Genome-Wide Identification, Evolution, and Expression Analysis of the DMP Gene Family in Peanut (Arachis hypogaea L.)
Abstract
1. Introduction
2. Results
2.1. The Characterization of the AhDMP Gene Family in Peanuts
2.2. Phylogenetic Analysis of AhDMPs
2.3. Chromosomal Localization, Collinearity Analysis, and Selection Pressure Analysis of AhDMP Genes
2.4. Conserved Motif and Domain Analysis of AhDMP
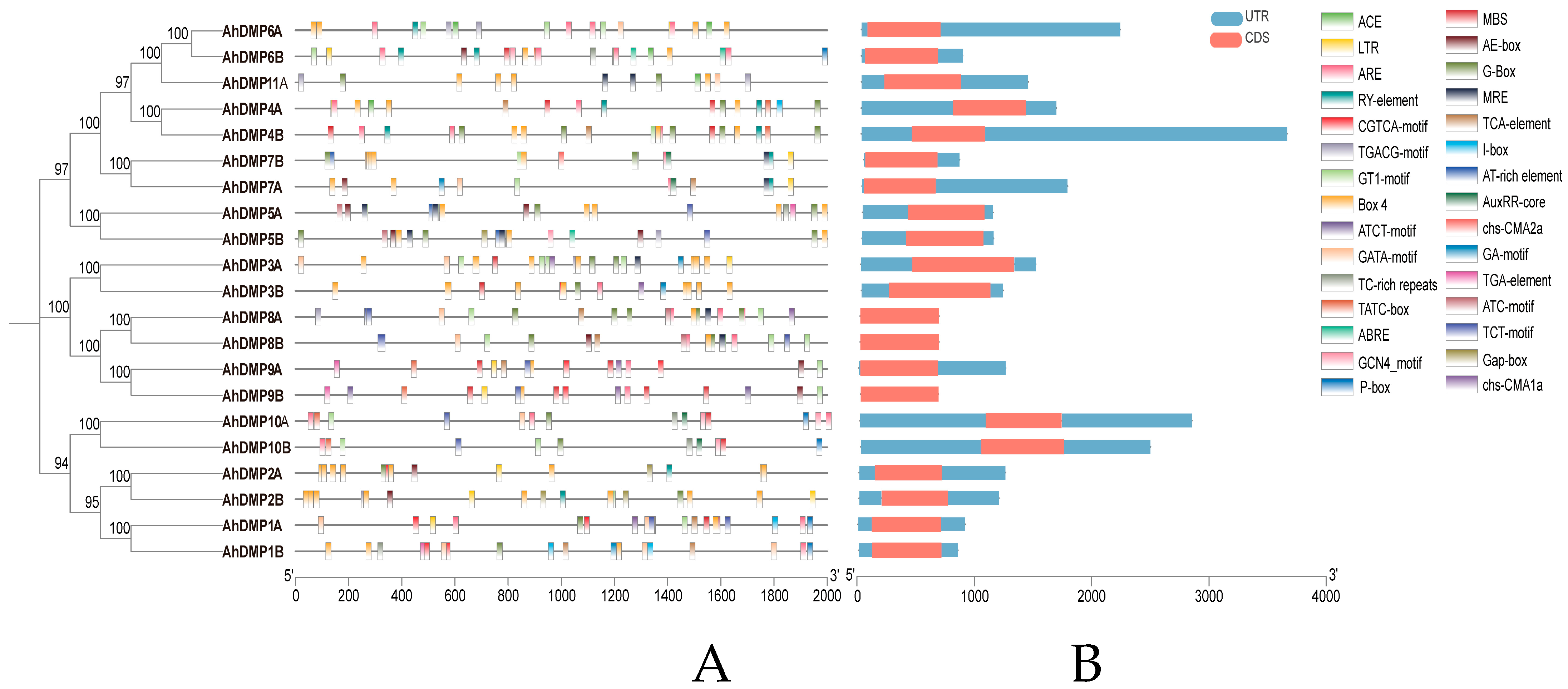
2.5. Gene Structure and Cis-Acting Element Analysis of the AhDMP Gene Family
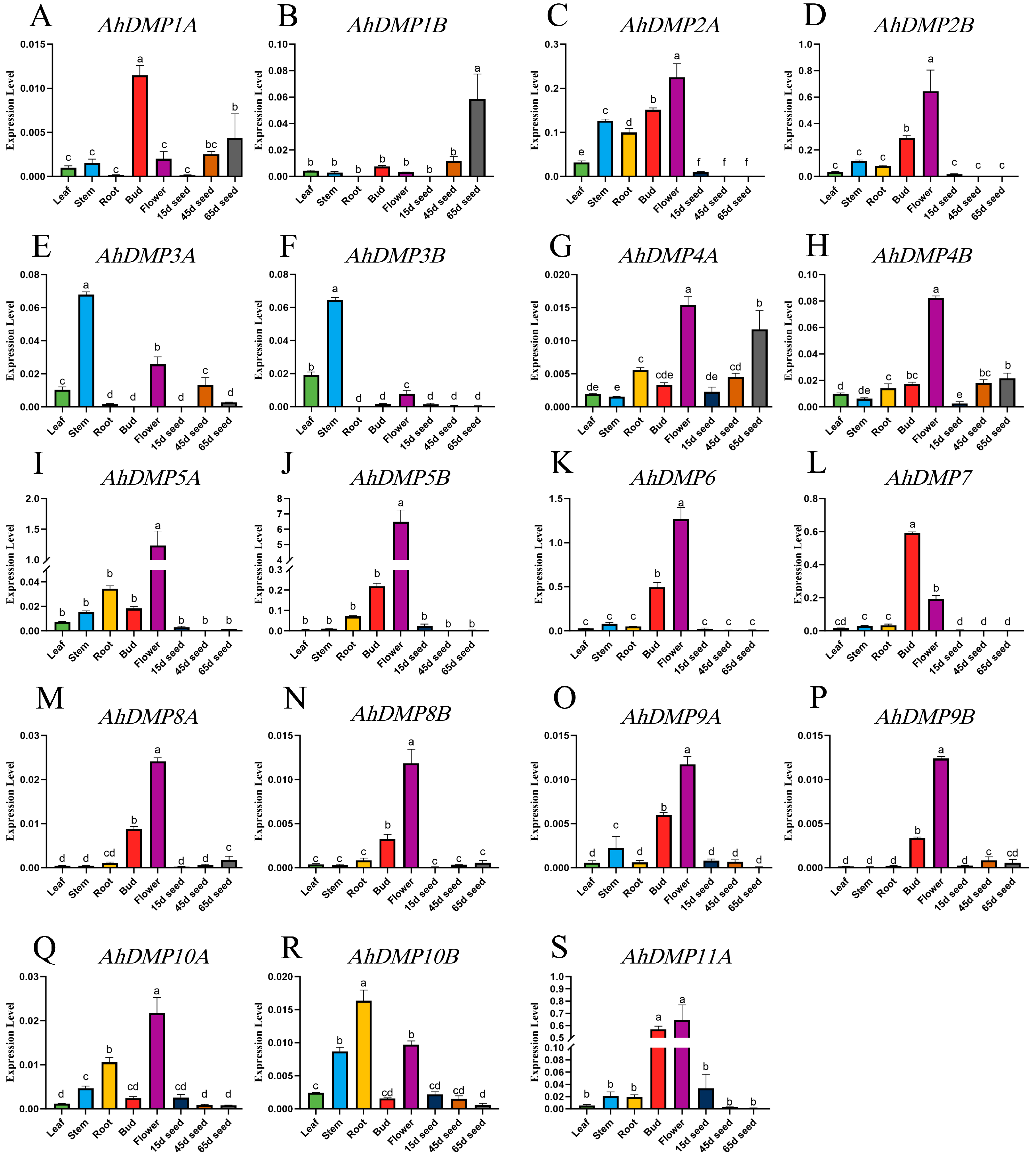
2.6. Expression Profiles of Peanut AhDMP Genes Across Different Tissues
2.7. Subcellular Localization of DMPs in Arachis hypogaea
3. Discussion
4. Materials and Methods
4.1. Identification and Analysis of the DMP Members
4.2. Multiple Sequence Alignment and Phylogenetic Analysis of DMP Genes
4.3. Conserved Domain and Motif Analysis of DMP Genes
4.4. Chromosomal Localization, Gene Duplication, and Selective Pressure Analyses
4.5. Analysis of Promoter Region and Gene Structure in AhDMP Genes
4.6. qRT-PCR Analysis of AhDMP Genes
4.7. Subcellular Localization of AhDMPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Xicluna, J.; Lacombe, B.; Dreyer, I.; Alcon, C.; Jeanguenin, L.; Sentenac, H.; Thibaud, J.-B.; Chérel, I. Increased Functional Diversity of Plant K+ Channels by Preferential Heteromerization of the Shaker-like Subunits AKT2 and KAT2. J. Biol. Chem. 2007, 282, 486–494. [Google Scholar] [CrossRef]
- Chen, Y.; Heazlewood, J.L. Organellar Proteomic Profiling to Analyze Membrane Trafficking Pathways. Trends Plant Sci. 2021, 26, 299–300. [Google Scholar] [CrossRef]
- Chen, Y.; Weckwerth, W. Mass Spectrometry Untangles Plant Membrane Protein Signaling Networks. Trends Plant Sci. 2020, 25, 930–944. [Google Scholar] [CrossRef]
- Mori, T.; Kawai-Toyooka, H.; Igawa, T.; Nozaki, H. Gamete Dialogs in Green Lineages. Mol. Plant 2015, 8, 1442–1454. [Google Scholar] [CrossRef]
- Yamada, K.; Osakabe, Y.; Mizoi, J.; Nakashima, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of an Arabidopsis thaliana Abiotic Stress-Inducible Facilitated Diffusion Transporter for Monosaccharides. J. Biol. Chem. 2010, 285, 1138–1146. [Google Scholar] [CrossRef]
- Cyprys, P.; Lindemeier, M.; Sprunck, S. Gamete Fusion Is Facilitated by Two Sperm Cell-Expressed DUF679 Membrane Proteins. Nat. Plants 2019, 5, 253–257. [Google Scholar] [CrossRef]
- Kasaras, A.; Melzer, M.; Kunze, R. Arabidopsis Senescence-Associated Protein DMP1 Is Involved in Membrane Remodeling of the ER and Tonoplast. BMC Plant Biol. 2012, 12, 54. [Google Scholar] [CrossRef]
- Olvera-Carrillo, Y.; Van Bel, M.; Van Hautegem, T.; Fendrych, M.; Van Durme, M.; Huysmans, M.; Šimášková, M.; Buscaill, P.; Rivas, S.; Coll, N.S.; et al. A Conserved Core of PCD Indicator Genes Discriminates Developmentally and Environmentally Induced Programmed Cell Death in Plants. Plant Physiol. 2015, 169, 2684–2699. [Google Scholar] [CrossRef]
- Kasaras, A.; Kunze, R. Expression, Localisation and Phylogeny of a Novel Family of Plant-specific Membrane Proteins. Plant Biol. 2010, 12, 140–152. [Google Scholar] [CrossRef]
- Takahashi, T.; Mori, T.; Ueda, K.; Yamada, L.; Nagahara, S.; Higashiyama, T.; Sawada, H.; Igawa, T. The Male Gamete Membrane Protein DMP9/DAU2 Is Required for Double Fertilization in Flowering Plants. Development 2018, 145, dev170076. [Google Scholar] [CrossRef]
- Bi, W.; Chen, Y.; Song, Y.; Liu, J.; Zhao, T.; Sun, C.; Qin, J.; Tu, Z.; Li, Y.; Wang, X.; et al. Potato DMP2 Positively Regulates Plant Immunity by Modulating Endoplasmic Reticulum Homeostasis. J. Integr. Plant Biol. 2025, 67, 1568–1581. [Google Scholar] [CrossRef]
- Ahmad, Z.; Tian, D.; Li, Y.; Aminu, I.M.; Tabusam, J.; Zhang, Y.; Zhu, S. Characterization, Evolution, Expression and Functional Divergence of the DMP Gene Family in Plants. Int. J. Mol. Sci. 2024, 25, 10435. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, H.; Wang, J.; Zhao, G.; Niu, K.; Zhou, X.; Zhang, R.; Yao, R. Genomic Identification and Expression Profiling of DMP Genes in Oat (Avena sativa) Elucidate Their Responsiveness to Seed Aging. BMC Genom. 2024, 25, 863. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, X.; Chen, W.; Yao, J.; Li, Y.; Fang, S.; Lv, Y.; Li, X.; Pan, J.; Liu, C.; et al. Cotton DMP Gene Family: Characterization, Evolution, and Expression Profiles during Development and Stress. Int. J. Biol. Macromol. 2021, 183, 1257–1269. [Google Scholar] [CrossRef]
- Nawade, B.; Bosamia, T.C.; Lee, J.H.; Jang, J.H.; Lee, O.R. Genome-Wide Characterization of the Soybean DOMAIN OF UNKNOWN FUNCTION 679 Membrane Protein Gene Family Highlights Their Potential Involvement in Growth and Stress Response. Front. Plant Sci. 2023, 14, 1216082. [Google Scholar] [CrossRef]
- Sarvamangala, C.; Gowda, M.V.C.; Varshney, R.K. Identification of Quantitative Trait Loci for Protein Content, Oil Content and Oil Quality for Groundnut (Arachis hypogaea L.). Field Crops Res. 2011, 122, 49–59. [Google Scholar] [CrossRef]
- Smith, B.W. Arachis hypogaea. Aerial flower and subterranean fruit. Am. J. Bot. 1950, 37, 802–815. [Google Scholar] [CrossRef]
- Bertioli, D.J.; Seijo, G.; Freitas, F.O.; Valls, J.F.M.; Leal-Bertioli, S.C.M.; Moretzsohn, M.C. An Overview of Peanut and Its Wild Relatives. Plant Genet. Res. 2011, 9, 134–149. [Google Scholar] [CrossRef]
- Jacquier, N.M.A.; Gilles, L.M.; Pyott, D.E.; Martinant, J.-P.; Rogowsky, P.M.; Widiez, T. Puzzling out Plant Reproduction by Haploid Induction for Innovations in Plant Breeding. Nat. Plants 2020, 6, 610–619. [Google Scholar] [CrossRef]
- Yongping, X.; Yunhuan, Z.; Yingduo, G.; Zhaocong, C. The first report on one haploid plant in cultivated peanut using anther culture technique. Chin. J. Oil Crop Sci. 2022, 44, 810–817, (In Chinese with English abstract). [Google Scholar]
- Croser, J.S.; Lülsdorf, M.M.; Davies, P.A.; Clarke, H.J.; Bayliss, K.L.; Mallikarjuna, N.; Siddique, K.H.M. Toward Doubled Haploid Production in the Fabaceae: Progress, Constraints, and Opportunities. Crit. Rev. Plant Sci. 2006, 25, 139–157. [Google Scholar] [CrossRef]
- Zhong, Y. Mutation of ZmDMP Enhances Haploid Induction in Maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Zhong, Y. A DMP-Triggered In vivo Maternal Haploid Induction System in the Dicotyledonous Arabidopsis. Nat. Plants 2020, 6, 466–472. [Google Scholar] [CrossRef]
- Establishment of a Dmp Based Maternal Haploid Induction System for Polyploid Brassica napus and Nicotiana tabacum. Available online: https://onlinelibrary.wiley.com/doi/epdf/10.1111/jipb.13244 (accessed on 28 May 2024).
- Chen, X.; Li, Y.; Ai, G.; Chen, J.; Guo, D.; Zhu, Z.; Zhu, X.; Tian, S.; Wang, J.; Liu, M.; et al. Creation of a Watermelon Haploid Inducer Line via ClDMP3-Mediated Single Fertilization of the Central Cell. Hortic. Res. 2023, 10, uhad081. [Google Scholar] [CrossRef]
- Long, L.; Feng, Y.-M.; Shang, S.-Z.; Zhao, J.-R.; Hu, G.-Y.; Xu, F.-C.; Song, C.-P.; Jin, S.-X.; Gao, W. In Vivo Maternal Haploid Induction System in Cotton. Plant Physiol. 2023, 194, 1286–1289. [Google Scholar] [CrossRef]
- Wang, N.; Xia, X.; Jiang, T.; Li, L.; Zhang, P.; Niu, L.; Cheng, H.; Wang, K.; Lin, H. In Planta Haploid Induction by Genome Editing of DMP in the Model Legume Medicago Truncatula. Plant Biotechnol. J. 2022, 20, 22–24. [Google Scholar] [CrossRef]
- Yin, S.; Li, S.; Sun, L.; Shi, K.; Fan, S.; Liu, X.; Ren, H. Mutating the Maternal Haploid Inducer Gene CsDMP in Cucumber Produces Haploids in Planta. Plant Physiol. 2024, 194, 1282–1285. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, J.; Luo, J.; Tang, D.; Zhu, X.; Wang, J.; Liu, Z.; Wang, P.; Zhong, Y.; Liu, C.; et al. Construction of Homozygous Diploid Potato through Maternal Haploid Induction. aBIOTECH 2022, 3, 163–168. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, M.; Cheng, D.; Liu, J.; Han, Q.; Liu, C.; Qi, X.; Yan, T.; Teng, L.; Xv, C.; et al. Mutation of GmDMP Genes Triggers Haploid Induction in Soybean. Available online: https://doi.org/10.1101/2024.03.24.585499 (accessed on 15 April 2025).
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The Origins of Genomic Duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef]
- Linster, E.; Layer, D.; Bienvenut, W.V.; Dinh, T.V.; Weyer, F.A.; Leemhuis, W.; Brünje, A.; Hoffrichter, M.; Miklankova, P.; Kopp, J.; et al. The Arabidopsis Nα -acetyltransferase NAA60 Locates to the Plasma Membrane and Is Vital for the High Salt Stress Response. New Phytol. 2020, 228, 554–569. [Google Scholar] [CrossRef]
- Coe, E.H. A Line of Maize with High Haploid Frequency. Am. Nat. 1959, 93, 381–382. [Google Scholar] [CrossRef]
- Liu, C.; Li, X.; Meng, D.; Zhong, Y.; Chen, C.; Dong, X.; Xu, X.; Chen, B.; Li, W.; Li, L.; et al. A 4-Bp Insertion at ZmPLA1 Encoding a Putative Phospholipase A Generates Haploid Induction in Maize. Mol. Plant 2017, 10, 520–522. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a Sperm-Specific Phospholipase, Triggers Maize Haploid Induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Gilles, L.M.; Khaled, A.; Laffaire, J.; Chaignon, S.; Gendrot, G.; Laplaige, J.; Bergès, H.; Beydon, G.; Bayle, V.; Barret, P.; et al. Loss of Pollen-specific Phospholipase NOT LIKE DAD Triggers Gynogenesis in Maize. EMBO J. 2017, 36, 707–717. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal Seeds from Hybrid Rice by Simultaneous Genome Engineering of Meiosis and Fertilization Genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef]
- Liu, H.; Wang, K.; Jia, Z.; Gong, Q.; Lin, Z.; Du, L.; Pei, X.; Ye, X. Efficient Induction of Haploid Plants in Wheat by Editing of TaMTL Using an Optimized Agrobacterium-Mediated CRISPR System. J. Exp. Bot. 2020, 71, 1337–1349. [Google Scholar] [CrossRef]
- Cheng, Z.; Sun, Y.; Yang, S.; Zhi, H.; Yin, T.; Ma, X.; Zhang, H.; Diao, X.; Guo, Y.; Li, X.; et al. Establishing in Planta Haploid Inducer Line by Edited SiMTL in Foxtail Millet (Setaria italica). Plant Biotechnol. J. 2021, 19, 1089–1091. [Google Scholar] [CrossRef]
- Tang, H.; Qiu, Y.; Wang, W.; Yu, M.; Chang, Y.; Han, Z.; Du, L.; Lin, Z.; Wang, K.; Ye, X. Development of a Haploid Inducer by Editing HvMTL in Barley. J. Genet. Genom. 2023, 50, 366–369. [Google Scholar] [CrossRef]
- La Camera, S.; Geoffroy, P.; Samaha, H.; Ndiaye, A.; Rahim, G.; Legrand, M.; Heitz, T. A Pathogen-Inducible Patatin-like Lipid Acyl Hydrolase Facilitates Fungal and Bacterial Host Colonization in Arabidopsis. Plant J. 2005, 44, 810–825. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Z.; Qi, F.; Zhou, Z.; Du, P.; Shi, L.; Dong, W.; Huang, B.; Han, S.; Pavan, S.; et al. A Telomere-to-Telomere Genome Assembly of the Cultivated Peanut. Mol. Plant 2025, 18, 5–8. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Big-Data Mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Gasteiger, E. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Cell-PLoc: A Package of Web Servers for Predicting Subcellular Localization of Proteins in Various Organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Almagro Armenteros, J.J.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM Predicts Alpha and Beta Transmembrane Proteins Using Deep Neural Networks. Available online: https://doi.org/10.1101/2022.04.08.487609 (accessed on 15 April 2025).
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Zhu, X.; Leng, X.; Sun, X.; Mu, Q.; Wang, B.; Li, X.; Wang, C.; Fang, J. Discovery of Conservation and Diversification of miR171 Genes by Phylogenetic Analysis Based on Global Genomes. Plant Genome 2015, 8, plantgenome2014-10. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A Web Application for Visualizing, Modifying and Annotating Phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Lescot, M. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shah, K.; Zhang, W.; Zhou, H.; Cheng, B.; Zhang, Z.; Yang, Z.; Moale, C.; Kamanova, S.; Han, M.; Ren, X.; et al. Identification of MdMED Family, Key Role of MdMED81, and Salicylic Acid at the Right Time of Year Triggers MdMED81 to Induce Flowering in Malus Domestica. Sci. Hortic. 2022, 304, 111341. [Google Scholar] [CrossRef]

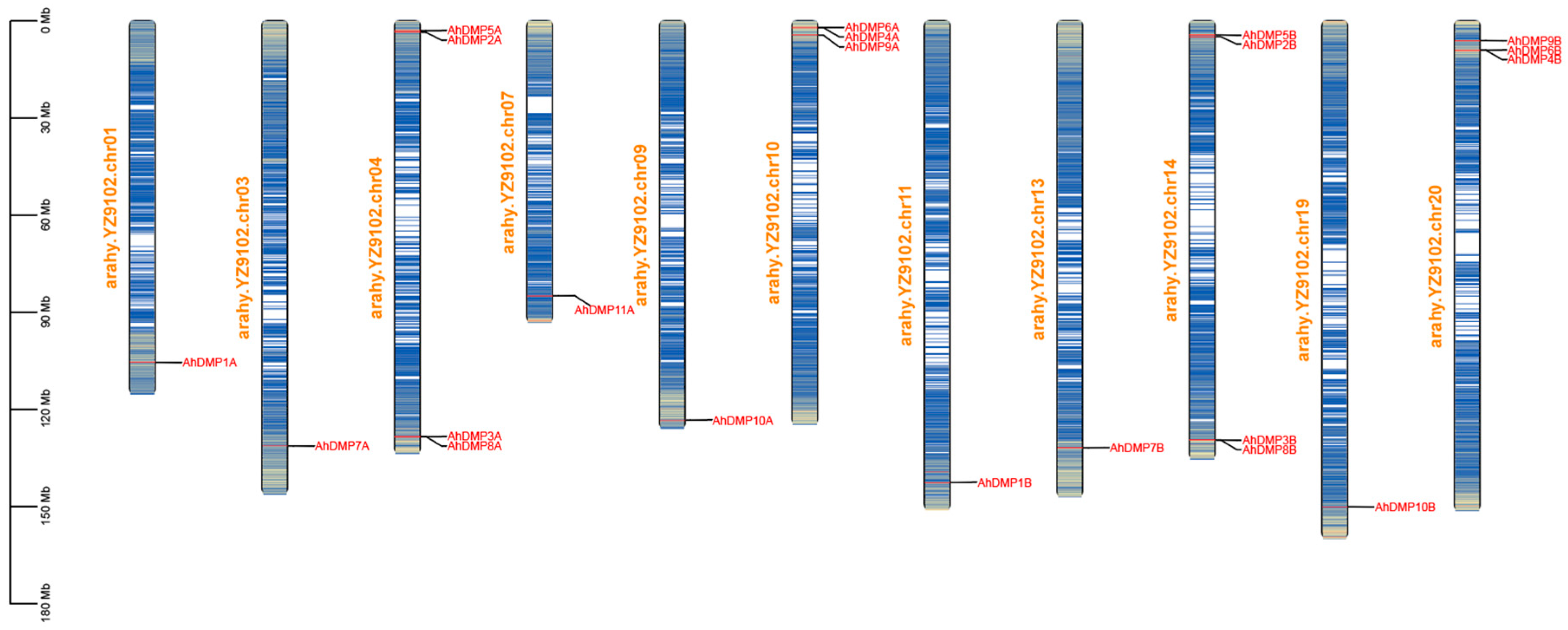
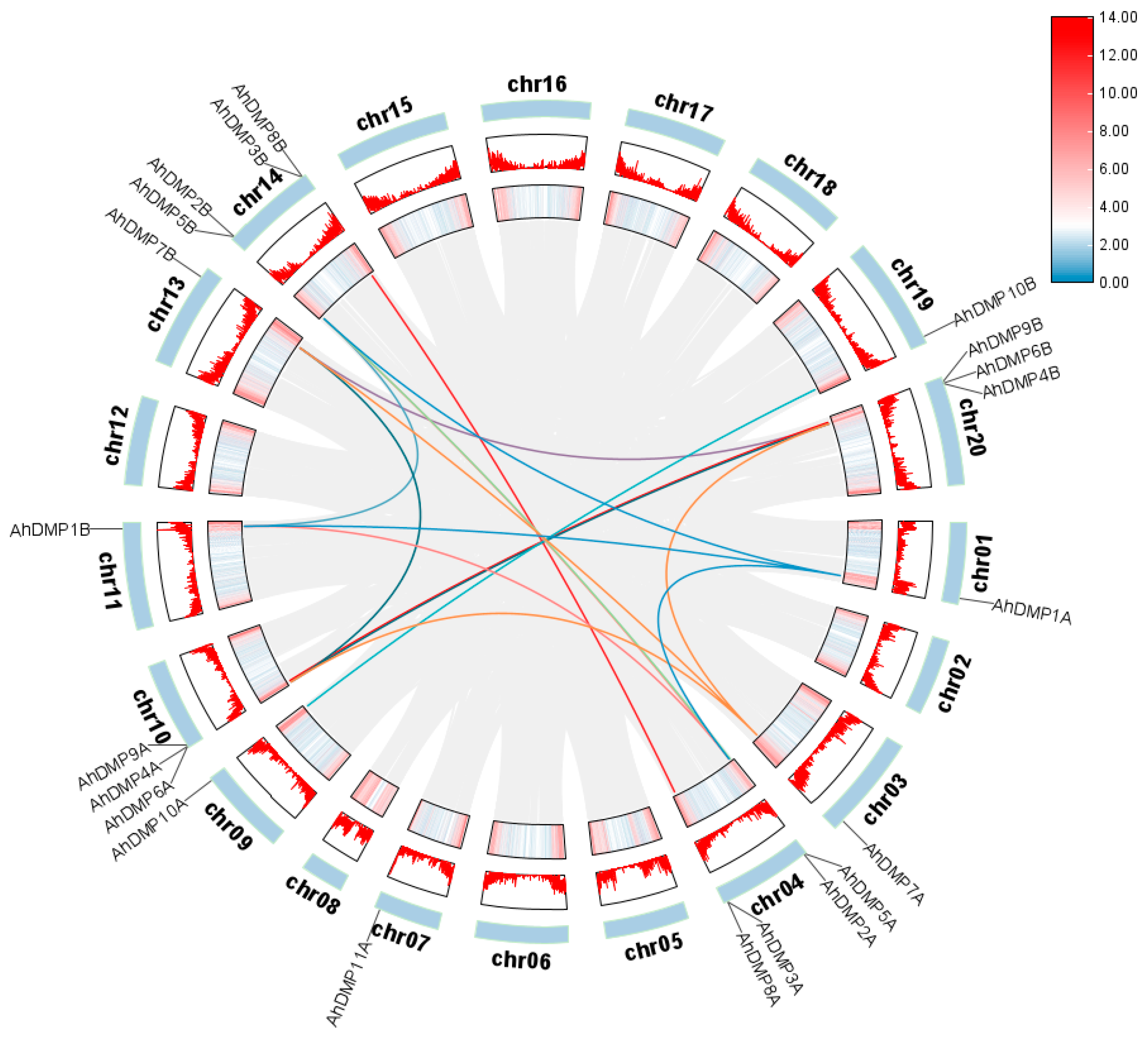
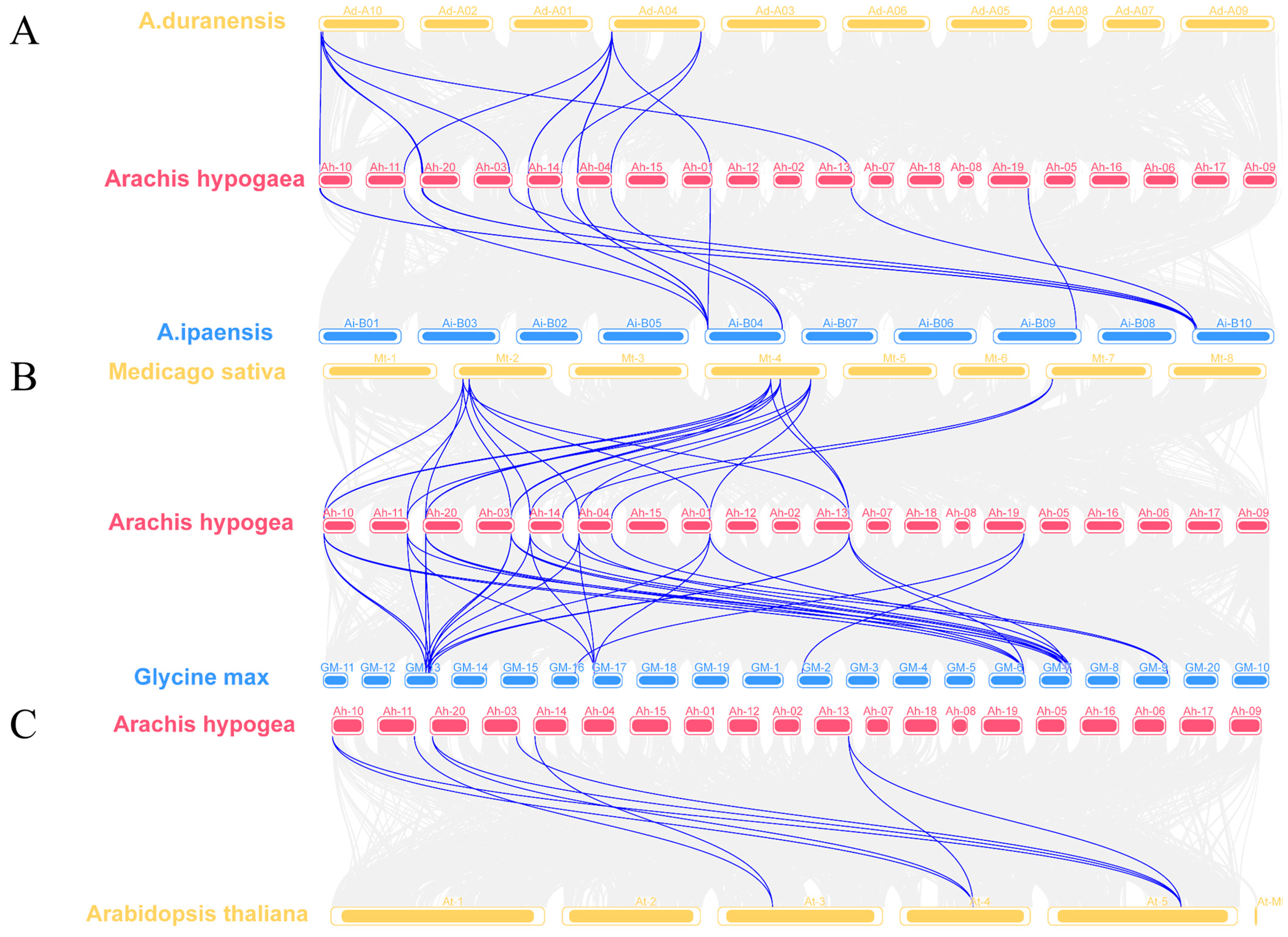
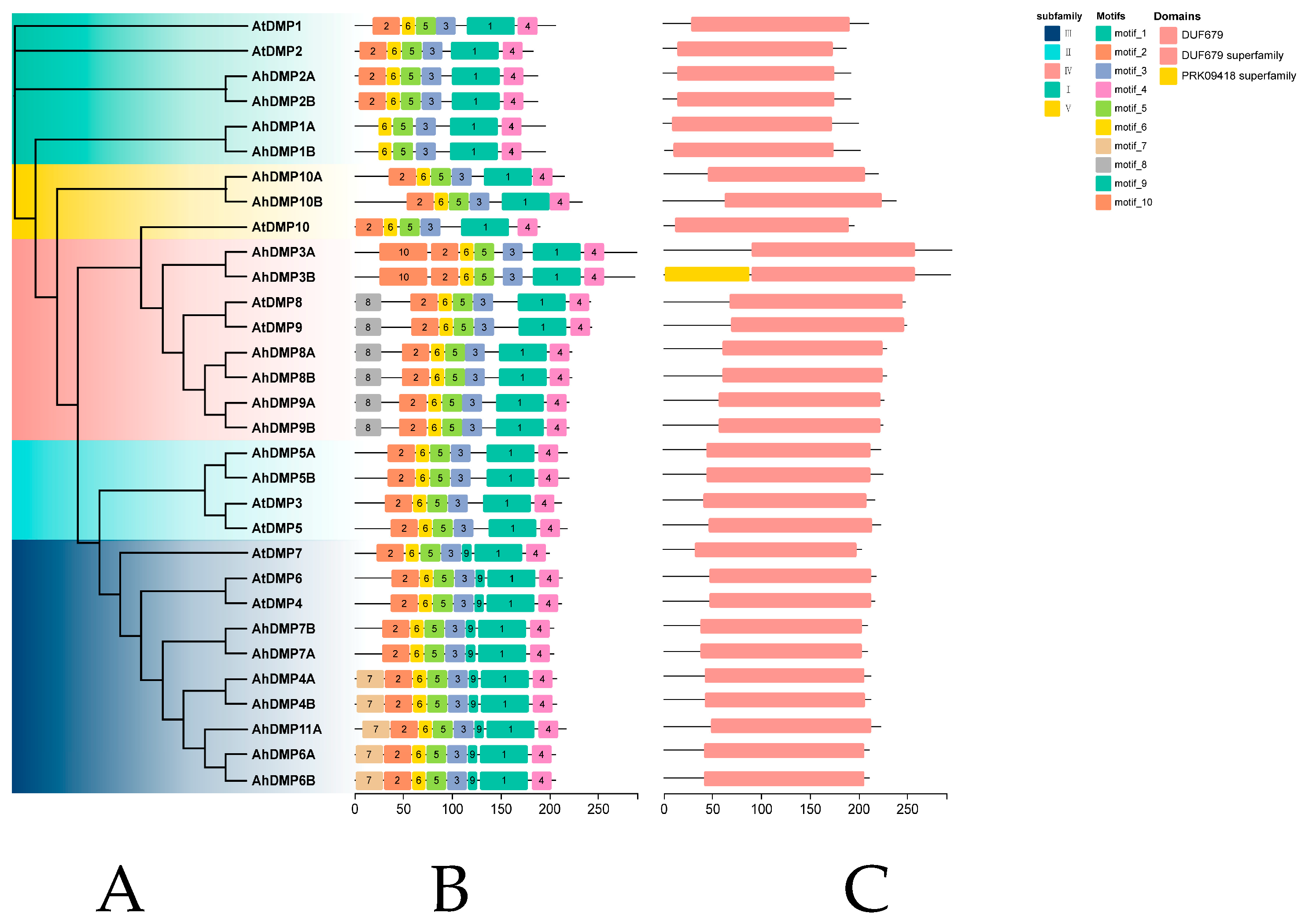

| Chr | Gene ID | Gene Name | Protein Length (aa) | Molecular Weight (MW)/kDa | Isoelectric Point (pi) | Transmembrane Domains | Subcellular Location |
|---|---|---|---|---|---|---|---|
| Chr01 | g2643.t1 | AhDMP1A | 196 | 21.34 | 9.17 | 4 | Plasma Membrane |
| Chr01 | g9634.t1 | AhDMP7A | 204 | 22.64 | 5.41 | 2 | Plasma Membrane |
| Chr04 | g11185.t1 | AhDMP2A | 188 | 20.30 | 7.68 | 4 | Plasma Membrane |
| Chr04 | g13490.t1 | AhDMP3A | 289 | 31.35 | 6.42 | 4 | Plasma Membrane/Extracellular |
| Chr04 | g11140.t1 | AhDMP5A | 218 | 23.41 | 8.64 | 3 | Plasma Membrane/Extracellular |
| Chr04 | g13491.t1 | AhDMP8A | 223 | 24.68 | 8.52 | 4 | Plasma Membrane |
| Chr07 | g23068.t1 | AhDMP11A | 217 | 24.05 | 4.99 | 4 | Plasma Membrane |
| Chr09 | g29711.t1 | AhDMP10A | 215 | 24.40 | 6.88 | 4 | Plasma Membrane |
| Chr10 | g30131.t1 | AhDMP4A | 207 | 22.30 | 4.87 | 3 | Plasma Membrane |
| Chr10 | g30130.t1 | AhDMP6A | 206 | 22.64 | 4.85 | 4 | Plasma Membrane |
| Chr10 | g30358.t1 | AhDMP9A | 220 | 24.26 | 8.73 | 4 | Plasma Membrane |
| Chr11 | g35860.t1 | AhDMP1B | 196 | 21.41 | 9.3 | 4 | Plasma Membrane |
| Chr13 | g43762.t1 | AhDMP7B | 204 | 22.64 | 5.41 | 2 | Plasma Membrane |
| Chr14 | g45489.t1 | AhDMP2B | 188 | 20.36 | 6.88 | 4 | Plasma Membrane |
| Chr14 | g48076.t1 | AhDMP3B | 287 | 31.16 | 6.94 | 4 | Plasma Membrane/Extracellular |
| Chr14 | g45442.t1 | AhDMP5B | 220 | 23.71 | 8.64 | 3 | Plasma Membrane/Extracellular |
| Chr14 | g48077.t1 | AhDMP8B | 223 | 24.68 | 8.52 | 4 | Plasma Membrane |
| Chr19 | g66369.t2 | AhDMP10B | 233 | 26.45 | 6.28 | 4 | Plasma Membrane |
| Chr20 | g67987.t1 | AhDMP4B | 207 | 22.46 | 4.8 | 4 | Plasma Membrane |
| Chr20 | g67988.t1 | AhDMP6B | 206 | 22.68 | 4.98 | 4 | Plasma Membrane |
| Chr20 | g67734.t1 | AhDMP9B | 220 | 24.23 | 8.73 | 4 | Plasma Membrane |
| Gene Name | Gene Name | Duplication Type | Ka | Ks | Ka/Ks |
|---|---|---|---|---|---|
| AhDMP1A | AhDMP2A | WGD or Segmental | 0.386466 | 0.879288 | 0.439522 |
| AhDMP1A | AhDMP1B | WGD or Segmental | 0.009065 | 0.028289 | 0.320457 |
| AhDMP1A | AhDMP2B | WGD or Segmental | 0.380863 | 0.846162 | 0.450107 |
| AhDMP1B | AhDMP2B | WGD or Segmental | 0.389245 | 0.875567 | 0.444564 |
| AhDMP2A | AhDMP1B | WGD or Segmental | 0.386909 | 0.905388 | 0.42734 |
| AhDMP2A | AhDMP2B | WGD or Segmental | 0.009438 | 0.044957 | 0.209928 |
| AhDMP3A | AhDMP3B | WGD or Segmental | 0.016748 | 0.063586 | 0.263386 |
| AhDMP5A | AhDMP5B | WGD or Segmental | 0.014868 | 0.058225 | 0.255349 |
| AhDMP6A | AhDMP7B | WGD or Segmental | 0.156265 | 1.180603 | 0.132361 |
| AhDMP6A | AhDMP4B | WGD or Segmental | 0.167732 | 0.587088 | 0.285701 |
| AhDMP7A | AhDMP6A | WGD or Segmental | 0.156328 | 1.14425 | 0.13662 |
| AhDMP7A | AhDMP7B | WGD or Segmental | 0 | 0.014035 | 0 |
| AhDMP7A | AhDMP4B | WGD or Segmental | 0.206464 | 0.966381 | 0.213647 |
| AhDMP7B | AhDMP4B | WGD or Segmental | 0.206379 | 0.968674 | 0.213053 |
| AhDMP8A | AhDMP8B | WGD or Segmental | 0.003908 | 0.032792 | 0.119161 |
| AhDMP9A | AhDMP9B | WGD or Segmental | 0.008008 | 0.039012 | 0.205272 |
| AhDMP1A | AhDMP1B | WGD or Segmental | 0.00381 | 0.047801 | 0.079708 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, P.; He, L.; Xue, L.; Liu, H.; Li, X.; Zhao, H.; Fu, L.; Han, S.; Dai, X.; Dong, W.; et al. Genome-Wide Identification, Evolution, and Expression Analysis of the DMP Gene Family in Peanut (Arachis hypogaea L.). Int. J. Mol. Sci. 2025, 26, 7243. https://doi.org/10.3390/ijms26157243
Qu P, He L, Xue L, Liu H, Li X, Zhao H, Fu L, Han S, Dai X, Dong W, et al. Genome-Wide Identification, Evolution, and Expression Analysis of the DMP Gene Family in Peanut (Arachis hypogaea L.). International Journal of Molecular Sciences. 2025; 26(15):7243. https://doi.org/10.3390/ijms26157243
Chicago/Turabian StyleQu, Pengyu, Lina He, Lulu Xue, Han Liu, Xiaona Li, Huanhuan Zhao, Liuyang Fu, Suoyi Han, Xiaodong Dai, Wenzhao Dong, and et al. 2025. "Genome-Wide Identification, Evolution, and Expression Analysis of the DMP Gene Family in Peanut (Arachis hypogaea L.)" International Journal of Molecular Sciences 26, no. 15: 7243. https://doi.org/10.3390/ijms26157243
APA StyleQu, P., He, L., Xue, L., Liu, H., Li, X., Zhao, H., Fu, L., Han, S., Dai, X., Dong, W., Shi, L., & Zhang, X. (2025). Genome-Wide Identification, Evolution, and Expression Analysis of the DMP Gene Family in Peanut (Arachis hypogaea L.). International Journal of Molecular Sciences, 26(15), 7243. https://doi.org/10.3390/ijms26157243






