. (2025). Mitochondrial Gene Regulation and Pain Susceptibility: A Multi-Omics Causal Inference Study. International Journal of Molecular Sciences, 26(17), 8690. https://doi.org/10.3390/ijms26178690





