Analysis of Wheat Pollen Ole E I Proteins Reveals Potential Roles in Fertility and Stress Adaptation
Abstract
1. Introduction
2. Results
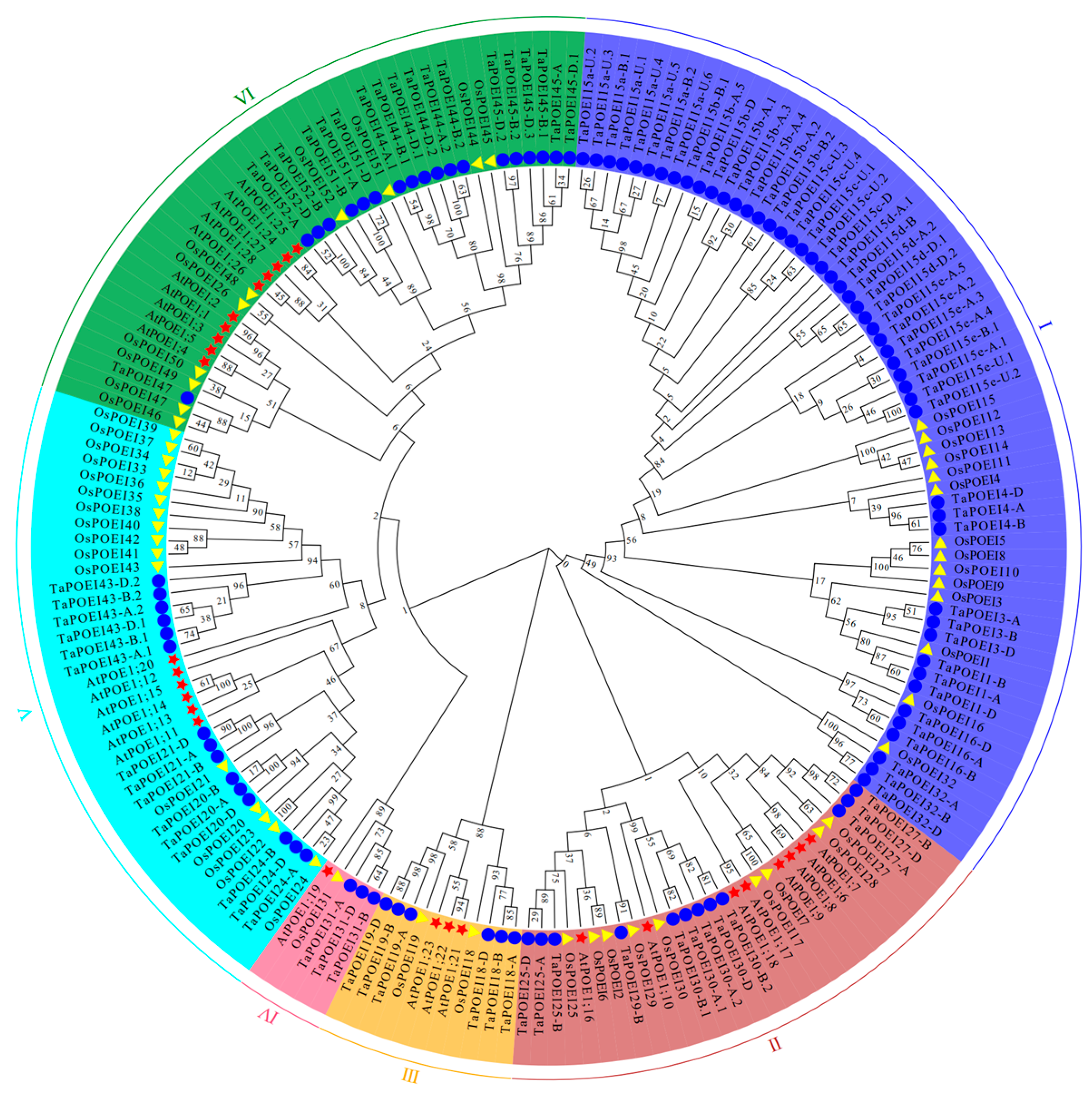
2.1. Genome-Wide Identification and Chromosomal Distribution, and Evolutionary Analysis of Putative POEI Genes in Wheat: Insights into Environmental Adaptation
2.2. Structural and Regulatory Features of TaPOEI Genes: Insights from Motif and Promoter Analysis
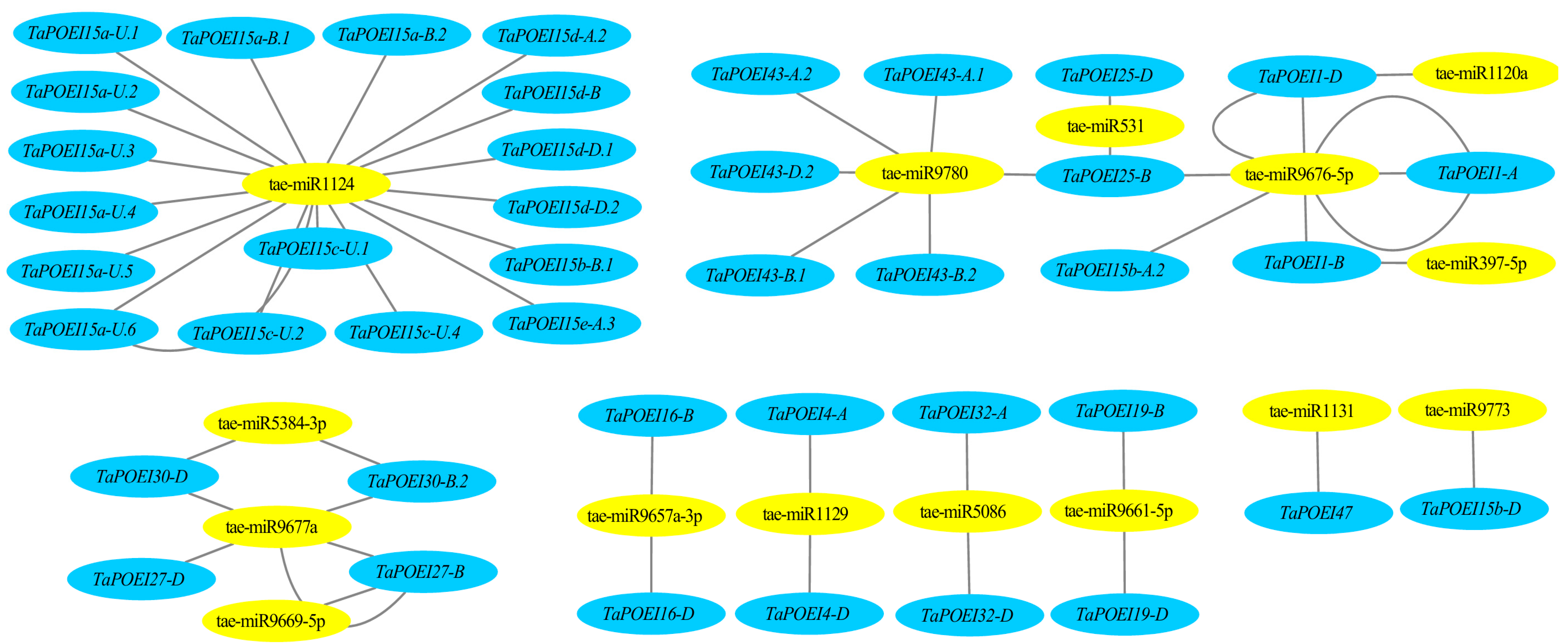
2.3. Interactions Between miRNAs and TaPOEI Genes: Implications for Wheat Reproductive Development
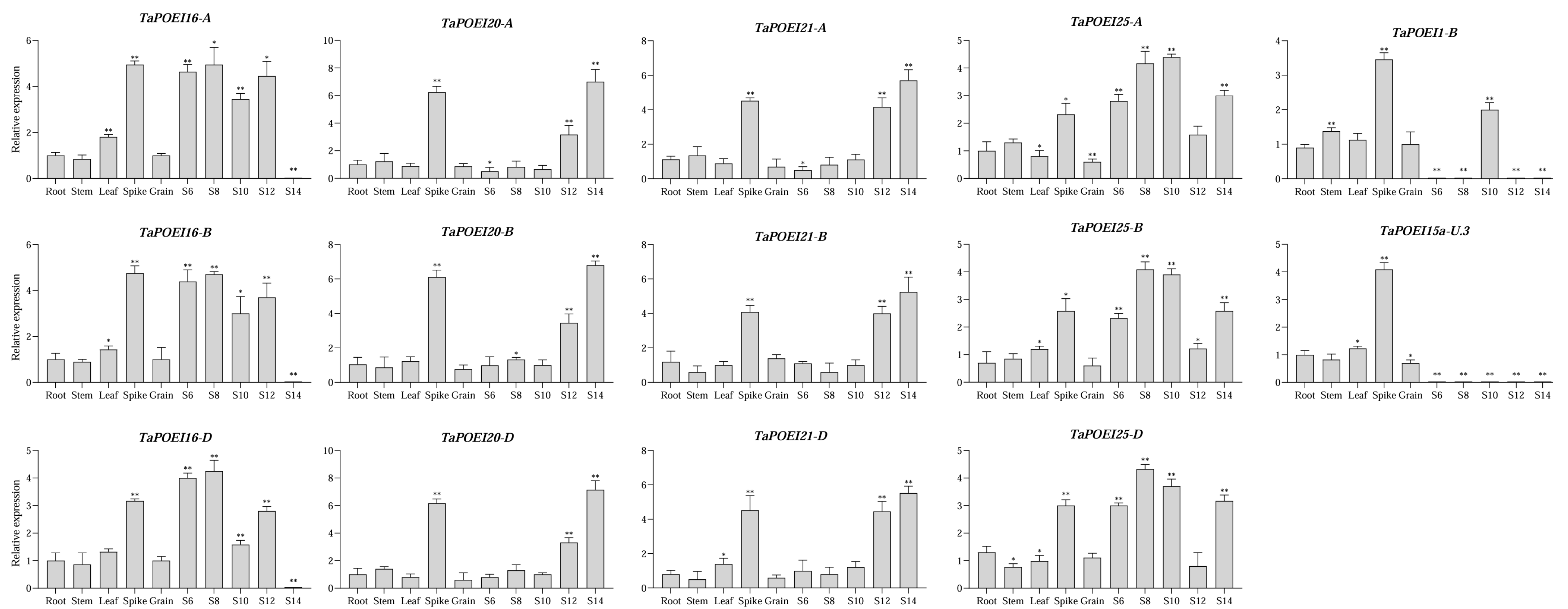
2.4. Tissue-Specific and Anther-Preferential Expression of TaPOEI Genes: Implications for Functional Roles in Reproductive Development
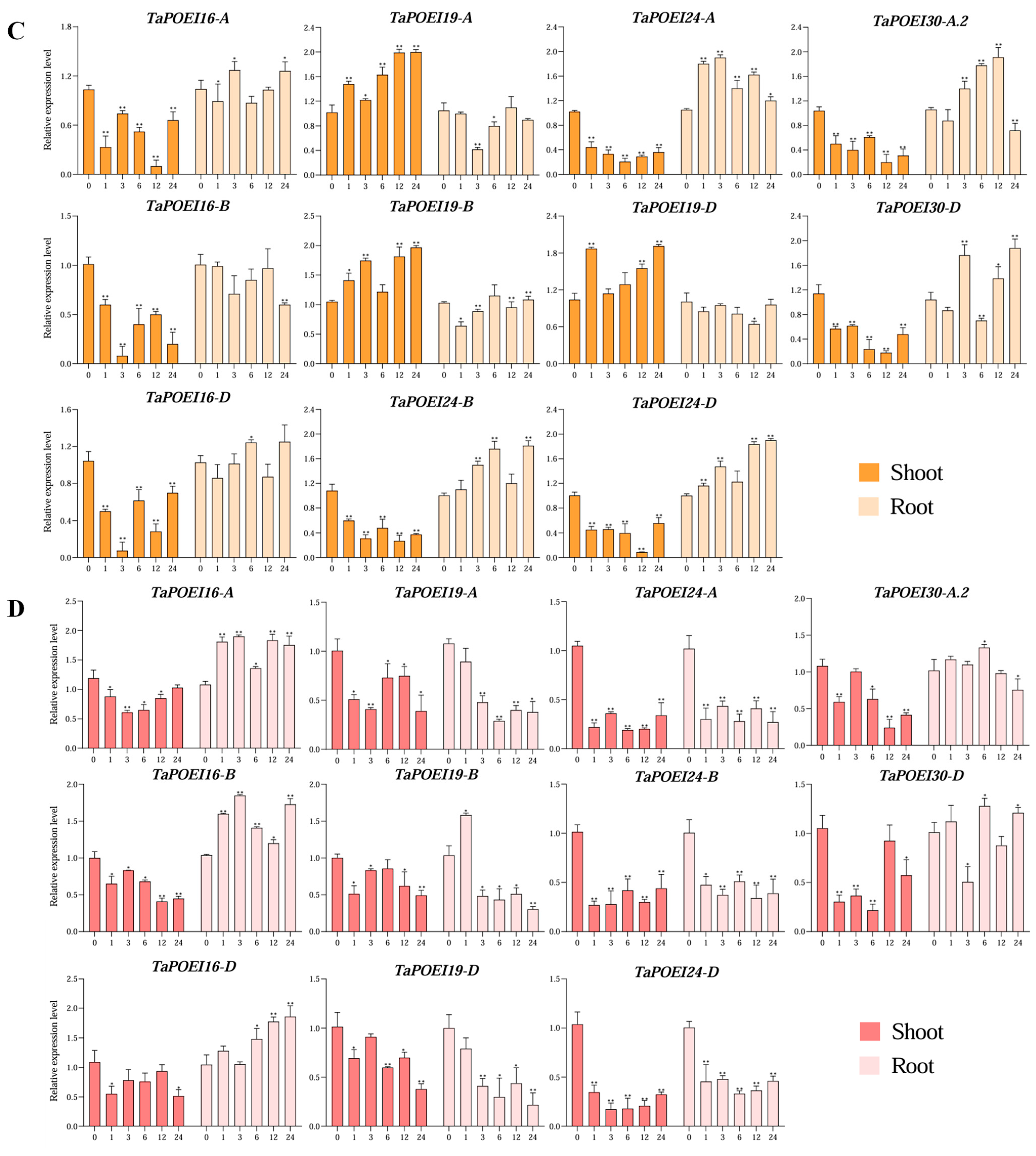
2.5. Expression Patterns of TaPOEI Genes Under Drought, Salinity, Cold, and Heat Stress: Insights into Stress-Responsive Mechanisms
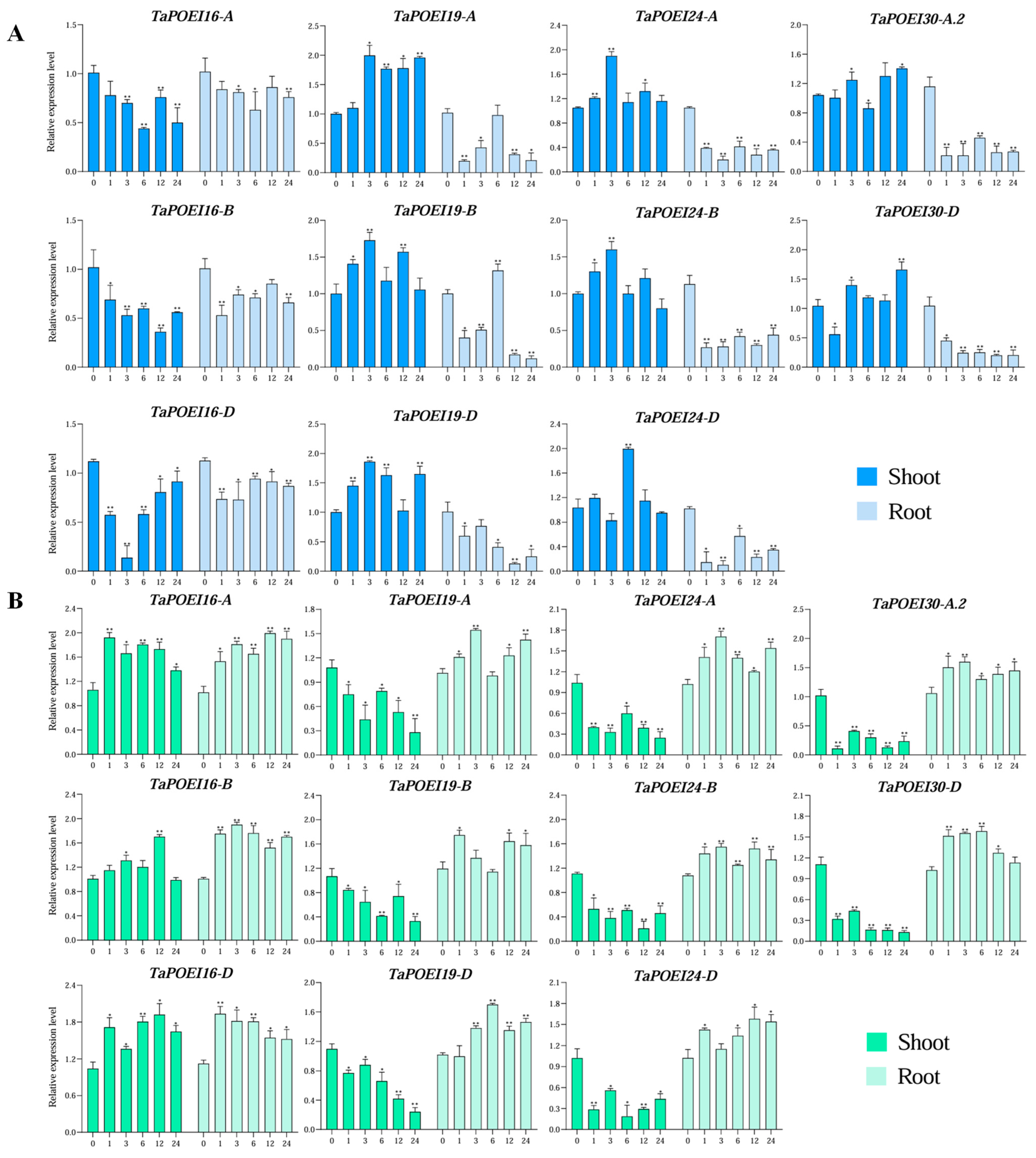
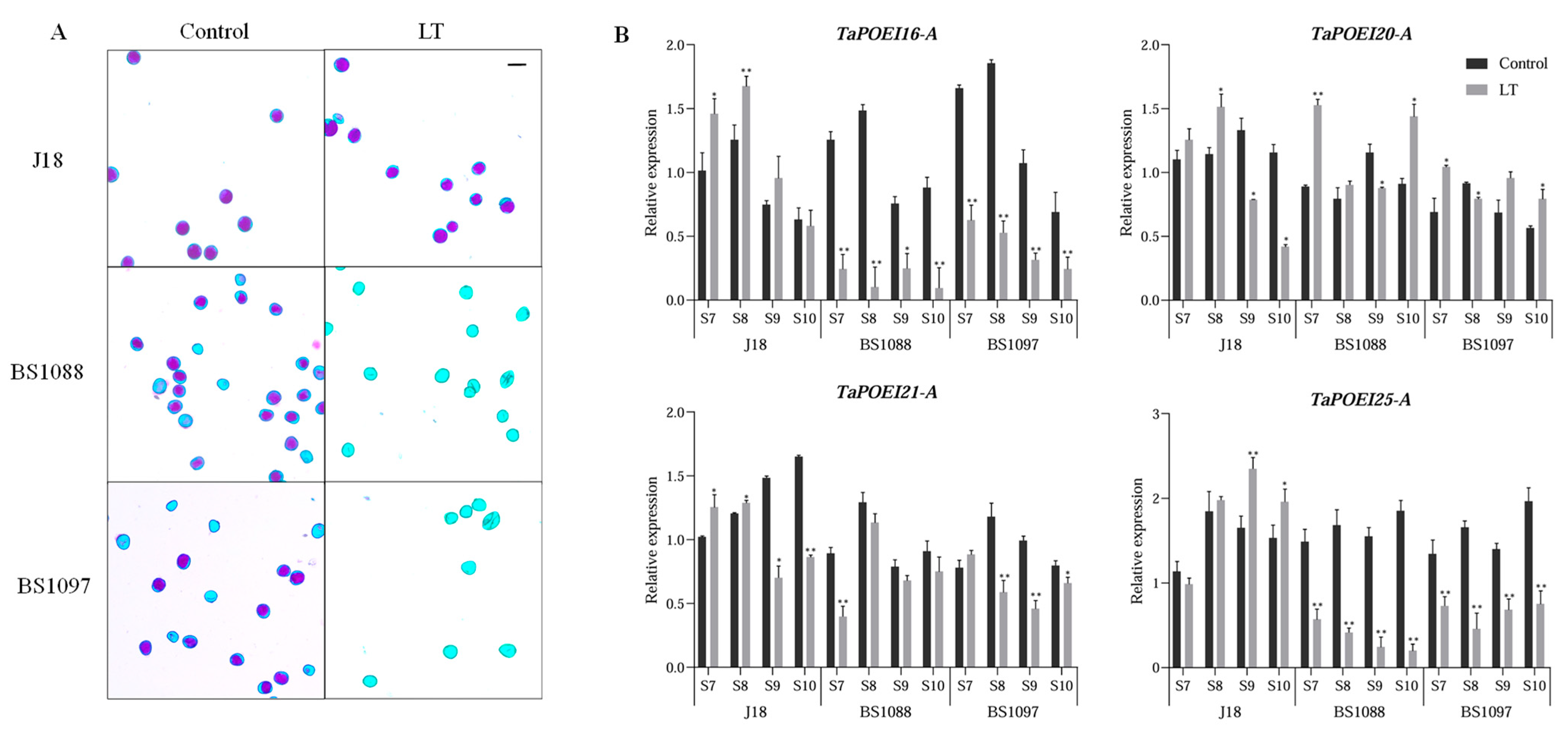
2.6. Expression Dynamics and Functional Insights of Anther-Preferential TaPOEI Genes in TGMS Wheat Lines Under Low-Temperature Conditions
3. Discussion
4. Materials and Methods
4.1. Identification and Characteristics of POEI Genes in Wheat
4.2. Chromosomal Location and Phylogenetic Relationships of POEI Genes
4.3. Analysis of Gene Structures, Conserved Motifs, Promoter Elements, and Expression Analysis
4.4. Prediction of microRNA Targets
4.5. Plant Materials and Stress Treatments
4.6. RNA Isolation and Reverse-Transcription Quantitative PCR (RT-qPCR) Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kopecká, R.; Kameniarová, M.; Černý, M.; Brzobohatý, B.; Novák, J. Abiotic Stress in Crop Production. Int. J. Mol. Sci. 2023, 24, 6603. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, H.; Li, H.; Gao, J.; Jiang, H.; Wang, C.; Guan, Y.; Yang, Z. Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008, 55, 266–277. [Google Scholar] [CrossRef]
- Alché, J.; Castro, J.; Olmedilla, A.; Fernández, M.; Rodríguez, R.; Villalba, M.; Rodríguez-García, M. The major olive pollen allergen (Ole e I) shows both gametophytic and sporophytic expression during anther development, and its synthesis and storage takes place in the RER. J. Cell Sci. 1999, 112, 2501–2509. [Google Scholar] [CrossRef]
- Li, X.; Yu, Q.; Hua, X.; He, J.; Liu, J.; Peng, L.; Wang, J.; Li, X.; Yang, Y. Phosphorylation of ADF7-mediated by AGC1.7 regulates pollen germination in Arabidopsis thaliana. Plant Cell Environ. 2025, 48, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Alché, J.; M’Rani-Alaoui, M.; Castro, J.; Rodríguez-García, M. Ole e 1, the major allergen from Olive (Olea europaea) pollen, increases its expression and is released to the culture medium during in vitro germination. Plant Cell Physiol. 2004, 45, 1149–1157. [Google Scholar] [CrossRef]
- Barral, P.; Suárez, C.; Batanero, E.; Alfonso, C.; Alché, J.; Rodríguez-García, M.; Villalba, M.; Rivas, G.; Rodríguez, R. An olive pollen protein with allergenic activity, Ole e 10, defines a novel family of carbohydrate-binding modules and is potentially implicated in pollen germination. Biochem. J. 2005, 390, 77–84. [Google Scholar] [CrossRef]
- Swapna, L.; Khurana, R.; Vijaya Kumar, S.; Tyagi, A.; Rao, K. Pollen-specific expression of Oryza sativa indica pollen allergen gene (OSIPA) promoter in rice and Arabidopsis transgenic systems. Mol. Biotechnol. 2010, 48, 49–59. [Google Scholar] [CrossRef]
- Robledo Retana, T.; Bradley-Clarke, J.; Croll, T.; Rose, R.; Hoti, I.; Stagg, A.; Villalba, M.; Pickersgill, R. Lig v 1 structure and the inflammatory response to the Ole e 1 protein family. Allergy 2020, 75, 2395–2398. [Google Scholar] [CrossRef]
- Alonso-Díaz de Durana, M.D.; Pérez-Fernández, E.; Villalba, M.; Martín-Pedraza, L.; Fernández-Rivas, M. Allergy to Cypress and Olive Pollen: Clinical Phenotypes and Allergen Recognition. J. Investig. Allergol. Clin. Immunol. 2024, 34, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.; Eberhardt, R.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2011, 40, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Stemeseder, T.; Freier, R.; Wildner, S.; Fuchs, J.; Briza, P.; Lang, R.; Batanero, E.; Lidholm, J.; Liedl, K.; Campo, P.; et al. Crystal structure of Pla l 1 reveals both structural similarity and allergenic divergence within the Ole e 1-like protein family. J. Allergy Clin. Immunol. 2017, 140, 277–280. [Google Scholar] [CrossRef]
- Fernández-González, M.; González-Fernández, E.; Fernández-González, D.; Rodríguez-Rajo, F. Secondary outcomes of the Ole e 1 proteins involved in pollen tube development: Impact on allergies. Front. Plant Sci. 2020, 11, 974. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Xu, L.; Tang, C.; Zhang, H.; Gao, H.; Cao, P.; Yin, H.; Wu, L.; Wu, J.; Gu, C.; et al. PbrPOE21 inhibits pear pollen tube growth in vitro by altering apical reactive oxygen species content. Planta 2020, 252, 43. [Google Scholar] [CrossRef]
- Carlos, J.; Isabel, M.; Alché, J. Systematic and phylogenetic analysis of the Ole e 1 pollen protein family members in plants. In Systems and Computational Biology-Bioinformatics and Computational Modeling; InTech: London, UK, 2011. [Google Scholar]
- Jiang, S.; Jasmin, P.; Ting, Y.; Ramachandran, S. Genome-wide identification and molecular characterization of Ole e I, Allerg_1, and Allerg_2 domain-containing pollen-allergen-like genes in Oryza sativa. DNA Res. 2005, 12, 167–179. [Google Scholar] [CrossRef]
- Song, W.; Duan, F.; Li, W.; Lin, Q.; Zhou, H.; Han, X.; Wang, J. GmPOI gene encoding a pollen_Ole_e_I conserved domain is involved in response of soybean to various stresses. Biol. Plantarum. 2013, 57, 85–90. [Google Scholar] [CrossRef]
- Hu, B.; Liu, B.; Liu, L.; Liu, C.; Xu, L.; Ruan, Y. Epigenetic control of pollen Ole e 1 allergen and extension family gene expression in Arabidopsis thaliana. Acta Physiol. Plant 2014, 36, 2203–2209. [Google Scholar] [CrossRef]
- Rubinstein, A.; Broadwater, A.; Lowrey, K.; Bedinger, P. Pex1, a pollen-specific gene with an extensin-like domain. Proc. Natl. Acad. Sci. USA 1995, 92, 3086–3090. [Google Scholar] [CrossRef]
- Stratford, S.; Barnes, W.; Hohorst, D.; Sagert, J.; Cotter, R.; Golubiewski, A.; Showalter, A.; McCormick, S.; Bedinger, P. A leucine-rich repeat region is conserved in pollen extensin-like (Pex) proteins in monocots and dicots. Plant Mol. Biol. 2001, 46, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Fabrice, T.; Vogler, H.; Draeger, C.; Munglani, G.; Gupta, S.; Herger, A.; Knox, P.; Grossniklaus, U.; Ringli, C. LRX proteins play a crucial role in pollen grain and pollen tube cell wall development. Plant Physiol. 2018, 176, 1981–1992. [Google Scholar] [CrossRef]
- Muschietti, J.; Dircks, L.; Vancanneyt, G.; McCormick, S. LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 2002, 6, 321–338. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, N.; Fu, N.; Li, X.; Guo, S.; Shen, H. Cloning and expression analysis of fertility-related genes for the genic male sterile line in pepper (Capsicum annuum L.). Sci. Agric. Sin. 2014, 47, 3264–3276. [Google Scholar]
- Dervisi, I.; Petropoulos, O.; Agalou, A.; Podia, V.; Papandreou, N.; Iconomidou, V.A.; Haralampidis, K.; Roussis, A. The SAH7 homologue of the allergen Ole e 1 interacts with the putative stress sensor SBP1 (Selenium-Binding Protein 1) in Arabidopsis thaliana. Int. J. Mol. Sci. 2023, 24, 3580. [Google Scholar] [CrossRef]
- Tang, Z.; Zhang, L.; Xu, C.; Yuan, S.; Zhang, F.; Zheng, Y.; Zhao, C. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012, 159, 721–738. [Google Scholar] [CrossRef]
- Bai, J.; Wang, Y.; Wang, P.; Duan, W.; Yuan, S.; Sun, H.; Yuan, G.; Ma, J.; Wang, N.; Zhang, F.; et al. Uncovering male fertility transition responsive miRNA in a wheat photo-thermosensitive genic male sterile line by deep sequencing and degradome analysis. Front. Plant Sci. 2017, 8, 1370. [Google Scholar] [CrossRef]
- Sun, L.; Sun, G.; Shi, C.; Sun, D. Transcriptome analysis reveals new microRNAs-mediated pathway involved in anther development in male sterile wheat. BMC Genomics. 2018, 19, 333. [Google Scholar] [CrossRef]
- Yue, H.; Zhang, H.; Su, N.; Sun, X.; Zhao, Q.; Weining, S.; Nie, X.; Yue, W. Integrating small RNA and degradome sequencing to reveal drought memory response in wheat (Triticum Aestivum L.). Int. J. Mol. Sci. 2022, 23, 5917. [Google Scholar] [CrossRef]
- Xu, L.; Wang, D.; Liu, S.; Fang, Z.; Su, S.; Guo, C.; Zhao, C.; Tang, Y. Comprehensive atlas of wheat (Triticum aestivum L.) auxin response factor expression during male reproductive development and abiotic stress. Front. Plant Sci. 2020, 11, 586144. [Google Scholar] [CrossRef]
- Wang, D.; Zuo, J.; Liu, S.; Wang, W.; Lu, Q.; Hao, X.; Fang, Z.; Liang, T.; Sun, Y.; Guo, C.; et al. BRI1 EMS SUPPRESSOR1 genes regulate abiotic stress and anther development in wheat (Triticum aestivum L.). Front. Plant Sci. 2023, 14, 1219856. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Zhang, L.; Yang, D.; Zhao, C.; Zheng, Y. Cold stress contributes to aberrant cytokinesis during male meiosis I in a wheat thermosensitive genic male sterile line. Plant Cell Environ. 2010, 34, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, M.; Qiu, C.; Naz, S.; Cao, F.; Wu, F. Genome-wide discovery of miRNAs with differential expression patterns in responses to salinity in the two contrasting wheat cultivars. Int. J. Mol. Sci. 2021, 22, 12556. [Google Scholar] [CrossRef]
- Guo, R.; Sun, D.; Tan, Z.; Rong, D.; Li, C. Two recessive genes controlling thermos photoperiod-sensitive male sterility in wheat. Theor. Appl. Genet. 2006, 112, 1271–1276. [Google Scholar] [CrossRef]
- Van Bel, M.; Silvestri, F.; Weitz, E.M.; Kreft, L.; Botzki, A.; Coppens, F.; Vandepoele, K. PLAZA 5.0: Extending the scope and power of comparative and functional genomics in plants. Nucleic Acids Res. 2022, 50, 1468–1474. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Kong, X.; Long, Y.; Liu, S.; Zhang, H.; Jia, J.; Cui, W.; Zhang, Z.; Song, X.; Qiu, L.; et al. Integrated single-nucleus and spatial transcriptomics captures transitional states in soybean nodule maturation. Nat. Plants 2023, 9, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Browne, R.; Iacuone, S.; Li, S.; Dolferus, R.; Parish, R. Anther morphological development and stage determination in Triticum aestivum. Front. Plant Sci. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, Y.; Cao, G.; Shao, L.; Ding, Q.; Ma, L. Dynamic expression of miRNAs and functional analysis of target genes involved in the response to male sterility of the wheat line YS3038. Plant Physiol. Biochem. 2021, 162, 363–377. [Google Scholar] [CrossRef]
- Goldberg, R.; Beals, T.; Sanders, P. Anther development: Basic principles and practical applications. Plant Cell. 1993, 5, 1217–1229. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, J.; Jia, Y.; Wang, W.; Guo, C.; Fang, Z.; Zhang, Y.; Fu, J.; Zhao, S.; Zhao, C.; Wang, D.; et al. Analysis of Wheat Pollen Ole E I Proteins Reveals Potential Roles in Fertility and Stress Adaptation. Int. J. Mol. Sci. 2025, 26, 7707. https://doi.org/10.3390/ijms26167707
Zuo J, Jia Y, Wang W, Guo C, Fang Z, Zhang Y, Fu J, Zhao S, Zhao C, Wang D, et al. Analysis of Wheat Pollen Ole E I Proteins Reveals Potential Roles in Fertility and Stress Adaptation. International Journal of Molecular Sciences. 2025; 26(16):7707. https://doi.org/10.3390/ijms26167707
Chicago/Turabian StyleZuo, Jinghong, Yanfeng Jia, Weiwei Wang, Chunman Guo, Zhaofeng Fang, Yujuan Zhang, Jinzhou Fu, Sijia Zhao, Changping Zhao, Dezhou Wang, and et al. 2025. "Analysis of Wheat Pollen Ole E I Proteins Reveals Potential Roles in Fertility and Stress Adaptation" International Journal of Molecular Sciences 26, no. 16: 7707. https://doi.org/10.3390/ijms26167707
APA StyleZuo, J., Jia, Y., Wang, W., Guo, C., Fang, Z., Zhang, Y., Fu, J., Zhao, S., Zhao, C., Wang, D., Yang, G., & Tang, Y. (2025). Analysis of Wheat Pollen Ole E I Proteins Reveals Potential Roles in Fertility and Stress Adaptation. International Journal of Molecular Sciences, 26(16), 7707. https://doi.org/10.3390/ijms26167707






