BlihIA—A Novel Type I Restriction-Modification System from Bacillus licheniformis Is Sensitive to In Vitro Inhibition by ArdB Antirestriction Protein
Abstract
1. Introduction
2. Results
2.1. BlihIA Restriction Activity In Vivo
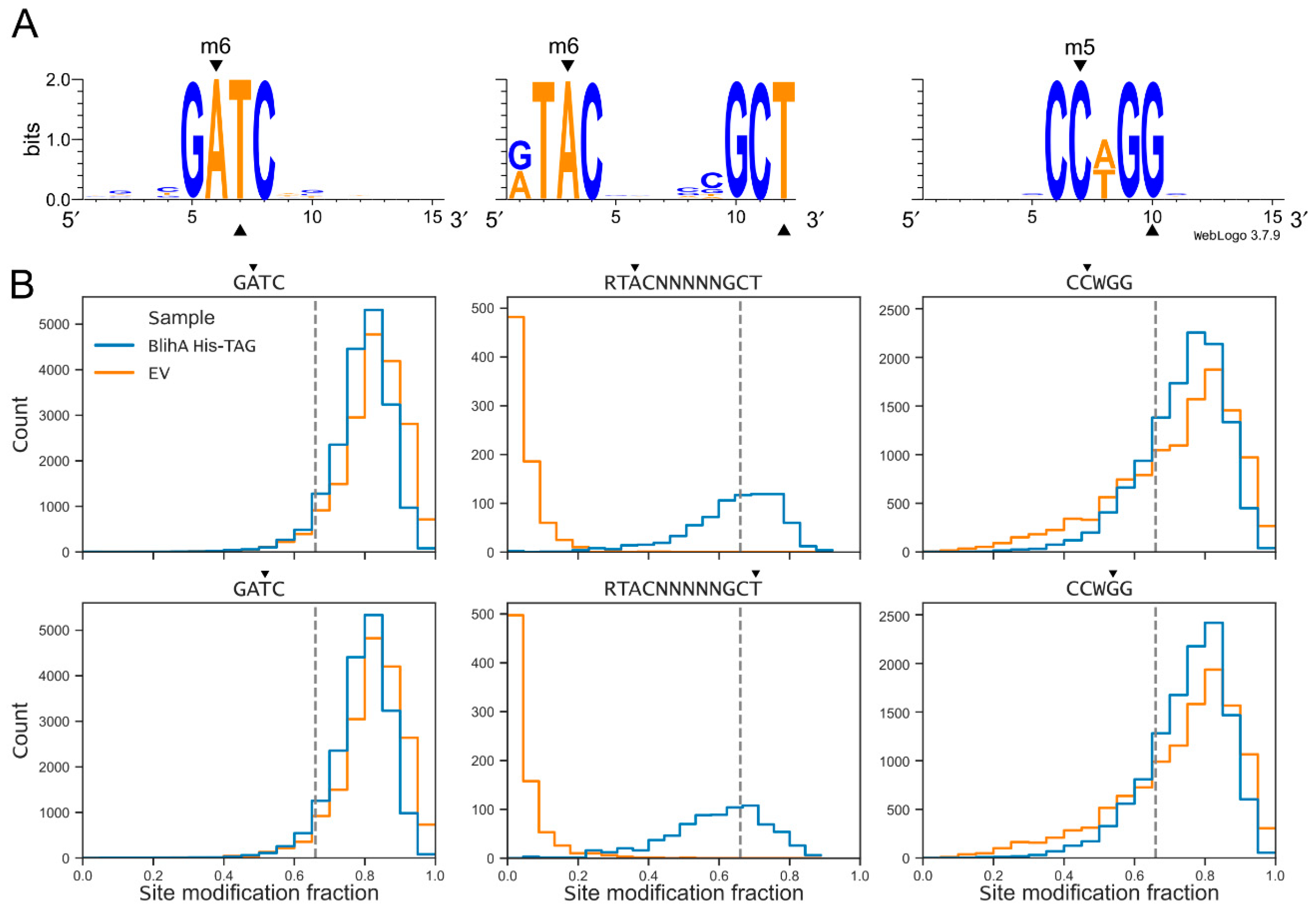
2.2. Identification of BlihIA Methylation Site
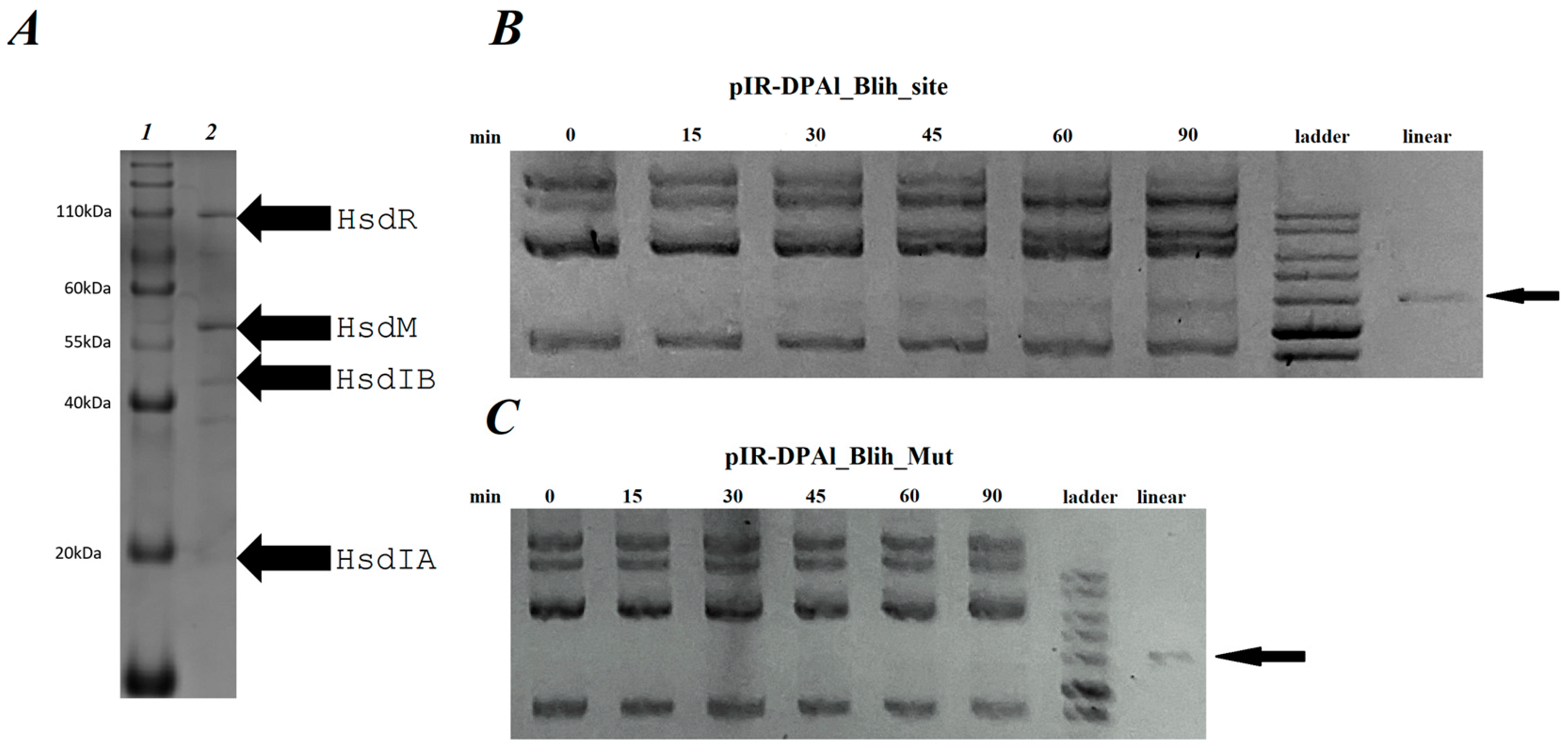
2.3. BlihIA Restriction Activity In Vitro
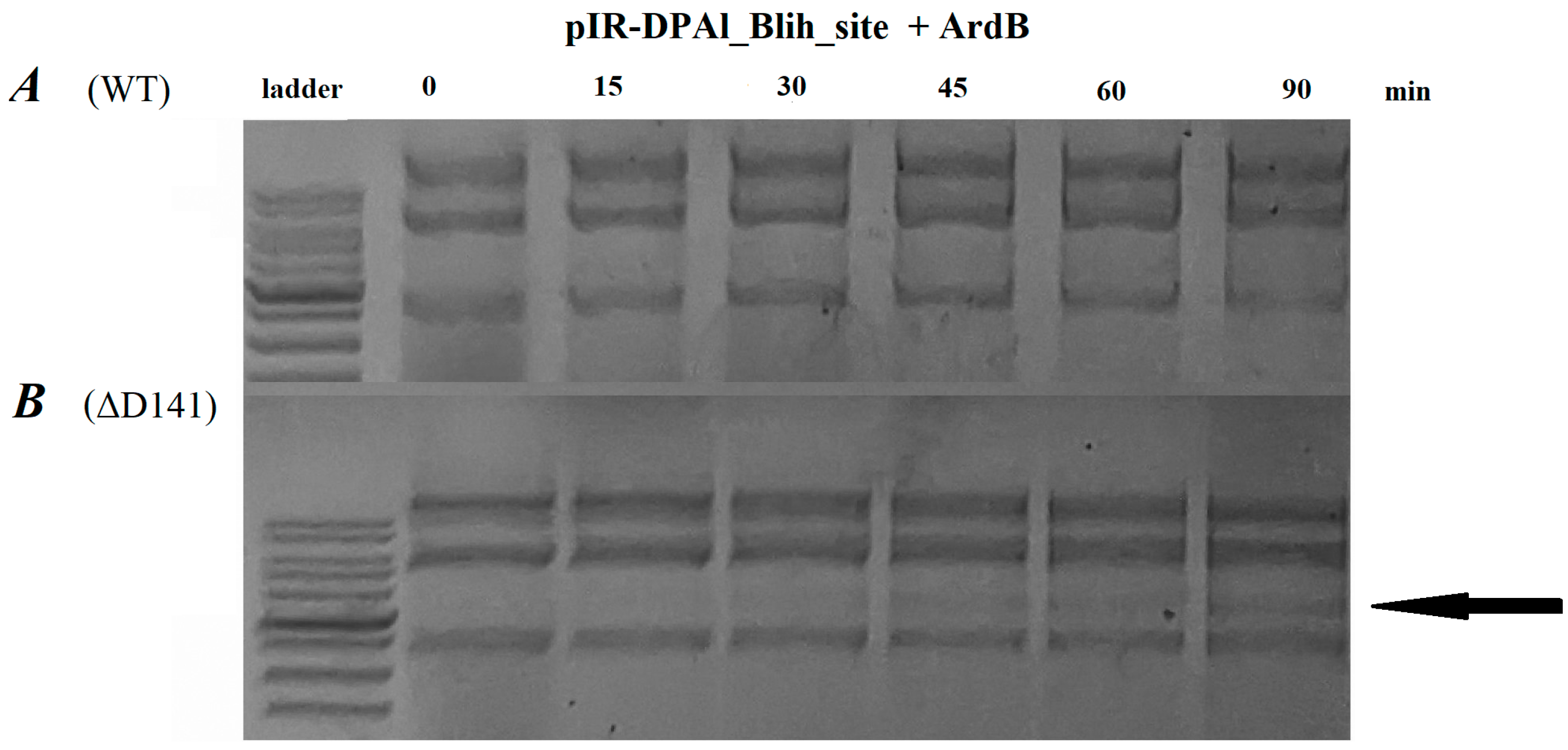
2.4. ArdB Protein Inhibits BlihIA Restriction Activity In Vitro
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Plasmids
4.2. The Cloning of the BlihIA Genes
4.3. BlihIA Restriction Activity In Vivo
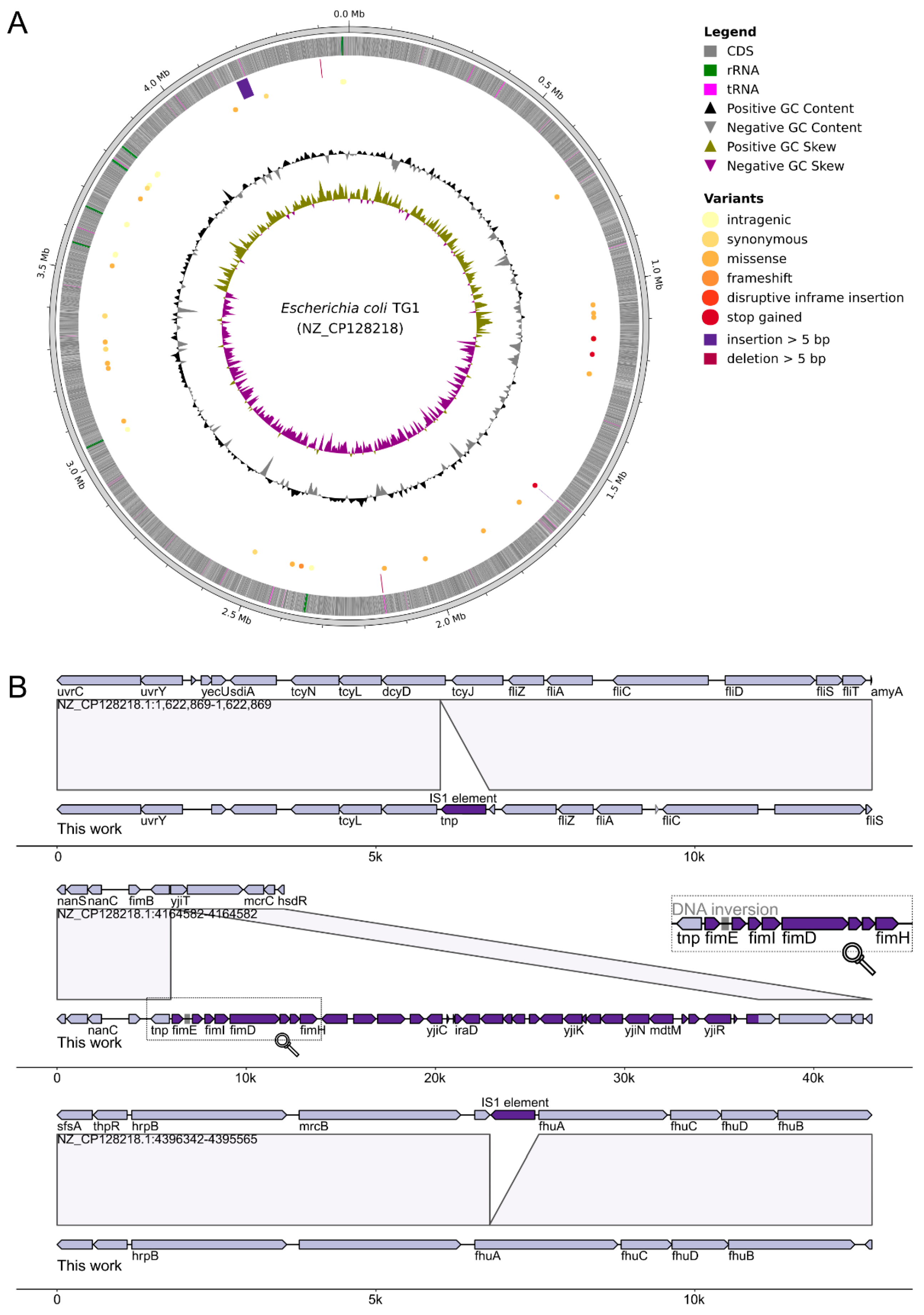
4.4. ONT Sequencing and Data Analysis
4.5. Analysis of Methylation Patterns
4.6. BlihIA Complex Purification
4.7. MALDI-TOF Mass Spectrometry
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Murray, N.E. Type I Restriction Systems: Sophisticated Molecular Machines (a Legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 2000, 64, 412–434. [Google Scholar] [CrossRef]
- Loenen, W.A.M.; Dryden, D.T.F.; Raleigh, E.A.; Wilson, G.G. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014, 42, 20–44. [Google Scholar] [CrossRef]
- Wilson, G.G.; Murray, N.E. Restriction and modification systems. Annu. Rev. Genet. 1991, 25, 585–627. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, P.H.; Touchon, M.; Rocha, E.P.C. Regulation of Genetic Flux between Bacteria by Restriction–Modification Systems. Proc. Natl. Acad. Sci. USA 2016, 113, 5658–5663. [Google Scholar] [CrossRef] [PubMed]
- Olijslager, L.H.; Weijers, D.; Swarts, D.C. Distribution of Specific Prokaryotic Immune Systems Correlates with Host Optimal Growth Temperature. NAR Genom. Bioinforma. 2024, 6, lqae105. [Google Scholar] [CrossRef] [PubMed]
- Atack, J.M.; Guo, C.; Yang, L.; Zhou, Y.; Jennings, M.P. DNA Sequence Repeats Identify Numerous Type I Restriction-Modification Systems That Are Potential Epigenetic Regulators Controlling Phase-Variable Regulons; Phasevarions. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2020, 34, 1038–1051. [Google Scholar] [CrossRef]
- Atack, J.M.; Guo, C.; Litfin, T.; Yang, L.; Blackall, P.J.; Zhou, Y.; Jennings, M.P. Systematic Analysis of REBASE Identifies Numerous Type I Restriction-Modification Systems with Duplicated, Distinct hsdS Specificity Genes That Can Switch System Specificity by Recombination. mSystems 2020, 5, e00497-20. [Google Scholar] [CrossRef]
- Shaw, L.P.; Rocha, E.P.C.; MacLean, R.C. Restriction-Modification Systems Have Shaped the Evolution and Distribution of Plasmids across Bacteria. Nucleic Acids Res. 2023, 51, 6806–6818. [Google Scholar] [CrossRef] [PubMed]
- Isaev, A.B.; Musharova, O.S.; Severinov, K. V Microbial Arsenal of Antiviral Defenses—Part I. Biochemistry 2021, 86, 319–337. [Google Scholar] [CrossRef] [PubMed]
- Serfiotis-Mitsa, D.; Roberts, G.A.; Cooper, L.P.; White, J.H.; Nutley, M.; Cooper, A.; Blakely, G.W.; Dryden, D.T.F. The Orf18 Gene Product from Conjugative Transposon Tn916 Is an ArdA Antirestriction Protein That Inhibits Type I DNA Restriction-Modification Systems. J. Mol. Biol. 2008, 383, 970–981. [Google Scholar] [CrossRef]
- Atanasiu, C.; Su, T.-J.; Sturrock, S.S.; Dryden, D.T.F. Interaction of the Ocr Gene 0.3 Protein of Bacteriophage T7 with EcoKI Restriction/Modification Enzyme. Nucleic Acids Res. 2002, 30, 3936–3944. [Google Scholar] [CrossRef] [PubMed]
- Serfiotis-Mitsa, D.; Herbert, A.P.; Roberts, G.A.; Soares, D.C.; White, J.H.; Blakely, G.W.; Uhrín, D.; Dryden, D.T.F. The Structure of the KlcA and ArdB Proteins Reveals a Novel Fold and Antirestriction Activity against Type I DNA Restriction Systems in Vivo but Not in Vitro. Nucleic Acids Res. 2009, 38, 1723–1737. [Google Scholar] [CrossRef]
- González-Montes, L.; Del Campo, I.; Garcillán-Barcia, M.P.; de la Cruz, F.; Moncalián, G. ArdC, a SsDNA-Binding Protein with a Metalloprotease Domain, Overpasses the Recipient HsdRMS Restriction System Broadening Conjugation Host Range. PLoS Genet. 2020, 16, e1008750. [Google Scholar] [CrossRef] [PubMed]
- Skutel, M.; Yanovskaya, D.; Demkina, A.; Shenfeld, A.; Musharova, O.; Severinov, K.; Isaev, A. RecA-Dependent or Independent Recombination of Plasmid DNA Generates a Conflict with the Host EcoKI Immunity by Launching Restriction Alleviation. Nucleic Acids Res. 2024, 52, 5195–5208. [Google Scholar] [CrossRef]
- Bickle, T.A. Restricting Restriction. Mol. Microbiol. 2004, 51, 3–5. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Yu, Q.; Ding, Z.; Zhang, L.; Shi, G.; Li, Y. Biotechnological and Food Synthetic Biology Potential of Platform Strain: Bacillus Licheniformis. Synth. Syst. Biotechnol. 2023, 8, 281–291. [Google Scholar] [CrossRef]
- Kudryavtseva, A.A.; Cséfalvay, E.; Gnuchikh, E.Y.; Yanovskaya, D.D.; Skutel, M.A.; Isaev, A.B.; Bazhenov, S.V.; Utkina, A.A.; Manukhov, I.V. Broadness and Specificity: ArdB, ArdA, and Ocr against Various Restriction-Modification Systems. Front. Microbiol. 2023, 14, 1133144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Zhang, Y.H.P. Simple, Fast and High-Efficiency Transformation System for Directed Evolution of Cellulase in Bacillus Subtilis. Microb. Biotechnol. 2011, 4, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Plague, G.R.; Boodram, K.S.; Dougherty, K.M.; Bregg, S.; Gilbert, D.P.; Bakshi, H.; Costa, D. Transposable Elements Mediate Adaptive Debilitation of Flagella in Experimental Escherichia Coli Populations. J. Mol. Evol. 2017, 84, 279–284. [Google Scholar] [CrossRef]
- Chanin, R.B.; West, P.T.; Wirbel, J.; Gill, M.O.; Green, G.Z.M.; Park, R.M.; Enright, N.; Miklos, A.M.; Hickey, A.S.; Brooks, E.F.; et al. Intragenic DNA Inversions Expand Bacterial Coding Capacity. Nature 2024, 634, 234–242. [Google Scholar] [CrossRef]
- Lim, J.K.; Gunther, N.W., 4th; Zhao, H.; Johnson, D.E.; Keay, S.K.; Mobley, H.L. In Vivo Phase Variation of Escherichia Coli Type 1 Fimbrial Genes in Women with Urinary Tract Infection. Infect. Immun. 1998, 66, 3303–3310. [Google Scholar] [CrossRef] [PubMed]
- Crits-Christoph, A.; Kang, S.C.; Lee, H.H.; Ostrov, N. MicrobeMod: A Computational Toolkit for Identifying Prokaryotic Methylation and Restriction-Modification with Nanopore Sequencing. bioRxiv 2023, 2023. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, A.A.; Osetrova, M.S.; Livinyuk, V.Y.; Manukhov, I.V.; Zavilgelsky, G.B. The Importance of C-Terminal Aspartic Acid Residue (D141) to the Antirestriction Activity of the ArdB (R64) Protein. Mol. Biol. 2017, 51, 724–727. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus Subtilis Group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef]
- Su, Y.; Liu, C.; Fang, H.; Zhang, D. Bacillus Subtilis: A Universal Cell Factory for Industry, Agriculture, Biomaterials and Medicine. Microb. Cell Fact. 2020, 19, 173. [Google Scholar] [CrossRef]
- Song, C.W.; Rathnasingh, C.; Park, J.M.; Kwon, M.; Song, H. CRISPR-Cas9 Mediated Engineering of Bacillus Licheniformis for Industrial Production of (2R,3S)-Butanediol. Biotechnol. Prog. 2021, 37, 1–10. [Google Scholar] [CrossRef]
- Vary, P.S.; Biedendieck, R.; Fuerch, T.; Meinhardt, F.; Rohde, M.; Deckwer, W.-D.; Jahn, D. Bacillus Megaterium--from Simple Soil Bacterium to Industrial Protein Production Host. Appl. Microbiol. Biotechnol. 2007, 76, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Küppers, T.; Steffen, V.; Hellmuth, H.; O’Connell, T.; Bongaerts, J.; Maurer, K.-H.; Wiechert, W. Developing a New Production Host from a Blueprint: Bacillus Pumilus as an Industrial Enzyme Producer. Microb. Cell Fact. 2014, 13, 46. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.; Wang, Q.; Wang, B.; Bian, X.; Zeng, Q.; Lai, D.; Wang, Q.; Li, Y. Efficient Transformation and Genome Editing in a Nondomesticated, Biocontrol Strain, Bacillus Subtilis GLB191. Phytopathol. Res. 2024, 6, 69. [Google Scholar] [CrossRef]
- She, Q.; Hunter, E.; Qin, Y.; Nicolau, S.; Zalis, E.A.; Wang, H.; Chen, Y.; Chai, Y. Negative Interplay between Biofilm Formation and Competence in the Environmental Strains of Bacillus Subtilis. mSystems 2020, 5, 10–1128. [Google Scholar] [CrossRef]
- Huff, F.; Muth, C.; Naumer, C.; Meinhardt, F. The Restriction Modification System of Bacillus Licheniformis MS1 and Generation of a Readily Transformable Deletion Mutant. Appl. Microbiol. Biotechnol. 2017, 101, 7933–7944. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtseva, A.A.; Okhrimenko, I.S.; Didina, V.S.; Zavilgelsky, G.B.; Manukhov, I.V. Antirestriction Protein ArdB (R64) Interacts with DNA. Biochem. 2020, 85, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Alves, J.; Dry, I.; White, J.H.; Dryden, D.T.F.; Lynskey, N.N. Generation of Tools for Expression and Purification of the Phage-Encoded Type I Restriction Enzyme Inhibitor, Ocr. Microbiology 2024, 170, 001465. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.-W.; Hofmann, A.; Kim, J.-H.; Hwangbo, S.-A.; Kim, J.-H.; Kim, J.-W.; Huynh, T.Y.L.; Choy, H.E.; Kim, S.-J.; Lee, J.; et al. Structural Features of a Minimal Intact Methyltransferase of a Type I Restriction-Modification System. Int. J. Biol. Macromol. 2022, 208, 381–389. [Google Scholar] [CrossRef]
- MacWilliams, M.P.; Bickle, T.A. Generation of New DNA Binding Specificity by Truncation of the Type IC EcoDXXI HsdS Gene. EMBO J. 1996, 15, 4775–4783. [Google Scholar] [CrossRef] [PubMed]
- Schiffer, C.J.; Grätz, C.; Pfaffl, M.W.; Vogel, R.F.; Ehrmann, M.A. Characterization of the Staphylococcus Xylosus Methylome Reveals a New Variant of Type I Restriction Modification System in Staphylococci. Front. Microbiol. 2023, 14, 946189. [Google Scholar] [CrossRef]
- Bazhenov, S.V.; Scheglova, E.S.; Utkina, A.A.; Kudryavtseva, A.A.; Al Ebrahim, R.; Manukhov, I.V. New Temperature-Switchable Acyl Homoserine Lactone-Regulated Expression Vector. Appl. Microbiol. Biotechnol. 2022, 107, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Delver, E.P.; Kotova, V.U.; Zavilgelsky, G.B.; Belogurov, A.A. Nucleotide Sequence of the Gene (Ard) Encoding the Antirestriction Protein of Plasmid ColIb-P9. J. Bacteriol. 1991, 173, 5887–5892. [Google Scholar] [CrossRef]
- Sidoruk, K.V.; Levitin, E.I.; Sviridov, B.V.; Piksasova, O.V.; Shustikova, T.E. A Method of DNA Extraction from a Wide Range of Objects via Treatment with Ammonium Salts. Appl. Biochem. Microbiol. 2021, 57, 899–906. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fan, J.; Sun, Z.; Liu, S. NextPolish: A Fast and Efficient Genome Polishing Tool for Long-Read Assembly. Bioinformatics 2020, 36, 2253–2255. [Google Scholar] [CrossRef]
- Nishimura, O.; Hara, Y.; Kuraku, S. GVolante for Standardizing Completeness Assessment of Genome and Transcriptome Assemblies. Bioinformatics 2017, 33, 3635–3637. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Berkeley, M.; Kriventseva, E.V. OrthoDB in 2020: Evolutionary and Functional Annotations of Orthologs. Nucleic Acids Res. 2021, 49, D389–D393. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and Standardized Annotation of Bacterial Genomes via Alignment-Free Sequence Identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise Alignment for Nucleotide Sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A Program for Annotating and Predicting the Effects of Single Nucleotide Polymorphisms, SnpEff: SNPs in the Genome of Drosophila Melanogaster Strain W1118; Iso-2; Iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Hackl, T.; Ankenbrand, M.; van Adrichem, B.; Wilkins, D.; Haslinger, K. Gggenomes: Effective and Versatile Visualizations for Comparative Genomics. arXiv 2024, arXiv:2411.13556. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Bailey, T.L. STREME: Accurate and Versatile Sequence Motif Discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Abdennur, N.; Fudenberg, G.; Flyamer, I.M.; Galitsyna, A.A.; Goloborodko, A.; Imakaev, M.; Venev, S. Bioframe: Operations on Genomic Intervals in Pandas Dataframes. Bioinformatics 2024, 40, btae088. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]

| Strain | EOP * | t-Test |
|---|---|---|
| E. coli TG1 | 1 | |
| E. coli TG1 pIRal-2_RM-Ia [18] | 0.011 ± 0.007 | The means are not significantly different at p < 0.05 |
| E. coli TG1pIR-DPAl-BlihIA-His-TAG | 0.022 ± 0.005 |
| Name | Resistance | Description |
|---|---|---|
| pIRal-2_RM-Ia [18] | Kn | Source of blihIA genes |
| pIR-DPAl [39] | Kn | Temperature-switchable acyl homoserine lactone-regulated expression vector |
| pIR-DPAl_Blich_site (this work) | Kn | pIR-DPAI based plasmid contains the single BlihIA recognition site |
| pIR-DPAl_Blich_site_Mut (this work) | Kn | pIR-DPAI-based plasmid contains the mutation in a single BlihIA recognition site |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kudryavtseva, A.; Berezov, R.; Utkina, A.; Kotovskaya, O.; Skutel, M.; Trofimova, A.; Isaev, A.; Manukhov, I. BlihIA—A Novel Type I Restriction-Modification System from Bacillus licheniformis Is Sensitive to In Vitro Inhibition by ArdB Antirestriction Protein. Int. J. Mol. Sci. 2025, 26, 8674. https://doi.org/10.3390/ijms26178674
Kudryavtseva A, Berezov R, Utkina A, Kotovskaya O, Skutel M, Trofimova A, Isaev A, Manukhov I. BlihIA—A Novel Type I Restriction-Modification System from Bacillus licheniformis Is Sensitive to In Vitro Inhibition by ArdB Antirestriction Protein. International Journal of Molecular Sciences. 2025; 26(17):8674. https://doi.org/10.3390/ijms26178674
Chicago/Turabian StyleKudryavtseva, Anna, Rodion Berezov, Anna Utkina, Oksana Kotovskaya, Mikhail Skutel, Anna Trofimova, Artem Isaev, and Ilya Manukhov. 2025. "BlihIA—A Novel Type I Restriction-Modification System from Bacillus licheniformis Is Sensitive to In Vitro Inhibition by ArdB Antirestriction Protein" International Journal of Molecular Sciences 26, no. 17: 8674. https://doi.org/10.3390/ijms26178674
APA StyleKudryavtseva, A., Berezov, R., Utkina, A., Kotovskaya, O., Skutel, M., Trofimova, A., Isaev, A., & Manukhov, I. (2025). BlihIA—A Novel Type I Restriction-Modification System from Bacillus licheniformis Is Sensitive to In Vitro Inhibition by ArdB Antirestriction Protein. International Journal of Molecular Sciences, 26(17), 8674. https://doi.org/10.3390/ijms26178674






