Liquid Biopsy as a Means of Assessing Prognosis and Identifying Novel Risk Factors in Multiple Myeloma
Abstract
1. Introduction
2. Results
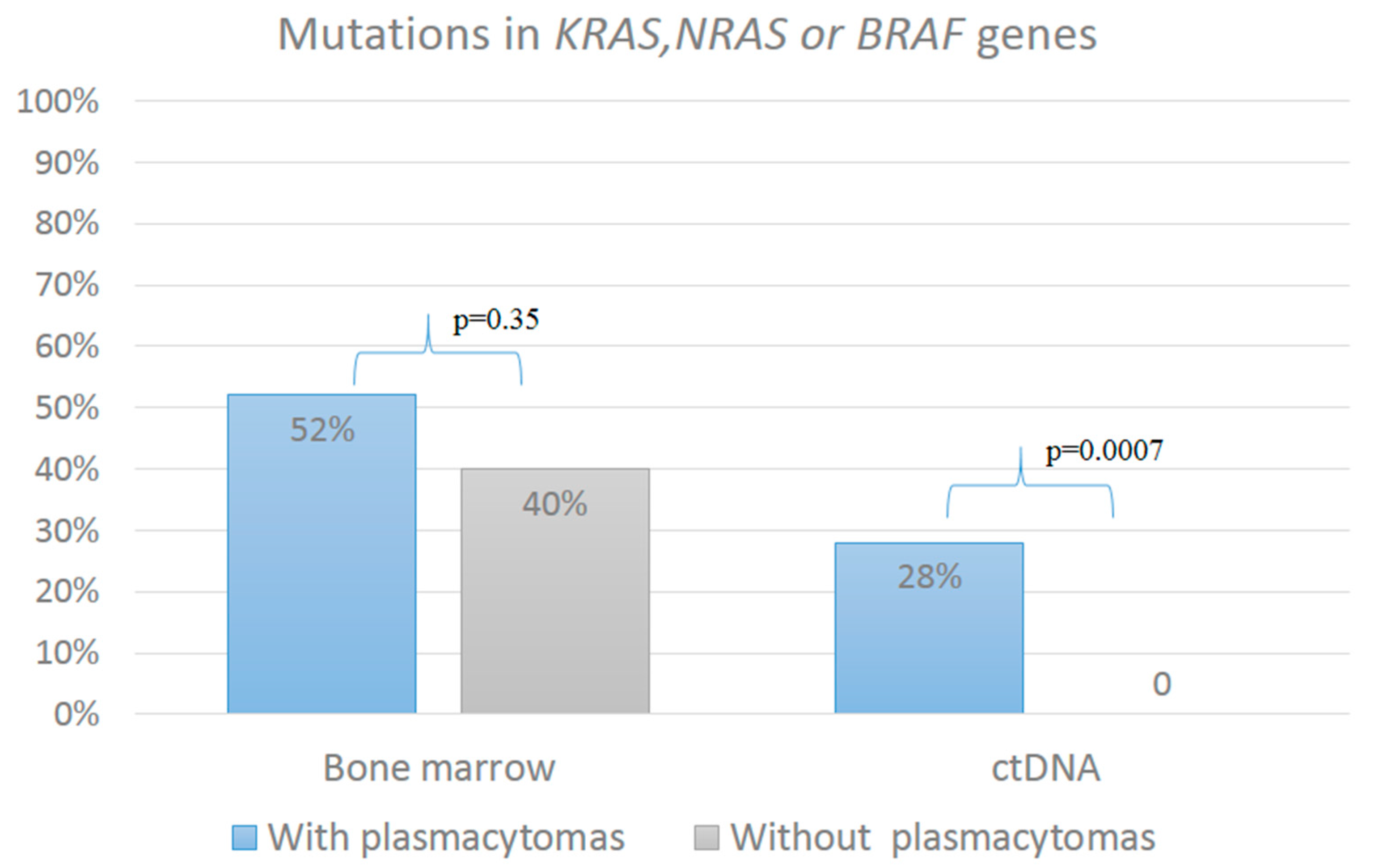
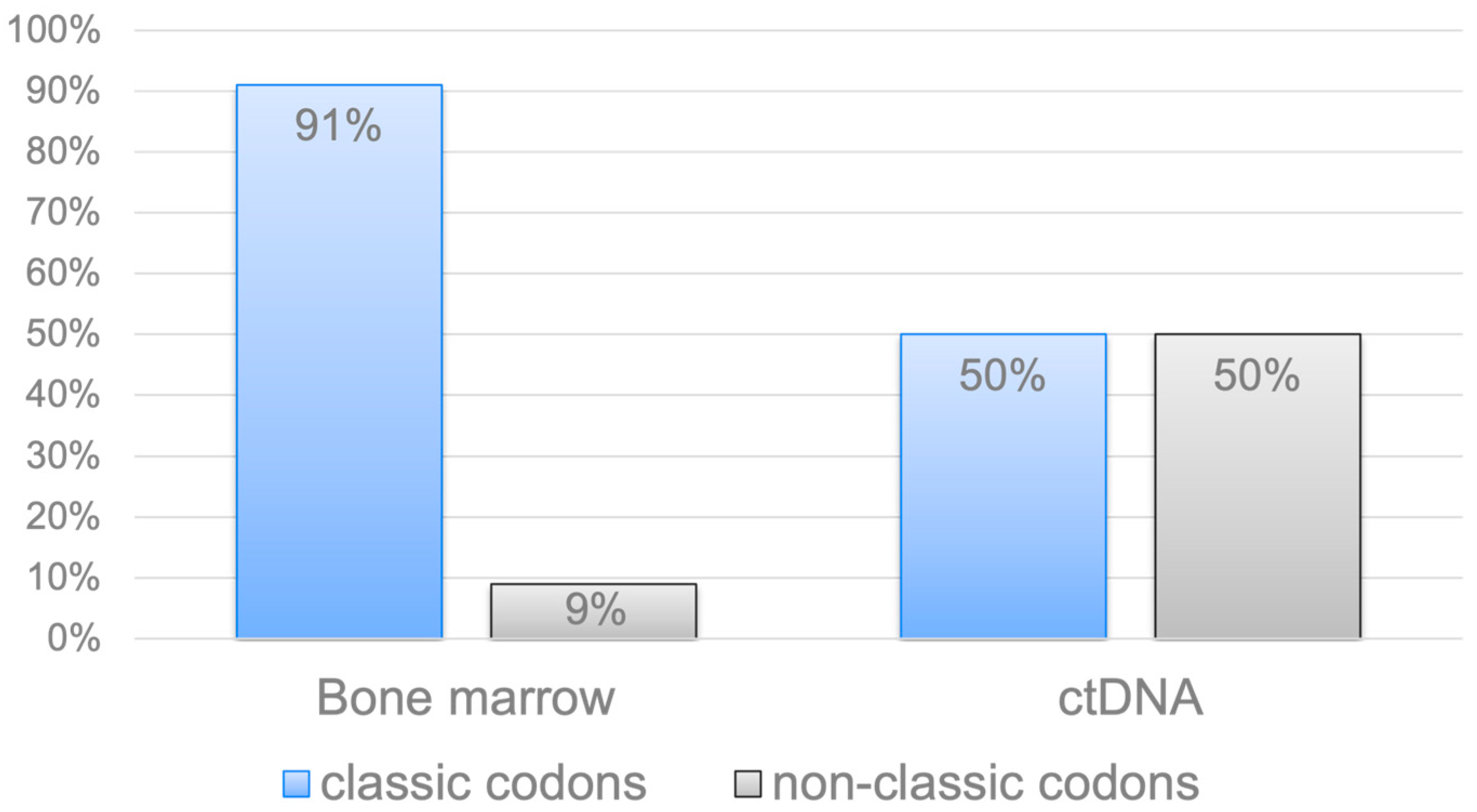
2.1. Analysis of the KRAS, NRAS, and BRAF Gene Mutations
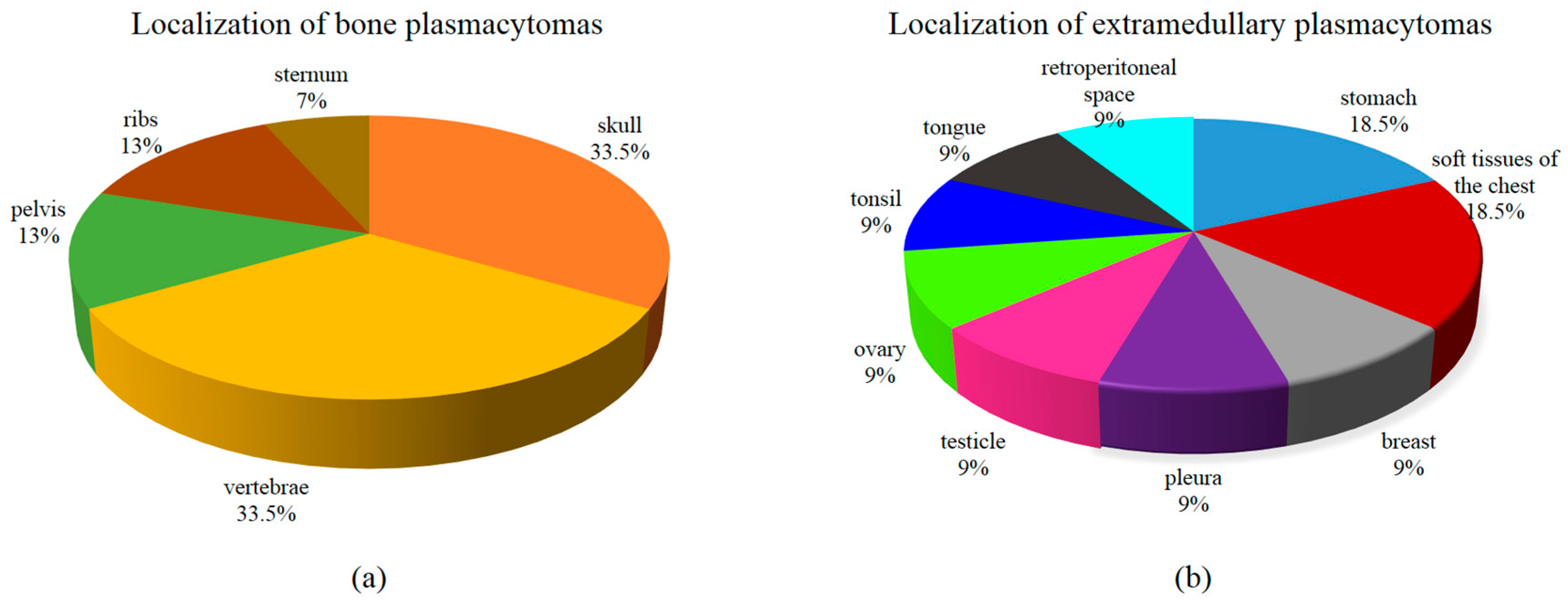
2.2. Analysis of the Clinical Course of MM Complicated with Plasmacytoma
3. Discussion
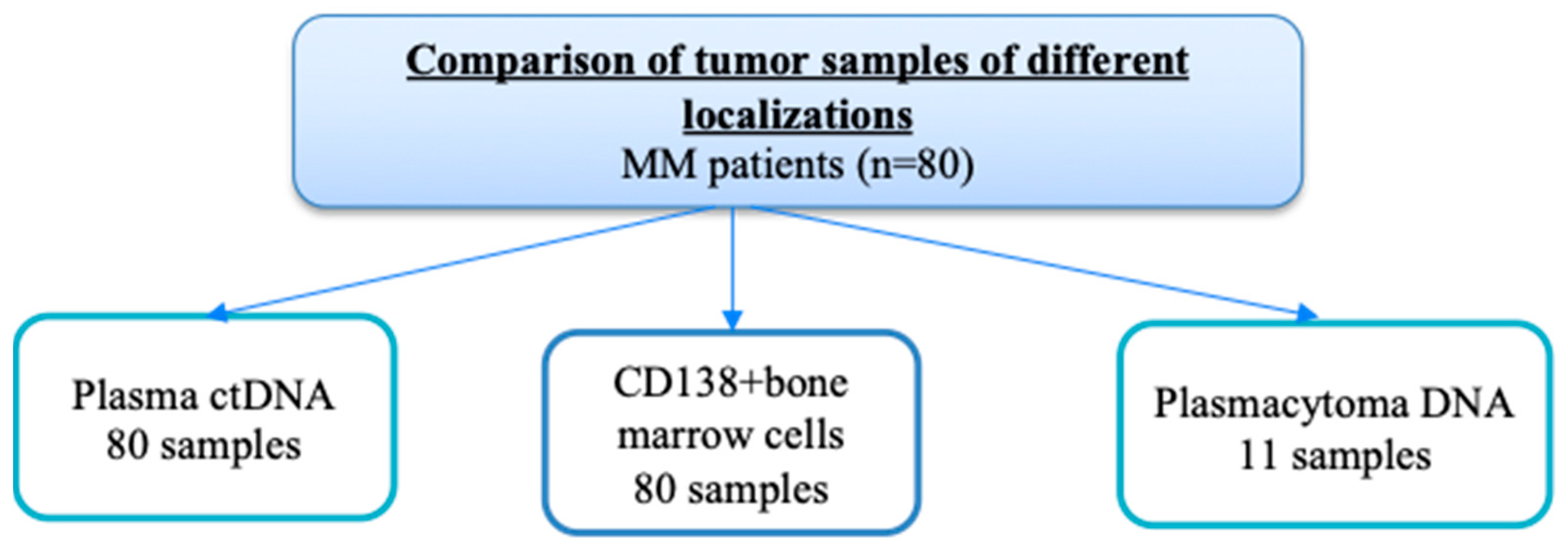
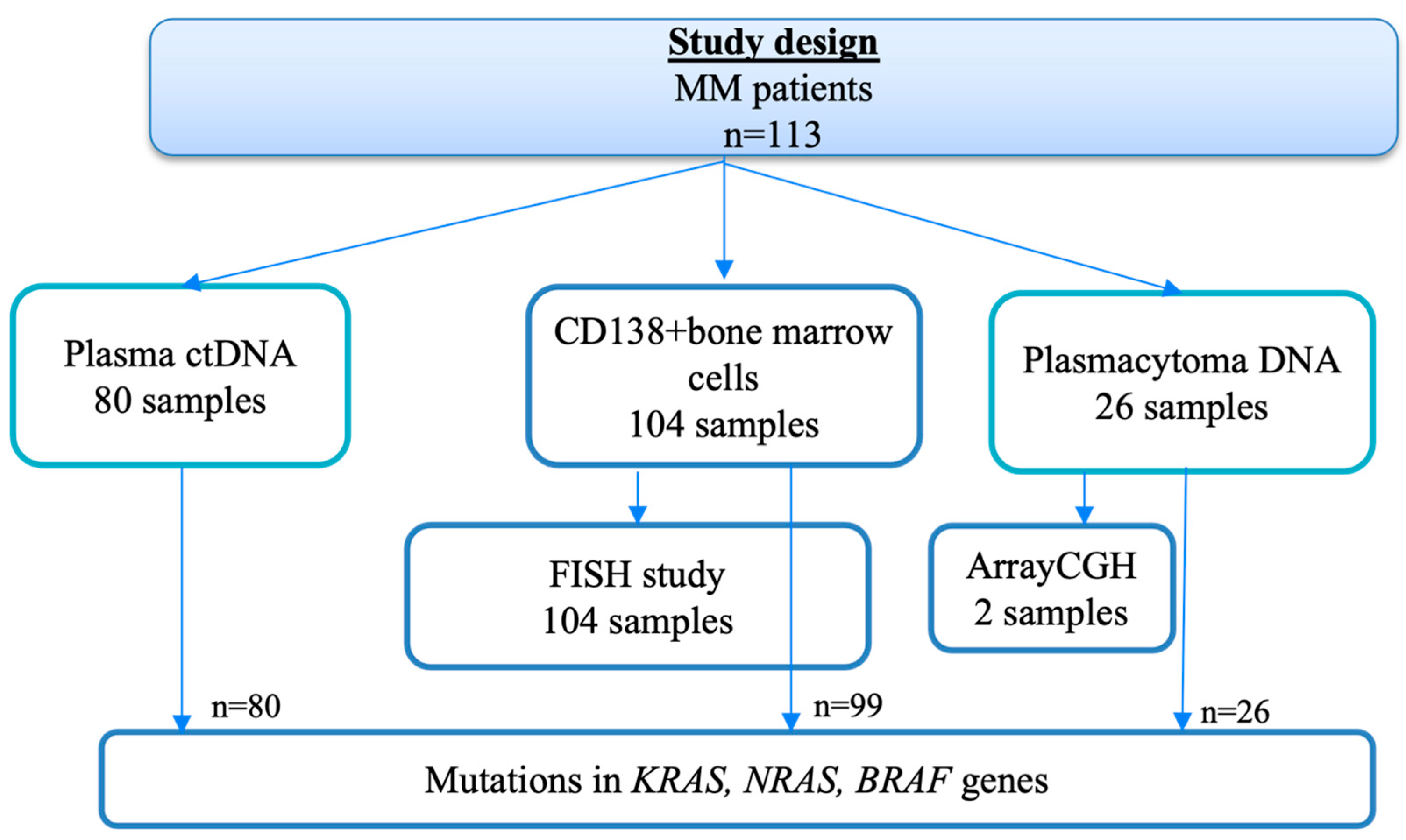
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ctDNA | Circulating tumor DNA |
| MM | Multiple myeloma |
| MGUS | Monoclonal gammapathy of undetermined significance |
| IMWG | International Myeloma Working Group |
| CT | Computer tomography |
| CNVs | Copy number variations |
| array-CGH | Comparative genomic hybridization on a microarray |
| FISH | Fluorescence in situ hybridization |
| D-S stage | Durie-Salmon stage |
| ISS | International Staging System |
| LDH | Lactate dehydrogenase |
| FLC | Free light chains |
| BJ | Bence Jones |
| HSCs | Hematopoietic stem cells |
| cnLOH | Copy number neutral loss of heterozygosity |
| Mb | Megabase |
References
- Bolli, N.; Avet-Loiseau, H.; Wedge, D.C.; Van Loo, P.; Alexandrov, L.B.; Martincorena, I.; Dawson, K.J.; Iorio, F.; Nik-Zainal, S.; Bignell, G.R.; et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat. Commun. 2014, 5, 2997. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 2017, 8, 268. [Google Scholar] [CrossRef]
- Rosiñol, L.; Beksac, M.; Zamagni, E.; Van de Donk, N.W.C.J.; Anderson, K.C.; Badros, A.; Caers, J.; Cavo, M.; Dimopoulos, M.A.; Dispenzieri, A.; et al. Expert review on soft-tissue plasmacytomas in multiple myeloma: Definition, disease assessment and treatment considerations. Br. J. Haematol. 2021, 194, 496–507. [Google Scholar] [CrossRef]
- Short, K.D.; Rajkumar, S.V.; Larson, D.; Buadi, F.; Hayman, S.; Dispenzieri, A.; Gertz, M.; Kumar, S.; Mikhael, J.; Roy, V.; et al. Incidence of extramedullary disease in patients with multiple myeloma in the era of novel therapy, and the activity of pomalidomide on extramedullary myeloma. Leukemia 2011, 25, 906–908. [Google Scholar] [CrossRef]
- Jagosky, M.H.; Usmani, S.Z. Extramedullary Disease in Multiple Myeloma. Curr. Hematol. Malig. Rep. 2020, 15, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Weinstock, M.; Aljawai, Y.; Morgan, E.A.; Laubach, J.; Gannon, M.; Roccaro, A.M.; Varga, C.; Mitsiades, C.S.; Paba-Prada, C.; Schlossman, R.; et al. Incidence and clinical features of extramedullary multiple myeloma in patients who underwent stem cell transplantation. Br. J. Haematol. 2015, 169, 851–858. [Google Scholar] [CrossRef]
- Dahl, I.M.; Rasmussen, T.; Kauric, G.; Husebekk, A. Differential expression of CD56 and CD44 in the evolution of extramedullary myeloma. Br. J. Haematol. 2002, 116, 273–277. [Google Scholar] [CrossRef]
- Firsova, M.V.; Mendeleeva, L.P.; Kovrigina, A.M.; Solovev, M.V.; Savchenko, V.G. Plasmacytoma in patients with multiple myeloma: Morphology and immunohistochemistry. BMC Cancer 2020, 20, 346. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, W.; Wang, J. Progress in the identification of gene mutations involved in multiple myeloma. Onco Targets Ther. 2019, 12, 4075–4080. [Google Scholar] [CrossRef]
- Chng, W.J.; Gonzalez-Paz, N.; Price-Troska, T.; Jacobus, S.; Rajkumar, S.V.; Oken, M.M.; Kyle, R.A.; Henderson, K.J.; Van Wier, S.; Greipp, P.; et al. Clinical and biological significance of RAS mutations in multiple myeloma. Leukemia 2008, 22, 2280–2284. [Google Scholar] [CrossRef]
- Mulligan, G.; Lichter, D.I.; Di Bacco, A.; Blakemore, S.J.; Berger, A.; Koenig, E.; Bernard, H.; Trepicchio, W.; Li, B.; Neuwirth, R.; et al. Mutation of NRAS but not KRAS significantly reduces myeloma sensitivity to single-agent bortezomib therapy. Blood 2014, 123, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, N.; Biersack, H.; Czerwinska, A.C.; Schemme, J.; Hardel, T.T.; Bernard, V.; Rades, D.; Lehnert, H.; Luley, K.B.; Thorns, C. Favorable prognostic impact of RAS mutation status in multiple myeloma treated with high-dose melphalan and autologous stem cell support in the era of novel agents: A single center perspective. Leuk. Lymphoma 2016, 57, 226–229. [Google Scholar] [CrossRef]
- Laganà, A.; Perumal, D.; Melnekoff, D.; Readhead, B.; Kidd, B.A.; Leshchenko, V.; Kuo, P.Y.; Keats, J.; DeRome, M.; Yesil, J.; et al. Integrative network analysis identifies novel drivers of pathogenesis and progression in newly diagnosed multiple myeloma. Leukemia 2018, 32, 120–130. [Google Scholar] [CrossRef]
- Perroud, C.; Thurian, D.; Andres, M.; Künzi, A.; Wiedemann, G.; Zeerleder, S.; Bacher, U.; Pabst, T.; Banz, Y.; Porret, N.; et al. Effect of MAPK activation via mutations in NRAS, KRAS and BRAF on clinical outcome in newly diagnosed multiple myeloma. Hematol. Oncol. 2023, 41, 912–921. [Google Scholar] [CrossRef]
- Shirazi, F.; Jones, R.J.; Singh, R.K.; Zou, J.; Kuiatse, I.; Berkova, Z.; Wang, H.; Lee, H.C.; Hong, S.; Dick, L.; et al. Activating KRAS, NRAS, and BRAF mutants enhance proteasome capacity and reduce endoplasmic reticulum stress in multiple myeloma. Proc. Natl. Acad. Sci. USA 2020, 117, 20004–20014. [Google Scholar] [CrossRef]
- Yang, Y.; Bolomsky, A.; Oellerich, T.; Chen, P.; Ceribelli, M.; Häupl, B.; Wright, G.W.; Phelan, J.D.; Huang, D.W.; Lord, J.W.; et al. Oncogenic RAS commandeers amino acid sensing machinery to aberrantly activate mTORC1 in multiple myeloma. Nat. Commun. 2022, 13, 5469, Erratum in Nat. Commun. 2022, 13, 5030. [Google Scholar] [CrossRef]
- Pasca, S.; Tomuleasa, C.; Teodorescu, P.; Ghiaur, G.; Dima, D.; Moisoiu, V.; Berce, C.; Stefan, C.; Ciechanover, A.; Einsele, H. KRAS/NRAS/BRAF Mutations as Potential Targets in Multiple Myeloma. Front. Oncol. 2019, 9, 1137. [Google Scholar] [CrossRef]
- Allegra, A.; Cancemi, G.; Mirabile, G.; Tonacci, A.; Musolino, C.; Gangemi, S. Circulating Tumour Cells, Cell Free DNA and Tumour-Educated Platelets as Reliable Prognostic and Management Biomarkers for the Liquid Biopsy in Multiple Myeloma. Cancers 2022, 14, 4136. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.; Caetano, J.; Barahona, F.; Lopes, R.; Carneiro, E.; Costa-Silva, B.; João, C. Liquid biopsies for multiple myeloma in a time of precision medicine. J. Mol. Med. 2020, 98, 513–525, Correction in J. Mol. Med. 2024, 102, 1527. [Google Scholar] [CrossRef] [PubMed]
- Heestermans, R.; De Brouwer, W.; Maes, K.; Vande Broek, I.; Vaeyens, F.; Olsen, C.; Caljon, B.; De Becker, A.; Bakkus, M.; Schots, R.; et al. Liquid Biopsy-Derived DNA Sources as Tools for Comprehensive Mutation Profiling in Multiple Myeloma: A Comparative Study. Cancers 2022, 14, 4901. [Google Scholar] [CrossRef]
- Vlachová, M.; Pečinka, L.; Gregorová, J.; Moráň, L.; Růžičková, T.; Kovačovicová, P.; Almáši, M.; Pour, L.; Štork, M.; Hájek, R.; et al. Liquid biopsy of peripheral blood using mass spectrometry detects primary extramedullary disease in multiple myeloma patients. Sci Rep. 2024, 14, 18777. [Google Scholar] [CrossRef]
- Soloveva, M.; Solovev, M.; Risinskaya, N.; Nikulina, E.; Yakutik, I.; Biderman, B.; Obukhova, T.; Chabaeva, Y.; Kulikov, S.; Sudarikov, A.; et al. Loss of Heterozygosity and Mutations in the RAS-ERK Pathway Genes in Tumor Cells of Various Loci in Multiple Myeloma. Int. J. Mol. Sci. 2024, 25, 9426. [Google Scholar] [CrossRef]
- Kadam Amare, P.; Nikalje Khasnis, S.; Hande, P.; Lele, H.; Wable, N.; Kaskar, S.; Nikam Gujar, N.; Gardi, N.; Prabhudesai, A.; Todi, K.; et al. Cytogenetic Abnormalities in Multiple Myeloma: Incidence, Prognostic Significance, and Geographic Heterogeneity in Indian and Western Populations. Cytogenet. Genome Res. 2022, 162, 529–540. [Google Scholar] [CrossRef]
- Zhan, F.; Colla, S.; Wu, X.; Chen, B.; Stewart, J.P.; Kuehl, W.M.; Barlogie, B.; Shaughnessy, J.D., Jr. CKS1B, overexpressed in aggressive disease, regulates multiple myeloma growth and survival through SKP2- and p27Kip1-dependent and -independent mechanisms. Blood 2007, 109, 4995–5001. [Google Scholar] [CrossRef] [PubMed]
- Hanamura, I. Gain/Amplification of Chromosome Arm 1q21 in Multiple Myeloma. Cancers 2021, 13, 256. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; de Ramón, C.; Corchete, L.A.; Krzeminski, P.; Rojas, E.A.; Isidro, I.; García-Sanz, R.; Martínez-López, J.; Oriol, A.; Bladé, J.; et al. Quantitative expression of Ikaros, IRF4, and PSMD10 proteins predicts survival in VRD-treated patients with multiple myeloma. Blood Adv. 2020, 4, 6023–6033. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessy, J.D., Jr.; Qu, P.; Usmani, S.; Heuck, C.J.; Zhang, Q.; Zhou, Y.; Tian, E.; Hanamura, I.; van Rhee, F.; Anaissie, E.; et al. Pharmacogenomics of bortezomib test-dosing identifies hyperexpression of proteasome genes, especially PSMD4, as novel high-risk feature in myeloma treated with Total Therapy 3. Blood 2011, 118, 3512–3524. [Google Scholar] [CrossRef]
- Menges, C.W.; Altomare, D.A.; Testa, J.R. FAS-associated factor 1 (FAF1): Diverse functions and implications for oncogenesis. Cell Cycle 2009, 8, 2528–2534. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.E.; Walker, B.A.; Jenner, M.W.; Chiecchio, L.; Dagrada, G.; Protheroe, R.K.; Johnson, D.C.; Dickens, N.J.; Brito, J.L.; Else, M.; et al. Deletions of CDKN2C in multiple myeloma: Biological and clinical implications. Clin. Cancer Res. 2008, 14, 6033–6041. [Google Scholar] [CrossRef]
- Shaughnessy, J.D., Jr.; Zhan, F.; Burington, B.E.; Huang, Y.; Colla, S.; Hanamura, I.; Stewart, J.P.; Kordsmeier, B.; Randolph, C.; Williams, D.R.; et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood 2007, 109, 2276–2284. [Google Scholar] [CrossRef]
- Mlynarcikova, M.; Balcarkova, J.; Mickova, P.; Scudla, V.; Pika, T.; Bacovsky, J.; Minarik, J.; Janousova, E.; Jarosova, M. Molecular Cytogenetic Analysis of Chromosome 8 Aberrations in Patients With Multiple Myeloma Examined in 2 Different Stages, at Diagnosis and at Progression/Relapse. Clin. Lymphoma Myeloma Leuk. 2016, 16, 358–365. [Google Scholar] [CrossRef]
- Firsova, M.V.; Risinskaya, N.V.; Solovev, M.V.; Obukhova, T.N.; Kislitsyna, M.A.; Nikulina, E.E.; Yakutik, I.A.; Abramova, T.V.; Sudarikov, A.B.; Kovrigina, A.M.; et al. Multiple myeloma with extramedullary plasmacytoma: Pathogenesis and clinical case. Oncohematology 2022, 17, 67–80. (In Russian) [Google Scholar] [CrossRef]
- Staniek, J.; Lorenzetti, R.; Heller, B.; Janowska, I.; Schneider, P.; Unger, S.; Warnatz, K.; Seidl, M.; Venhoff, N.; Thiel, J.; et al. TRAIL-R1 and TRAIL-R2 Mediate TRAIL-Dependent Apoptosis in Activated Primary Human B Lymphocytes. Front. Immunol. 2019, 10, 951. [Google Scholar] [CrossRef]
- Dib, A.; Gabrea, A.; Glebov, O.K.; Bergsagel, P.L.; Kuehl, W.M. Characterization of MYC translocations in multiple myeloma cell lines. J. Natl. Cancer Inst. Monogr. 2008, 2008, 25–31. [Google Scholar] [CrossRef]
- Kuzyk, A.; Mai, S. c-MYC-induced genomic instability. Cold Spring Harb. Perspect. Med. 2014, 4, a014373. [Google Scholar] [CrossRef]
- Muñoz-Bellvis, L.; Fontanillo, C.; González-González, M.; Garcia, E.; Iglesias, M.; Esteban, C.; Gutierrez, M.L.; Abad, M.M.; Bengoechea, O.; De Las Rivas, J.; et al. Unique genetic profile of sporadic colorectal cancer liver metastasis versus primary tumors as defined by high-density single-nucleotide polymorphism arrays. Mod. Pathol. 2012, 25, 590–601. [Google Scholar] [CrossRef]
- Brandt, V.P.; Holland, H.; Wallenborn, M.; Koschny, R.; Frydrychowicz, C.; Richter, M.; Holland, L.; Nestler, U.; Sander, C. SNP array genomic analysis of matched pairs of brain and liver metastases in primary colorectal cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 18173–18183. [Google Scholar] [CrossRef]
- Wang, S.Y.; Hao, H.L.; Deng, K.; Li, Y.; Cheng, Z.Y.; Lv, C.; Liu, Z.M.; Yang, J.; Pan, L. Expression levels of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) and focal adhesion kinase in patients with multiple myeloma and their relationship to clinical stage and extramedullary infiltration. Leuk. Lymphoma 2012, 53, 1162–1168. [Google Scholar] [CrossRef]
- Kienast, J.; Berdel, W.E. c-maf in multiple myeloma: An oncogene enhancing tumor-stroma interactions. Cancer Cell 2004, 5, 109–110. [Google Scholar] [CrossRef]
- Chang, H.; Qi, Q.; Xu, W.; Patterson, B. c-Maf nuclear oncoprotein is frequently expressed in multiple myeloma. Leukemia 2007, 21, 1572–1574. [Google Scholar] [CrossRef]
- Righolt, C.; Mai, S. Shattered and stitched chromosomes-chromothripsis and chromoanasynthesis-manifestations of a new chromosome crisis? Genes Chromosomes Cancer 2012, 51, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Magrangeas, F.; Avet-Loiseau, H.; Munshi, N.C.; Minvielle, S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 2011, 118, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Rustad, E.H.; Yellapantula, V.D.; Glodzik, D.; Maclachlan, K.H.; Diamond, B.; Boyle, E.M.; Ashby, C.; Blaney, P.; Gundem, G.; Hultcrantz, M.; et al. Revealing the impact of structural variants in multiple myeloma. Blood Cancer Discov. 2020, 1, 258–273. [Google Scholar] [CrossRef] [PubMed]



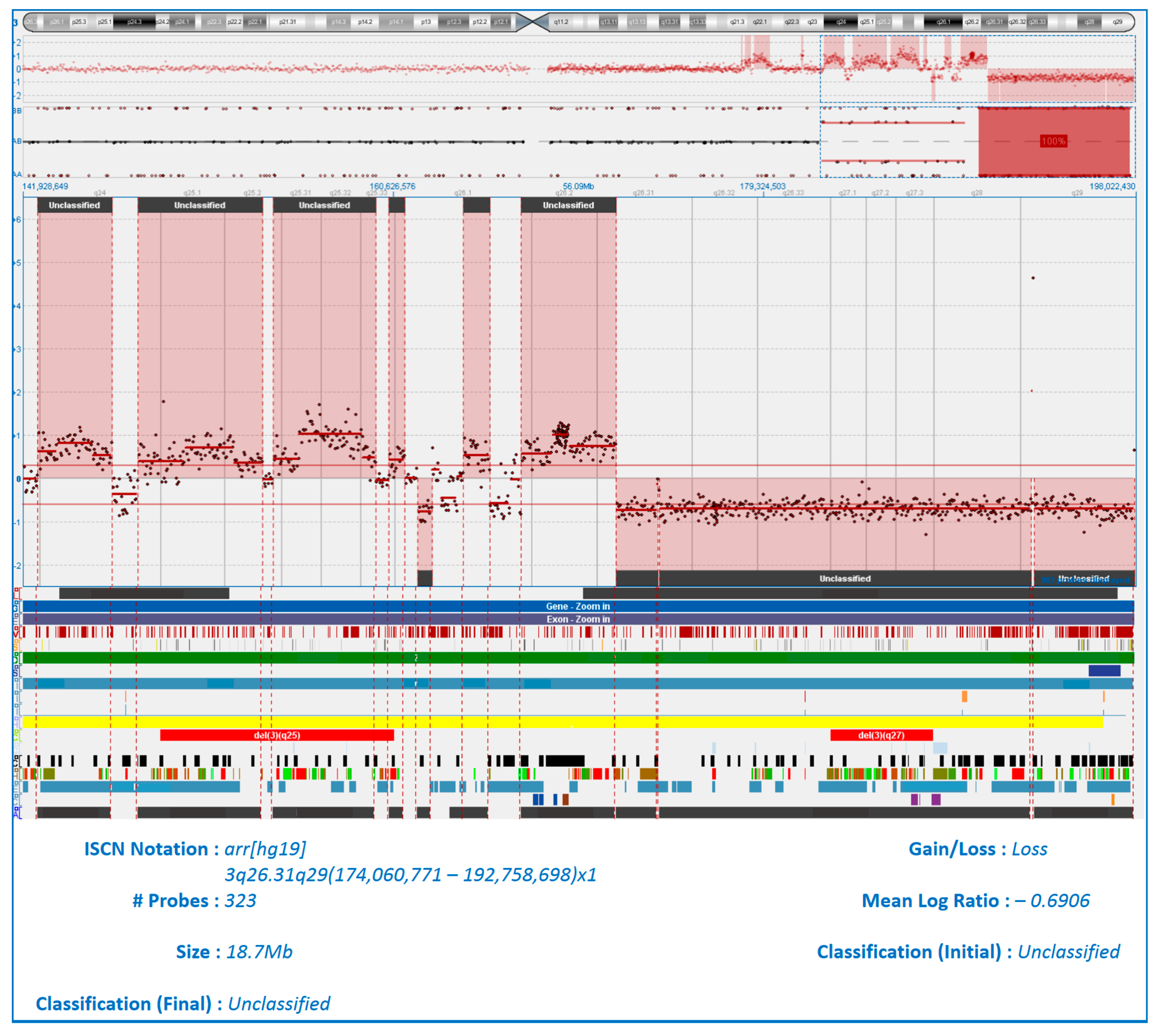
| Parameters | Patients with Plasmacytomas | Patients Without Plasmacytomas | p |
|---|---|---|---|
| (n = 50) | (n = 30) | ||
| Age, years, median, and range | 56.5 (41–73) | 55 (29–83) | 0.85 |
| Male/female | 22 (44%)/28 (56%) | 14 (47%)/16 (53%) | 0.82 |
| Type of secretion | 0.88 | ||
| G | 30 (60%) | 16 (54%) | |
| A | 10(20%) | 9 (30%) | |
| BJ | 5 (10%) | 3 (10%) | |
| D | 3 (6%) | 1 (3%) | |
| FLC | 2 (4%) | 1 (3%) | |
| BJ protein excretion | 0.63 | ||
| Yes | 31 (62%) | 21 (70%) | |
| No | 19 (38%) | 9 (30%) | |
| Type of FLC | 0.1 | ||
| κ | 32 (64%) | 17 (57%) | |
| λ | 18 (36%) | 13 (43%) | |
| D-S stage | <0.0001 | ||
| IA, IB | 1 (2%) | 5 (17%) | |
| IIA | 3 (6%) | 8 (27%) | |
| IIIA | 37 (74%) | 14 (47%) | |
| IIIB | 9 (18%) | 3 (10%) | |
| ISS stage | 0.94 | ||
| I | 16 (32%) | 10 (33%) | |
| II | 11 (22%) | 8 (27%) | |
| III | 9 (18%) | 4 (13%) | |
| ND | 14 (28%) | 8 (27%) | |
| Hemoglobin (g/L), median, and range | 115 (66–148) | 108.5 (72–156) | 0.39 |
| LDH (U/L), median, and range | 182.5 (65 -694) | 166.5 (71–385) | 0.25 |
| Plasma cells in bone marrow aspiration, %, median, and range | 17.6 (0–92.0) | 15.1 (4.0–56.0) | 0.95 |
| FISH | 0.68 | ||
| Standard risk | 21 (42%) | 16 (53%) | |
| High risk | 28 (56%) | 14 (46%) | |
| ND | 1 (2%) | 0 | |
| Double/Triple hit (n = 42) | 0.69 | ||
| Yes | 7 (25%) | 2 (14%) | |
| no | 21 (75%) | 12 (86%) |
| Mutations in Genes | Patients with Plasmacytomas (n = 50) | Patients Without Plasmacytomas (n = 30) | p |
|---|---|---|---|
| In bone marrow | |||
| KRAS | 6 (12%) | 7 (23%) | 0.2 |
| NRAS | 11 (22%) | 2 (7%) | 0.12 |
| BRAF | 7 (14%) | 2 (7%) | 0.5 |
| KRAS + NRAS | 1 (2%) | 0 | 1 |
| NRAS + BRAF | 1 (2%) | 1 (3%) | 1 |
| In ctDNA | |||
| KRAS | 5 (10%) | - | 0.15 |
| NRAS | 5 (10%) | - | 0.15 |
| BRAF | 4 (8%) | - | 0.29 |
| Chromosomal Localization | Type of Aberration | Size, Mb 1 |
|---|---|---|
| 1p34 | del | 4.67–25.59 |
| 1q21 | gain | 8.41–22.36 |
| 10q11 | del | 92.79 |
| 16q11.2 | del | 32.55–43.54 |
| 17p13 | del | 8.05–18.89 |
| Xp22 | del | 58.37 |
| Xq11 | del | 25.18–92.89 |
| Parameters | Patients with MM (n = 113) |
|---|---|
| Age, years, median, and range | 55 (29–83) |
| Male/female | 48/65 |
| Type of secretion | |
| G | 69 (61%) |
| A | 23 (20.4%) |
| BJ | 15 (13.3%) |
| D | 4 (3.5%) |
| non-secretory | 2 (1,8%) |
| Type of FLC | n = 111 |
| κ | 68 (61%) |
| λ | 43(39%) |
| D-S stage | n = 112 |
| IA, IB | 6 (5.3%) |
| IIA | 19 (17%) |
| IIIA | 68 (60.7%) |
| IIIB | 19 (17%) |
| ISS stage | n = 79 |
| I–II–III | 46%–28%–26% |
| Hemoglobin (g/L), median, and range | 110 (55–156) |
| LDH (U/L), median, and range | 174 (65–694) |
| Plasma cells in bone marrow aspiration, %, median, and range | 16 (0.4–58) |
| FISH | n = 104 |
| Standard risk | 48 (46%) |
| High risk | 56 (54%) |
| Plasmacytomas | |
| yes | 74 (65%) |
| no | 39 (35%) |
| n = 74 | |
| bone (B) | 59 (80%) |
| extramedullary (E) | 5 (7%) |
| B + E | 10 (13%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soloveva, M.; Solovev, M.; Yakutik, I.; Biderman, B.; Nikulina, E.; Risinskaya, N.; Obukhova, T.; Gladysheva, M.; Kovrigina, A.; Chabaeva, Y.; et al. Liquid Biopsy as a Means of Assessing Prognosis and Identifying Novel Risk Factors in Multiple Myeloma. Int. J. Mol. Sci. 2025, 26, 8505. https://doi.org/10.3390/ijms26178505
Soloveva M, Solovev M, Yakutik I, Biderman B, Nikulina E, Risinskaya N, Obukhova T, Gladysheva M, Kovrigina A, Chabaeva Y, et al. Liquid Biopsy as a Means of Assessing Prognosis and Identifying Novel Risk Factors in Multiple Myeloma. International Journal of Molecular Sciences. 2025; 26(17):8505. https://doi.org/10.3390/ijms26178505
Chicago/Turabian StyleSoloveva, Maiia, Maksim Solovev, Igor Yakutik, Bella Biderman, Elena Nikulina, Natalya Risinskaya, Tatiana Obukhova, Maria Gladysheva, Alla Kovrigina, Yulia Chabaeva, and et al. 2025. "Liquid Biopsy as a Means of Assessing Prognosis and Identifying Novel Risk Factors in Multiple Myeloma" International Journal of Molecular Sciences 26, no. 17: 8505. https://doi.org/10.3390/ijms26178505
APA StyleSoloveva, M., Solovev, M., Yakutik, I., Biderman, B., Nikulina, E., Risinskaya, N., Obukhova, T., Gladysheva, M., Kovrigina, A., Chabaeva, Y., Kulikov, S., Sudarikov, A., & Mendeleeva, L. (2025). Liquid Biopsy as a Means of Assessing Prognosis and Identifying Novel Risk Factors in Multiple Myeloma. International Journal of Molecular Sciences, 26(17), 8505. https://doi.org/10.3390/ijms26178505






