Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century
Abstract
1. Introduction
2. Chromosome Identification and Karyotype Evolution
| Research Object | Research Approach | Aims and Main Findings | References |
|---|---|---|---|
| Thlaspideae (Brassicaceae): Alliaria petiolata (2x, 6x), Didymophysa fenestrata, Graellsia saxifragifolia, G. stylosa, Parlatoria cakiloidea, Peltaria turkmena, Peltariopsis grossheimii, P. planisiliqua, Pseudocamelina glaucophylla, P. szowitsii, Thlaspi arvensa | Multiple FISH approaches: CCP with Arabidopsis thaliana BAC contigs; GISH with gDNA of Pa. cakiloidea and A. petiolata; oligo-FISH with probes localising satDNAs, BAC-FISH with clones targeting 5S and 35S rDNA | Reconstruction of chromosomal organisation in the ancestral genome and analysis of karyotype structure and evolution in 12 Thlaspideae representatives; detection of genus- and species-specific CRs (e.g., pericentric inversions); evidence for allohexaploid origin of A. petiolata (6x) from diploid A. petiolata and Pa. cakiloidea | [72] |
| Catolobus pendulus | CCP with Arabidopsis thaliana BAC contigs | Karyotype organisation of an understudied species; the hypotetraploid C. pendulus genome originated from a whole-genome duplication in a genome resembling the ACK, followed by chromosomal rediploidisation | [59] |
| B. distachyon, B. stacei, B. hybridum | BAC-FISH with chromosome-specific B. distachyon clones | Reconstruction of chromosome evolution between genome D and S in annual Brachypodium diploids and their derived allopolyploid; complete chromosomal synteny observed between B. hybridum and its progenitors | [73] |
| Brachypodium boissieri, B. mexicanum, B. phoenicoides, B. retusum, B. rupestre | BAC-FISH with chromosome-specific B. distachyon clones | Chromosome identification in all species; reconstruction of chromosome evolution among the A1.1, A1.2, A2, E1, E2, and G genome in perennial Brachypodium polyploids; demonstration of ‘orphan’ genomes in the model grass genus | [74] |
| Phaseolus vulgaris, Vigna aconitifolia, V. unguiculata | BAC-FISH with chromosome-specific P. vulgaris and V. unguiculata clones; FISH with 5S and 35S rDNA-targeting probes | Intergeneric analyses of karyotype organisation; detection of macrosynteny breaks between Vigna and Phaseolus; CRs (duplications, inversions, and translocations) contribute to karyotype divergence in Vigna | [75] |
| Phaseolus leptostachyus, P. macvaughii | BAC-FISH with chromosome-specific Phaseolus vulgaris clones; FISH with 5S and 35S rDNA-targeting probes | Interspecific analyses of karyotype organisation; CRs observed in P. leptostachyus are not shared with P. macvaughii; only one nested chromosome fusion (chromosomes 10 and 11), is common to both; pericentric inversions detected in chromosomes 3 and 4 exclusively in P. macvaughii | [76] |
| Macroptilium atropurpureum, M. bracteatum, M. erythroloma, M. gracile, M. lathyroides, M. martii | BAC-FISH with chromosome-specific Phaseolus vulgaris clones; FISH with 5S and 35S rDNA-targeting probes | Interspecific analyses of karyotype organisation; BAC markers show synteny on orthologous chromosomes, while karyotype differentiation primarily driven by the number and distribution of rDNA loci | [77] |
| Passiflora alata, P. watsoniana | BAC-FISH with Passiflora edulis clones; Ty1-Copia and Ty3-Gypsy elements from P. edulis; FISH with 5S and 35S rDNA-targeting probes | Interspecific analyses of karyotype organisation; despite karyotype variability, no synteny breaks were observed in the chromosomal distribution of BACs and rDNA sites, except for an additional 35S rDNA locus on chromosome 3 of P. watsoniana; LTRs were uniformly dispersed, with occasional slight accumulation in proximal chromosome regions | [78] |
| Phaseolus vulgaris, Vigna angularis, V. unguiculata | BAC-FISH with V. unguiculata clones; oligo-FISH with CCP probes designed from the P. vulgaris genome sequence; FISH with a probe targeting 35S rDNA | Intergeneric analyses of karyotype organisation; first oligo-FISH CP in legumes; combination of BAC- and oligo-FISH resources, establishing a cytogenetic map of V. angularis; detection of CRs (translocations, inversions); chromosomes 2 and 3 identified as hotspots for CRs and de novo centromere formation | [79] |
| Phaseolus vulgaris, Vigna unguiculata | Oligo-FISH with barcode probes designed from the Vigna unguiculata genome sequence | Intergeneric analyses of karyotype organisation; in silico integration of previously established BAC-based chromosome-specific and rDNA markers with a newly developed oligo-FISH-based chromosome identification system; alignment of cytogenetic data with genome sequence data for representatives of two distinct genera; confirmation of known and detection of novel CRs (translocations, peri- and paracentric inversions) | [104] |
| Phaseoleae (Fabaceae): Phaseolus vulgaris, Vigna unguiculata, Lablab purpureus, Macroptilium atropurpureum | Oligo-FISH with painting probes specific to Pv2 and Pv3 chromosomes of P. vulgaris | Intergeneric analyses of karyotype organisation; inference of basic chromosome number and number of genomic blocks in the APK; the translocation between APK2 and APK3 is exclusive to Phaseolus, as chromosomes 2 and 3 of L. purpureus and M. atropurpureum resemble the orthologous chromosomes of V. unguiculata and are more closely related to the APK | [103] |
| Phaseolus acutifolius, P. coccineus, P. dumosus, P. filiformis, P. leptostachyus, P. lunatus, P. macvaughii, P. microcarpus, P. vulgaris, P. vulgaris | CCP with oligo probes designed from the Vigna unguiculata genome sequence and oligo probes specific for chromosomes 2 and 3 of P. vulgaris; FISH with 5S and 35S rDNA-targeting probes and the CentPv1 repeat probe | Karyotype evolution in the genus Phaseolus; detection of chromosomal rearrangements (translocations, inversions, duplications, deletions), primarily in species of the Leptostachyus group; P. leptostachyus experienced rapid genome reshuffling without whole-genome duplication, resulting in a reduction of its chromosome number from 11 to 10 pairs | [101] |
| Vigna subgenera: Vigna; Plectrotropis, Haydonia, Lasiospron, Ceratotropis | CCP with oligo probes specific for chromosomes Pv2 and Pv3 of Phaseolus vulgaris; barcode oligo probes designed from the Vigna unguiculata genome sequence; BAC clones derived from P. vulgaris and V. unguiculata; FISH with 5S and 35S rDNA-targeting probes | Karyotype evolution in the genus Vigna; chromosome identification across eight taxa; macrosynteny observed for chromosomes 2, 3, 4, 6, 7, 8, 9 and 10 in all taxa except V. vexillata, which possesses the most divergent karyotype; only minor differences in painting patterns observed among the subgenera | [81] |
| Vigna lasicarpa, V. unguiculata | CCP with oligo probes specific for chromosomes Pv1, Pv2, Pv3, and Pv5 of Phaseolus vulgaris; barcode oligo probes designed from the Vigna unguiculata reference genome; BAC clones derived from V. unguiculata and P. vulgaris; FISH with 5S and 35S rDNA-targeting probes and the T3AG3 telomeric repeat | Karyotype evolution in the genus Vigna; demonstration of conserved oligo-FISH patterns on chromosomes 2, 6, 8, 10 and 11 between V. unguiculata and V. lasicarpa; paracentric inversions in Vla3 and Vla9; descending dysploidy in V. lasicarpa driven by end-to-end fusion of homoeologous chromosomes 5 and 7 | [82] |
| Lupinus angustifolius, L. cryptanthus, L. micranthus, L. cosentinii, L. pilosus | BAC-FISH with L. angustifolius clones; oligo-FISH with probes specific for chromosome Lang06 of L. angustifolius | Karyotype evolution among five wild Lupinus species; demonstration of putative CRs within the Lang06 region, altering synteny and associated with speciation | [80] |
| Cicer arietinum | Oligo-FISH with chromosome-specific probes designed from the C. arietinum kabuli morphotype genome sequence; BAC-FISH with single-copy clones from the desi morphotype of C. arietinum; FISH with 5S and 35S rDNA-targeting probes and the T3AG3 telomeric repeat | Comparative analysis of closely related chickpea genotypes; individual chromosome identification and karyotype development; identification of CRs contributing to genome diversification among chickpea cultivars | [58] |
| Tripidium arundinaceum | Oligo-FISH with maize-derived painting probes; FISH with 5S and 35S rDNA-targeting probes | Chromosome identification and karyotyping in T. arundinaceum (sugarcane relative), effective despite 18 MY divergence from maize; conserved synteny with sorghum over 9 MYs | [100] |
| Saccharum complex (Erianthus fulvus, E. rockii, Miscanthus sinensis, Narenga porphyrocoma, Saccharum officinarum, S. spontaneum, S. robustum) | Oligo-FISH with painting probes designed from the S. officinarum genome sequence; FISH with 5S and 35S rDNA-targeting probes | Development of a set of 10 chromosome-specific landmarks effective for comparative karyotype analysis within the Saccharum complex; detection of CRs and novel cytotypes; chromosome fusions are common in various polyploids of the complex and alter the basic chromosome numbers | [90] |
| Aegilops markgrafii, Ae. tauschii, Ae. umbellulata, Ae. uniaristata, Ae. speltoides, Triticum aestivum | Oligo-FISH with D genome chromosome-specific painting probes designed from the Triticum aestivum genome sequence | Identification of the D genome in diploid (Aegilops) and polyploid (T. aestivum) species; detection of translocations involving D chromosomes, including two novel translocations (3D–7D and 4D–5D–7D) in three Ae. tauschii accessions; determination of the precise positions of chromosomal breakpoints in Ae. tauschii accessions; painting probes produce signals in four different genomes (U, C, M, N); CRs were identified in Ae. umbellulata, Ae. markgrafii, and Ae. uniaristata | [88] |
| Triticeae (Poaceae) genera: Aegilops, Campeiostachys, Dasypyrum, Elymus, Hordeum, Roegneria, Thinopyrum | Oligo-FISH with painting probes specific for chromosomes 1St to 7St, based on the Pseudoroegneria libanotica and Triticum aestivum reference genomes | Analysis of Triticeae karyotype organisation; identification of individual chromosomes of the St genome; conservation of St chromosomes in St-containing Triticeae representatives; weak St hybridisation signals observed in Y-genome chromosomes suggest an origin from the St genome | [84] |
| Thinopyrum elongatum, Th. bessarabicum wheat–tetraploid Th. elongatum substitution lines, Triticum durum–Th. elongatum amphidiploid | Oligo-FISH with painting probes designed from the Th. elongatum and Triticum aestivum genome sequences; tandem-repeat oligo probes pSc119.2 and pTa535 | Development of a complete set of the E-genome painting probes to facilitate the detection of alien material in wheat breeding; chromosome identification; Th. bessarabicum (2x) shows a close genetic relationship with diploid Th. elongatum; five of the seven E-genome chromosomes exhibit complete synteny in both diploids, except for a reciprocal translocation between 4E and 5Eb; a reciprocal translocation between 5E and 7E is present in one of the diploid Th. elongatum accessions | [83] |
| Avena eriantha, A. fatua, A. nuda, A. sativa, A. ventricosa, A. wiestii | Oligo-FISH with painting probes based on syntenic regions between wheat and barley; tandem-repeat oligo probes | Comparative karyotyping of eleven hexaploid and diploid Avena accessions; a high-resolution standard karyotype of A. sativa was established based on distinct FISH signals from multiple oligo probes | [86] |
| Elymus dahuricus, Hordeum vulgare | Oligo-FISH with painting probes based on syntenic regions between wheat and barley; oligo-FISH with repeat-based probes: pSc119.2, pTa535, Po5, 7E-716, 7E-599, 5S rDNA, 18S rDNA, 3A1, 13-J1011, 7E-744, d01-135, Ae334, and (GAA)7; GISH using Pseudoroegneria spicata gDNA; CENH3 immunolocalisation | Establishment of a universal karyotyping nomenclature system for E. dahuricus; precise determination of the linkage groups and sub-genomes of individual chromosomes; detection of a novel intergenomic rearrangement between the 2H and 5Y chromosomes in this allopolyploid | [85] |
| Rhynchospora (Cyperaceae): representative species from sections: Albae, Dichromena, Cephalotae, Pauciflorae, Polycephalae, Pseudocapitatae, Tenues | Oligo-FISH with two barcode probes (Rbv-I and Rbv-II) designed from the R. breviuscula genome sequence | Investigation of karyotype evolution and chromosomal variations in highly dynamic holocentric karyotypes; identification of all chromosomes in R. breviuscula; probes mapped in 13 other Rhynchospora representatives reveal CRs, including fusions, fissions, inversions, and translocations, as well as whole-genome duplication in R. pubera | [87] |
| Musa acuminata, M. balbisiana: wild subspecies, cultivars, and hybrids | Oligo-FISH with chromosome-specific probes designed using the M. acuminata genome sequence | Comparative karyotype analysis; detection of numerous accession-specific chromosome translocations correlated with banana speciation; demonstration of the complexity of banana genome evolution; identification of putative progenitors of banana cultivars | [60,89] |
| Elaeis guineensis, E. oleifera, Cocos nucifera, Phoenix dactylifera | Oligo-FISH with chromosome-specific probes designed using the E. guineensis genome sequence; FISH with a 5S rDNA-targeting probe | Establishment of a reference karyotype for E. guineensis and E. oleifera, identification of homoeologous regions in related species | [91] |
| Citrus maxima, C. medica, C. mangshanensis, C. reticulata, Microcitrus australasica, Poncirus trifoliata | Oligo-FISH with painting probes designed using the Citrus maxima genome; FISH with the 180 bp satellite repeat and with probes targeting 5S and 35S rDNA | Identification of all chromosomes in the studied species; complete chromosomal synteny was observed among six Citrus species over approximately 9 MYs of divergence, with no interchromosomal rearrangements identified in any species | [92] |
| Fragaria | Oligo-FISH with painting probes designed using the Fragaria vesca genome; FISH with 5S and 45S rDNA-targeting probes | Identification of individual chromosomes in 11 Fragaria representatives with different ploidy levels; comparative karyotyping; Fragaria species exhibit conserved karyotypes, with no interchromosomal rearrangements observed; differences in rDNA loci organisation patterns were found among polyploids; variations in signal intensity of oligo probes among homologous chromosomes in Fragaria polyploids provides new insights into their origins | [93] |
| Ipomoea | Oligo-FISH with barcode probes designed using I. nil; FISH with 5S and 35S rDNA-targeting probes | Comparative chromosome analysis in I. batatas and its wild relatives with different ploidy levels; I. trifida is the most closely related diploid to I. batatas; providing cytogenetic evidence for the segmental allopolyploid hypothesis of sweet potato origin | [94] |
| Sixteen diploid Ipomoea species representing all seven minor clades | Oligo-FISH with painting probes designed using I. nil chromosomes 7 and 15; FISH with 5S and 35S rDNA-targeting probes | Comparative chromosome analysis across the genus; significant cytogenetic divergence between 2n = 28 and 2n = 30 species, questioning molecular phylogeny-based classifications that group them into the same clade; significant interspecific variation in rDNA loci distribution complements CCP-based analyses | [99] |
| Cucumis sativus var. sativus, C. sativus var. hardwickii, C. hystrix, C. melo, C. metuliferus, C. subsericeus, C. dipsaceus, C. zeyheri, C. anguria | Oligo-FISH with painting probes designed using the C. sativus genome sequence | Designing chromosome painting oligo probe libraries; reconstruction of the ancestral karyotype for the genus; comparative analysis reveals the genome structure of all studied species and complex CRs that occurred during Cucumis karyotype evolution; compared to African species, Asian-origin species possess genomes that are highly reshuffled due to large-scale inversions, centromere repositioning, and chromothripsis-like events | [97,98] |
| Glycyrrhiza eglandulosa, G. glanndulosa, G. eurycarpa, G. glabra, G. inflata, G. prostrata, G. uralensis | Oligo-FISH with painting probes designed using the Glycyrrhiza uralensis genome sequence; FISH with 5S and 45S rDNA-targeting probes | Chromosome identification in G. uralensis and its relatives; exceptionally conserved chromosomal synteny was observed after 3–12 MYs of divergence, with no cytologically visible interchromosomal rearrangements detected by CP | [95] |
| Gossypium hirsutum | Oligo-FISH with probes designed using the Gossypium hirsutum genome sequence and targeting single and multiple chromosomes; FISH with oligo probes targeting telomeric sites and 5S and 45S rDNA loci | Developing robust markers for chromosome identification in previously intractable species | [96] |
| Pulmonaria officinalis group (P. obscura, P. officinalis s. str.) | Multicolour FISH with tandem-repeat probes (five newly identified satellite DNAs, 5S and 45S rDNA) | Designing a new set of chromosome-specific landmarks; comparative karyotyping; chromosome structure in P. officinalis s. str. is more variable than in P. obscura; confirmation of the hybrid status of 2n = 15 putative hybrids collected from mixed populations of P. obscura and P. officinalis s. str. | [105] |
| Silene latifolia, S. dioica, S. vulgaris, S. maritima | Comparative oligo-FISH with an X-chromosome-scaffold-originated probe designed from the S. latifolia genome sequence; Silene STAR-C centromeric and X43.1 repeat DNA probes | Development of a more robust probe for visualising Silene sex chromosomes than any previous markers, and investigation of their evolution; the hybridisation of this probe to the short arms of several autosomes in S. vulgaris and S. maritima suggests that extensive CRs played a role in the evolution of Silene sex chromosomes | [102] |
| Twelve wild representatives of the Hordeum genus | Comparative BAC-FISH with the Hbog_46L9 clone from H. bogdanii, which contains a Panicum-derived chromosomal segment; FISH with a 45S rDNA-targeting probe | Demonstration of horizontal gene transfer by identifying and characterising a foreign chromosomal segment from a Panicum-like donor in wild barley species, highlighting its evolution and dynamics within host genomes | [106] |
| Nearly 30 species of the Onobrychis genus | FISH with 5S and 35S rDNA-targeting probes | Determination of the number and chromosomal distribution of rDNA loci, along with the identification of selected chromosomes; demonstration of polymorphism in rDNA chromosomal patterns in diploids, contrasted with its absence in polyploids; inference of the ancestral basic chromosome number, rDNA loci counts, and mechanisms such as polyploidisation and descending dysploidy that have shaped chromosome number evolution in the genus | [107] |
3. Chromatin and Interphase Nuclear Organisation
| Research Object | Research Approach | Aims and Main Findings | References |
|---|---|---|---|
| Arabidopsis thaliana, Hordeum vulgare | 3D FISH using centromeric, 35S rDNA-targeting, telomeric, subtelomeric, and H5L-specific painting oligo probes; immunodetection of RNA polymerase II, ASY1, ZYP1, DMC1, HEI10, SSSU, and H3K27me3; visualisation of a GFP-tagged protein associated with the nuclear envelope; imaging performed using diffraction-limited confocal microscopy and super-resolution microscopy | A compendium of strategies to analyse the spatial distribution of nuclear and chromosomal signals from 3D image stacks | [66] |
| Arabidopsis thaliana (Col-0 and ddm1-2) | Image analysis using the semi-automatic ImageJ plug-in iCRAQ (https://github.com/gschivre/iCRAQ, accessed on 15 July 2025) and the DL-based tool Nucl.Eye.D (https://zenodo.org/records/7075507, accessed on 15 July 2025) | Detection and quantification of A. thaliana nuclear features using two segmentation methods: iCRAQ (semi-automated) and Nucl.Eye.D (deep learning), enabling precise analysis | [112] |
| Populus trichocarpa | 3D-FISH on frozen root tip sections; FISH with a 45S rDNA-targeting probe and oligopainting probes for chromosomes 17 and 19 | Improved signal quality compared to paraffin sections; chromosome-specific oligo probes enabled 3D analysis of chromosome territories; autosome pair 17 associated more frequently than sex chromosome 19 | [113] |
| Arabidopsis thaliana WT and crwn1-1, crwn4-1, and kaku4-2 mutants | BAC-FISH using clones specific to A. thaliana chromosomes 1 and 3 | Plant chromatin organisation is flexible, adapting to developmental and environmental cues; under heat stress, the nuclear lamina disassembles, and chromatin domains relocate from the nuclear envelope to the inner nucleus while remaining associated with CRWN1; CRWN1 plays a key role in genome folding dynamics during stress | [63] |
| Arabidopsis thaliana Columbia-0 and cap-d3 T-DNA insertion mutants | FISH using 180 bp centromeric repeat, 5S and 45S rDNA-targeting probes; immunostaining with antibodies against histone modifications H3K27me3, H3K9me1, H3K9me2, H3K4me3, H3K9ac, H3K14ac, H3K18ac or H3K9 + 14 + 18 + 23 + 27ac, and 5-methylcytosine | Evaluation of the role of CAP-D3 in interphase chromatin organisation and function; in cap-d3 mutants, heterochromatic sequences show increased association, while nuclear size and the general histone and DNA methylation patterns remain unchanged | [62] |
| Arabidopsis thaliana, Arabis cypria, Bunias orientalis, Cardamine amara, Descurainia preauxiana, Euclidium syriacum, Hesperis sylvestris | 2D and 3D FISH using centromere-specific oligo probes: pAL, ArCy1, CARCEN, HeSy1, and de novo identified 156-bp repeat of D. preauxiana; telomeric repeat and A. thaliana BAC clone T15P10 (AF167571) containing 35S rRNA genes | The CC-loop model in Arabidopsis thaliana links telomeres to the nucleolus; in crucifers, small genomes exhibit nucleolus-associated telomere clustering, whereas large genomes display a Rabl-like configuration or a dispersed chromosomal distribution | [116] |
| Avena sativa, Brachypodium distachyon, Hordeum vulgare, Oryza sativa, Secale cereale, Triticum aestivum, Zea mays | Nuclei sorting by flow cytometry; 5-ethynyl-2′-deoxyuridine labelling; FISH using a telomere oligo probe; centromere immunovisualisation with an antibody against OsCenH3 (rice centromeric histone H3 variant) | Conserved DNA replication dynamics and chromosome positioning across seven Poaceae species with varying genome sizes | [117] |
| Limnanthaceae, Brassicales | 2D and 3D FISH using centromere-specific oligo probes, telomeric repeat, Arabidopsis thaliana BAC clone T15P10 (AF167571) containing 35S rRNA genes, and clone pCT4.2 (M65137) corresponding to the 5S rDNA repeat | Five chromosome pairs in the interphase nuclei of Limnanthes species adopt a Rabl-like configuration | [118] |
| Oryza sativa | Oligo-FISH using painting probes specific for chromosome 9 and the S and L arms of chromosome 2; FISH with probes targeting centromeric, telomeric, and 45S rDNA sites | Six chromosome territory (CT) configurations were identified in O. sativa root meristematic nuclei and four in leaf nuclei, showing variations in CT volume and association frequency; the association of chromosome 9 CTs was influenced by 45S rDNA activity, linking nuclear organisation to the position and size of the nucleolus | [61] |
| Hordeum vulgare | Nuclei sorting by flow cytometry; oligo-FISH using a barley centromere-specific probe; FISH with telomeric repeats, 5S and 45S rDNA-targeting probes; immunostaining with antibodies against H. vulgare CENH3 | Analysis of nuclear morphology and chromosome organisation in cycling and endoreduplicated nuclei isolated from embryo and endosperm tissues of developing barley seeds; endoreduplicated nuclei exhibit irregular shapes, show reduced sister chromatid cohesion at 5S rDNA loci, and decreased CENH3 levels; progressive endoreduplication leads to intermingling centromeres and telomeres | [121] |
4. Chromosome Structure
| Research Object | Research Approach | Aims and Main Findings | References |
|---|---|---|---|
| Hordeum vulgare | Immunostaining with antibodies against Topo IIα and grass CENH3 (centromeric histone H3 variant); structured illumination microscopy (SIM) and photoactivated localisation microscopy | Topo IIα is dispersed along chromosome arms but accumulates at centromeres, telomeres, and NORs; at centromeres, Topo IIα intermingles with CENH3-containing chromatin | [126] |
| Hordeum vulgare | Oligo-FISH with probes specific to the 5HL chromosome; 5-ethynyl-2′-deoxyuridine (EdU) labelling; analysis of purified metaphase chromosomes; biopolymer modelling; spatial SIM of large fluorescently labelled chromosome segments | Direct differential visualisation of a condensed chromatin fibre confirms the helical model; revealing chromonemas—helically wound, 400-nm-thick chromatin threads that form the chromatids of mitotic chromosomes | [122] |
| Agave tequilana, Hesperaloe funifera, H. parviflora, Hesperoyucca whipplei, Yucca carnerosana, Y. constricta, Y. elata | Immunostaining with antibodies against agavoid CENH3; 3D super-resolution microscopy; scaling relationship of kinetochore size to chromosome size in the karyotype | A positive intra-karyotype relationship between kinetochore and chromosome size, similar to that observed in other eucaryotes; the scaling of total kinetochore size to genome size may originate from the mechanics of cell division | [133] |
| Prionium serratum | Immunostaining with antibodies against P. serratum CENH3, α-tubulin, histone H3S28ph, and histone H2A120ph | P. serratum exhibits a monocentric chromosome organisation, in contrast to the holocentricity observed in other species of the Cyperid clade (Thurniceae-Juncaceae-Cyperaceae) | [65] |
| Chionographis japonica | Oligo-FISH with probes for C. japonica satellite repeats (centromeric Chio1 and Chio2), LTR transposable elements, telomeric repeat, and 45S rDNA FISH; immunostaining with antibodies against C. japonica CENH3, MIS12, NDC80, and α-tubulin, as well as histone modifications including H3K4me2, H3K9me2, H3S10ph, H3S28ph, H3T3ph, and H2AT120ph; EdU labelling | Holocentric chromatids of C. japonica consist of 7–11 evenly spaced, megabase-sized centromere-specific histone H3-positive units, which contain satellite arrays of 23- and 28-bp-long monomers; the large-scale eu- and heterochromatin arrangement differs between C. japonica and other known holocentric species | [134] |
| Luzula sylvatica | Oligo-FISH with probes for satellite repeats Lusy1 and Lusy2; immunostaining with antibodies against L. elegans CENH3, KNL1, NDC80, and α-tubulin | L. sylvatica holocentromeres are predominantly associated with two satellite DNA repeats, Lusy1 and Lusy2, while CENH3 also binds to satellite-free gene-poor regions; Lusy1 plays a crucial role in centromere function across most Luzula species; holocentric chromosomes in Luzula may have originated from chromosome fusions of ancestral monocentric chromosomes and the expansion of CENH3-associated satDNA | [139] |
| Pisum sativum and related representatives of the Fabeae tribe (Pisum, Lathyrus, Vicia) | Oligo-FISH with painting probes PS6, for P. sativium chromosome 6, satDNA-based FabTR probes; immunostaining with anti-CENH3 antibodies | Assembly and analysis of a 177.6 Mb region of P. sativum chromosome 6 which includes 81.6 Mb centromere region (CEN6) and adjacent segments of both chromosome arms; three satellite repeats were associated with CENH3-enriched chromatin, while five others were not; comparative analysis revealed that the evolution of metapolycentromeres is driven by the expansion of centromeric chromatin into neighbouring chromosomal regions, accompanied by the accumulation of novel satellite repeats, which are complemented by CRs in some species | [64] |
| Cuscuta europaea, C. epithymum, C. australis, C. campestris, C. reflexa | Immunostaining with antibodies against Cuscuta CENH3, KNL1 and KLN2, CENP-C, MIS12, NDC80, BUB3;1/2, borealin, and α-tubulin | The transition from monocentricity to holocentricity in the genus Cuscuta was accompanied by dramatic changes in the kinetochore, including the loss of centromeric localisation of CENH3, CENP-C, KNL1, MIS12, and NDC80 proteins, as well as and the degeneration of the spindle assembly checkpoint (SAC); these changes indicate that holocentric Cuscuta species have lost the ability to form a standard kinetochore and no longer utilise the SAC to regulate microtubule attachment to chromosomes | [135] |
| Arabidopsis thaliana, Chionographis japonica, Cuscuta reflexa, Dionaea muscipula, Drosera capensis, Juncus effusus, Luzula nivea, Nelumbo nucifera, Nymphaea alba, Ocimum basilicum, Picea abies, Pisum sativum, Raphanus sativus, Rhynchospora pubera, Triticum aestivum | Immunostaining with antibodies against KNL1, NDC80, and α-tubulin | The KNL1 and NDC80 antibodies effectively labelled centromeres in condensed chromosomes during cell division, as well as the interphase nuclei of most species tested; KNL1 and NDC80 antibodies are better suited for immunolabeling centromeres than CENH3 antibodies, providing greater versatility across different plant species and enabling the study of centromere organisation in non-model species | [137] |
| Gossypium anomalum, G. arboreum, G. hirsutum, G. raimondii | Immunostaining with antibodies against cotton CENH3; FISH with probes for centromeric repeats of G. anomalum | Characterisation of G. anomalum centromeric sequences using chromatin immunoprecipitation against CENH3 antibodies; G. anomalum centromeres contained only retrotransposon-like repeats and lacked long arrays of satellite DNA | [136] |
| Petunia axillaris subsp. axillaris, P. axillaris subsp. parodi, P. integrifolia subsp. inflata, P. × hybrida | FISH with PSAT1, PSAT3, PSAT4, PSAT5, PSAT6, and PSAT7 satellite repeat probes | Seven repeat families (PSAT1, PSAT3, PSAT4, PSAT5, PSAT6, PSAT7, PSAT8) exhibited high sequence similarity and organisation across the four Petunia genomes; these repeat families occupy distinct chromosomal niches, differing in copy number and organisation | [140] |
| Rosa arvensis, R. multiflora, R. rugosa, R. majalis, R. nitida, R. persica | FISH with 18S rDNA and probes derived from the 5S rDNA genic region, as well as 5S_B and 5S_A IGS subregions | Locus-specific probes determined the number and chromosomal position of 5S rDNA families; two major 5S rDNA families (5S_A and 5S_B) were identified in Rosa diploids and polyploids; the 5S_B family often co-localised with 35S rDNA at NORs, while such co-localisation of the 5S_A family was rare | [141] |
| 41 woody plants representing 37 species and 27 genera, and 18 families, Zea mays | Oligo-FISH with the (AG3T3)3 probe | The AG3T3 sequence was observed at chromosome termini in 38 plants; its non-telomeric signals were detected in 23 plants, being particularly abundant in Chimonanthus campanulatus | [142] |
| Hordeum vulgare fluorescent marker lines | Time-lapse confocal microscopy imaging | Development of unique materials enabling detailed live-cell imaging of mitosis and cytokinesis; determination of the duration of mitosis and its stages in barley; demonstration that chromosome condensation in barley often precedes the mitotic preprophase | [129] |
| Hordeum vulgare | Electron tomography | Dissecting the 3D higher-order structure of metaphase chromosomes using a thin carbon film in electron tomography revealed periodic structures with a 300–400 nm pitch along the barley chromosome axis; their periodicity was twice that of the corresponding structures found in human chromosomes | [123] |
| Hordeum vulgare | Immunogold labelling; immunostaining with barley-specific antibody against Topo II; high-voltage transmission electron microscopy (HVTEM) and ultra-high-voltage transmission electron microscopy (UHVTEM) | HVTEM and UHVTEM combined with immunogold labelling are effective for detecting structural proteins such as Topo II; Topo II molecules are distributed along barley chromosomes in a non-specific pattern, with distinct accumulation at the chromosome termini, nucleolus organiser, and centromeric regions | [127] |
| Hordeum vulgare | Immunostaining with barley-specific antibodies against Topo IIα applied to flow-sorted chromosomes; sub-diffraction variants of fluorescence super-resolution microscopy, such as structured illumination, stimulated emission depletion, and single-molecule localisation microscopy | Protein imaging in barley metaphase chromosomes: comparing selected super-resolution approaches with conventional wide-field and confocal microscopy in terms of mapping resolution and accuracy | [125] |
| Hordeum vulgare | Scanning electron microscopy (SEM) | Investigating the role of calcium ions (Ca2+) in the chromosome structure of barley; BAPTA treatment led to a less condensed, dispersed chromosome structure due to Ca2+ chelation; high-resolution SEM provided detailed visualisation of chromosome ultrastructure under different calcium ion conditions | [124] |
| A human–Arabidopsis thaliana hybrid cell line containing a neo-chromosome | FISH using fifteen probes targeting A. thaliana single-copy genome regions, A. thaliana centromere repeat (Atcen, 180 bp), and telomeric short repeats of human (T2AG3) and A. thaliana (T3AG3) | The structure and function of plant and animal chromosomes are largely conserved, enabling the creation of a human–A. thaliana hybrid cell line; a neo-chromosome was formed by inserting segments of A. thaliana chromosomes 2–5 into human chromosome 15; the neo-chromosome contained A. thaliana centromeric repeats and human telomeres; however the A. thaliana centromere was not functional; most A. thaliana DNA was eliminated during culture | [143] |
| Secale cereale, Triticum aestivum, Aegilops speltoides | FISH with 5S rDNA, S. cereale genome-specific repeat Revolver, B-chromosome-specific repeats (D1100, E3900, Sc9c130, Sc26c38), and the DCR28 gene family | Based on a newly assembled ~430 Mb rye B chromosome pseudomolecule, five candidate genes were identified as trans-acting moderators influencing targeted B chromosome nondisjunction during the first pollen mitosis; among them is DCR28, a microtubule-associated gene; the DCR28 gene family appears to be neo-functionalised and is uniquely highly expressed during the first pollen mitosis in rye | [144] |
| Sorghum purpureosericeum | FISH on embryo sections, pollen grains, and meiocytes using the B-specific repeat CL135 and the centromeric probe CL29 | B chromosome occurrence is tissue- and organ-specific, primarily due to extensive elimination during embryo development, which continues throughout plant growth; accumulation of B chromosomes results either from nondisjunction during the first pollen mitosis or from additional nuclear divisions during pollen development | [145] |
| Aegilops speltoides | FISH with repetitive DNA probes Spelt1, Spelt52, pSc119.2, pTa71 (45S rDNA), As5SDNAE (5S rDNA), CCS1 (centromeric), and T3AG3 (telomeric) | Ectopic associations between B and A chromosomes were observed, along with cell-specific rearrangements of B chromosomes in both mitosis and microgametogenesis; the copy numbers of selected transposable elements and tandem repeats varied with genotype and tissue type, but were unaffected by the presence or absence of B chromosomes | [146] |
5. Natural and Induced Hybridisation and Polyploidy
| Research Object | Research Approach | Aims and Main Findings | References |
|---|---|---|---|
| Saccharum spontaneum | Oligo-FISH with haplotypic probes of S. spontaneum specific to chromosomes 8A, 8B, 8C, and 8D | Whole genome duplications in autopolyploid sugarcane AP85–441; no chromosomal aberrations found between autotetraploid AP85–441 and its spontaneously doubled version indicate strict regulation of chromosome duplication | [162] |
| Hieracium intybaceum, H pallidiflorum. H. picroides, H. prenanthoides | GISH with gDNA of H. intybaceum and H. prenanthoides; FISH with 5S and 35S rDNA-targeting probes | One of the first multiapproach studies of apomictic Hieracium allopolyploids; multiple origins of hybridogenous H. pallidiflorum and H. picroides from the same diploid–polyploid parental species H. intybaceum and H. prenanthoides; new insight into the taxonomic delineation of the species | [155] |
| Opuntia | FISH with 5S and 35S rDNA-targeting probes | O. × cristalensis appears to be a hybrid between the native Argentine species O. rioplatensis and the North American introduced species O. ficus-indica; the number of 5S rDNA sites in O. × cristalensis reflects its ploidy level, whereas the number of 35S rDNA sites does not; probably the first documented case of hybridisation between North and South American Opuntia species | [157] |
| Triticeae | FISH with (1) single copy oligos associated with each of the A-, B-, and D-genome chromosomes of Triticum aestivum; (2) oligos from 1H to 7H chromosomes designed using Hordeum vulgare genome; (3) the conserved oligos based on a wheat reference genome; (4) Synt1 to Synt7 oligos from the syntenic region with > 96% homology in wheat-barley linkage groups; (5) Synt7SL barcoding oligos | Development of a chromosome-specific painting using oligo pools for large-genome Triticeae species; high-throughput karyotyping of Triticeae and some wheat-alien derivatives; tracking interspecific chromosome homologous relationships and non-homologous CRs | [36] |
| Commelina benghalensis, C. communis (Cc), C. communis f. ciliata (Ccfc) | GISH with gDNA of Ccfc | Investigation of the role of polyploidisation in the distribution and survival of Cc and its subspecies Ccfc across urban-rural gradients; urban areas were dominated by Cc, whereas both Cc and Ccfc coexisted in rural areas; polyploidy and an additional genome provide Cc with enhanced survival in urban environments | [158] |
| Synthetic Triticum turgidum–Aegilops umbellulata hybrids | GISH with gDNA of Ae. umbellulata; FISH with following probes: oligo-pTa-535 (pTa535), oligo-pSc119.2 (pSC119.2), oligo-pTa71 (pTa71—35S rDNA-targeting probe), and (AAC)5 | Investigation of unreduced gamete formation mechanisms in T. turgidum–Ae. umbellulata triploid F1 hybrid crosses and the chromosome compositions in their F2 generations; chromosome numbers in F2 plants ranged from 35 to 43, with variations in chromosome loss/gain among genomes, chromosome loss was highest in the U genome; three types of chromosome translocations and polymorphic FISH karyotypes were identified | [161] |
| Camelina intermedia, C. hispida, C. laxa, C. neglecta, C. sativa | GISH with gDNA of C. hispida, C. laxa, C. neglecta, and C. intermedia; CCP with Arabidopsis thaliana BAC contigs as painting probes | The identification of the maternal genome of the allohexaploid C. sativa; a tetraploid C. neglecta-like genome (C. intermedia) is hypothesised to be the likely maternal ancestor of the C. sativa based on its high collinearity with two maternally inherited subgenomes; the study contributes to completing the image of the evolution of the Camelina genus | [156] |
| Synthetic and natural allotetraploid wheat hybrids | FISH with centromere-specific retrotransposon of wheat and the telomere repeat; multicolour GISH with gDNA of Triticum urartu, Aegilops longissima and Ae. tauschii | A series of nascent allotetraploid wheats from three diploid genomes (A, S*, and D) was synthesised; most progeny had consistent chromosome numbers, with each genome containing 14 chromosomes, suggesting stable chromosome number inheritance due to diploidisation; detected aneuploids have affected centromere pairing and clustering in early meiosis | [163] |
| Autopolyploid (4x, 8x, 10x) clones of Saccharum spontaneum | Oligo-FISH with painting probes specific for chromosomes 1 (Chr. 1), 7 (Chr. 7), and 8 (Chr. 8) of S. spontaneum | All clones showed stable, diploid-like chromosome behaviour during meiosis; in the 4x clone, two and two copies of Chr. 8 are of different size, and the pairing likely occurs between the homologs of similar size; considering high sequence similarity among Chr. 8 homologues, some unknown mechanisms are responsible for their peculiar pairing behaviour in the 4x clone | [67] |
| Brachypodium hybridum | FISH with 5S and 35S rDNA-targeting probes | Investigation of nucleolar dominance (ND) stability in B. hybridum genotype 3-7-2 compared to the reference genotype ABR113 revealed differences in tissue-specific expression; in ABR113, ND remained stable across all tissues, including primary and adventitious roots, leaves, and spikes; genotype 3-7-2 exhibited a strong upregulation of S-subgenome units in adventitious roots, but not in other tissues | [164] |
| Brachypodium hybridum, B. distachyon, B. stacei | FISH with 5S and 35S rDNA-targeting probes | Analysis of the structure, expression, and epigenetic landscape of 35S rDNA in allopolyploid B. hybridum and its diploid progenitors, B. distachyon and B. stacei; in B. hybridum, the copy number of B. stacei 35S rDNA homoeologues was reduced, accompanied by their transcriptional inactivation; DNA methylation played a role in the silencing of 35S rDNA loci in the S-subgenome | [165] |
| Brachypodium hybridum, B. distachyon, B. stacei | FISH with 5S and 35S rDNA-targeting probes | Comparative analysis of repetitive DNA, focusing on rDNA, in two B. hybridum genotypes of significantly different evolutionary ages; in the younger genotype, ABR113, partial elimination of 35S rDNA units was detected; the older genotype, Bhyb26, exhibited a tendency toward diploidisation, with a reduction in the number of both 35S and 5S rDNA loci | [166] |
| Multiple genotypes of Festuca × Lolium hybrids, Festuca × Festuca interspecific hybrids | FISH with a 45S rDNA-targeting probe; GISH with gDNA of F. pratensis and F. glaucescens | Investigating ND in Festulolium and fescue hybrids; providing new evidence that this phenomenon is maternity-independent, aligns with genome dominance, and occurs early after hybrid genome merging, being completed in the F2 generation | [167] |
| Two Tragopogon porrifolius lines, por1 and por2, which significantly differ in their 35S rDNA copy number | FISH with 5S and 35S rDNA-targeting probes | A positive correlation between the lower 35S rDNA copy number in por1 and the size of NORs on chromosomes D; both L- and S-variants of 35S rDNA were detected in por2,whereas only the S-rDNA variant was found in por1; in por1, the expression of S-rDNA was linked to secondary constrictions (SCs) of NORs located on both chromosomes A; in por2, silencing of S-rDNA was accompanied by NOR condensation on chromosomes A, the presence of SCs on D-NORs, and the expression of L-rDNA, suggesting bidirectional ND | [168] |
6. Cytogenetics-Assisted Crop Improvement
| Research Object | Research Approach | Trait(s) of Interest, Aims, and Key Findings | References |
|---|---|---|---|
| ×Triticosecale introgression lines | GISH with gDNA of Aegilops sharonensis and Ae. taushii | Leaf rust caused by Puccinia triticina: identification of Ae. kotschyi and Ae. tauschii chromosome segments in triticale translocation lines carrying resistance genes | [178] |
| Triticum aestivum–Secale cereale introgression lines | GISH with gDNA of S. cereale; FISH with the repetitive sequence pAs1 and pSc119.2 probes; the 6c6 wheat-specific centromeric probe; the pMD-CEN3 S. cereale-specific centromeric probe; and an Arabidopsis thaliana-type (T3AG3) telomeric probe | Stripe rust caused by Puccinia striiformis: cytomolecular characterisation of the wheat–rye T1RS.1BL translocation line, including the presence of complex chromosome translocations | [179,180] |
| ×Triticosecale × wheat derivatives | GISH with gDNA of S. cereale; FISH with pSc119.2, pTa71 (35S rDNA), and pAs1 probes | Yellow rust resistance: identification of the 1RS.1BL translocation in triticale × wheat progenies | [181] |
| Triticum aestivum–Agropyron cristatum introgression lines | GISH with gDNA of A. cristatum; oligo-FISH with pSc119.2-1, pTa531-1, pAcCR1, and CCS1 probes | Plant height and leaf size: identification of spontaneous T1AL.1PS and T1AS.1PL Robertsonian translocations in the wheat–A. cristatum translocation lines | [68] |
| Triticum aestivum–Agropyron cristatum introgression line | GISH with gDNA of A. cristatum | Multiple elite agronomic traits, including high resistance to powdery mildew and leaf rust: characterisation of the wheat–A. cristatum disomic 6P addition line | [182] |
| Triticum aestivum–Aegilops biuncialis introgression line | GISH with gDNA of Ae. umbelulata, Ae. comosa, and T. turgidum; oligo-FISH with pAs1 and pSc119.2 probes | Glume properties: characterisation of the wheat–Ae. biuncialis 5Mb disomic addition line | [183] |
| Triticum aestivum–Aegilops geniculata introgression lines | GISH with gDNA of Ae. geniculata; oligo-FISH with pAs1 and pSc119.2 probes | Fusarium head blight, powdery mildew, and stripe rust resistance: characterisation of substitution lines with high resistance to these diseases, derived from hybrid progeny between Ae. geniculata and hexaploid wheat | [69] |
| Triticum aestivum–Aegilops geniculata introgression lines | GISH with gDNA of Ae. geniculata; oligo-FISH with pSc119.2 and pTa535 probes | Stripe rust (3Mg DAL) and powdery mildew (7Mg DAL) resistance: characterisation of wheat–Ae. geniculata disomic addition lines | [184,185] |
| Triticum aestivum–Elymus sibiricus introgression line | GISH with gDNA of E. sibiricus; FISH with 35S rDNA-targeting probe | Leaf rust resistance: characterisation of the novel wheat–E. sibiricus 3St addition line | [186] |
| Triticum aestivum–Leymus mollis introgression line | GISH with gDNA of L. mollis; oligo-FISH with pSc119.2 and pTa535 probes | Stripe rust resistance, spike length: characterisation of a novel wheat–L. mollis 2Ns (2D) disomic substitution line | [187] |
| Interspecific derivatives between Triticum aestivum and Psathyrostachys huashanica | GISH with gDNA of P. huashanica; oligo-FISH with pSc119.2 and oligo-pTa535 probes | Fusarium head blight resistance: identifying and characterising two pathogen-resistant interspecific derivatives: wheat–P. huashanica 1Ns long arm ditelosomic addition line and 2Ns substitution line | [188] |
| Triticum aestivum–Psathyrostachys huashanica introgression line | GISH with gDNA of P. huashanica; oligo-FISH with pSc119.2 and pTa535 probes | Several elite agronomic traits, including elongated glumes, longer spikes, larger grains, and resistance to Fusarium head blight: characterisation of the wheat–P. huashanica 3Ns disomic 6P addition line | [189] |
| Triticum aestivum × Thinopyrum intermedium derivatives | GISH with gDNA of Th. bessarabicum; oligo-FISH with probes pAs1-1, pAs1-3, AFA-4, (GAA)10, and pSc119.2-1 | Fusarium head blight resistance: examining the chromosome composition of five wheat–Th. intermedium partial amphiploids with J-genome chromosomes | [190] |
| Triticum aestivum–Thinopyrum intermedium and Triticum aestivum–Th. ponticum introgression lines | GISH with gDNA of Th. bessarabicum, Th. intermedium, and Th. ponticum; oligo-FISH with pSc119.2 and pTa535 probes | Stripe rust resistance: characterisation of wheat–Thinopyrum disomic substitution lines | [191] |
| Brassica juncea–B. fruticulosa introgression lines | GISH with gDNA of B. fruticulosa and B. nigra; oligo-FISH with probes designed using the B. rapa genome and the repetitive sequence CentBr2 probe | Mustard aphid resistance: identification of introgressions from wild species into the crop and tracking the stability of introgressed fragments of interest across generations | [192] |
| Hibiscus cannabinus | FISH with 18S-1 (35S rDNA), pXV1 (5S rDNA), and pLT11 (telomeric) probes | Initial comparative cytogenetic characterisation of kenaf landrace and breeding lines | [193] |
| Phaseolus vulgaris | FISH with 5S and 35S rDNA-targeting probes | Cytogenetic characterisation of 154 common bean accessions: high polymorphism in the number of 45S rDNA sites among the five accessions studied by FISH | [194] |
| Camelina sativa | FISH with 5S and 35S rDNA-targeting probes | Cytogenetic characterisation of nine C. sativa genotypes: high polymorphism in the number of 5S and 45S rDNA sites | [195] |
| Veronica species/cultivars and their progenies | FISH with 5S and 35S rDNA-targeting probes | Improving Veronica breeding programmes: pre-screening of hybrids, identification of true hybrids, self-pollinated progenies, and false hybrids | [196] |
| Gentiana cruciata and G. tibetica somatic hybrids | GISH with gDNA of G. tibetica; FISH with 5S and 35S rDNA-targeting probes | Cytogenetic characterisation of interspecific somatic hybrids: relatively high chromosomal stability with a predominance of G. cruciata chromosomes | [197] |
| Lilium davidii var. unicolor, L. regale and Lilium intersectional hybrids | GISH with gDNA of L. longiflorum and L. speciosum ‘gloriosoides’; oligo-FISH with pTA794 (5S rDNA), telomeric, and pITS probes | Improving lily breeding: characterising the genomic composition of hybrid progeny and determining the parental origin of specific chromosomes; non-denaturing FISH provides an advantage when reprobing slides | [198] |
| Gossypium hirsutum–G. anomalum chromosome segment substitution lines | Oligo-FISH with painting probes specific for chromosomes 6 (Chr. 06), 9 (Chr. 9), and 11 (Chr. 11) of G. anomalum | Chromosome-specific identification of G. anomalum introgressions in a G. hirsutum background, supporting the SSR and resequencing data | [199] |
7. Further Current Fields of Plant Cytomolecular Research
8. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 2D | Two-dimensional |
| 3D | Three-dimensional |
| 3C | Chromosome conformation capture |
| ACK | Ancestral Crucifer Karyotype |
| AI | Artificial intelligence |
| APK | Ancestral Phaseoleae Karyotype |
| arabidopsis | Arabidopsis thaliana |
| BAC | Bacterial artificial chromosome |
| Brachypodium | Brachypodium distachyon |
| Cas | CRISPR-associated protein |
| CCP | Comparative chromosome painting |
| CENH3 | Centromeric histone H3 variant |
| ChIA-PET | Chromatin interaction analysis by paired-end tag sequencing |
| CP | Chromosome painting |
| CR | Chromosome rearrangement |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| CT | Chromosome territory |
| CWR | Crop wild relative |
| dCas | Nuclease-deficient (‘dead’) CRISPR-associated protein |
| EdU | 5-ethynyl-2′-deoxyuridine |
| FISH | Fluorescence in situ hybridisation |
| FITC | Fluorescein isothiocyanate |
| gDNA | Total genomic DNA |
| GISH | Genomic in situ hybridisation |
| Hi-C | High-throughput chromatin conformation capture |
| HiChIP | In situ Hi-C followed by chromatin immunoprecipitation |
| HVTEM | High-voltage transmission electron microscopy |
| ISH | In situ hybridisation |
| LTR | Long terminal repeat |
| MY | Million years |
| ND | Nucleolar dominance |
| Oligo-FISH | Oligonucleotide fluorescence in situ hybridisation |
| rDNA | Ribosomal DNA |
| REXdb | Database of retrotransposon protein domains |
| rRNA | Ribosomal RNA |
| SEM | Scanning electron microscopy |
| SIM | Structured illumination microscopy |
| TEM | Transmission electron microscopy |
| Topo | Topoisomerase |
| UHVTEM | Ultra-high-voltage transmission electron microscopy |
| WT | Wild type |
References
- Fransz, P.; van de Belt, J.; de Jong, H. Extended DNA fibers for high-resolution mapping. Methods Mol. Biol. 2023, 2672, 351–363. [Google Scholar] [CrossRef]
- Schwarzacher, T.; Heslop-Harrison, J.S. Practical In Situ Hybridization; BIOS Scientific Publishers: Oxford, UK, 2000. [Google Scholar]
- Pardue, M.L.; Gall, J.G. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc. Natl. Acad. Sci. USA 1969, 64, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Wobus, U. In situ hybridization confirms jumping nucleolus organizing regions in Allium. Chromosoma 1985, 92, 143–148. [Google Scholar] [CrossRef]
- Weiss, H.; Pasierbek, P.; Maluszynska, J. An improved nonfluorescent detection system for in situ hybridization in plants. Biotech. Histochem. 2000, 75, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Maluszynska, J.; Schweizer, D. Ribosomal RNA genes in B chromosomes of Crepis capillaris detected by non-radioactive in situ hybridization. Heredity 1989, 62 Pt 1, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Gill, B.S.; Wang, G.L.; Ronald, P.C.; Ward, D.C. Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 1995, 92, 4487–4491. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.S.; Jiang, J.; Gill, B.S.; Paterson, A.H.; Wing, R.A. Construction and characterization of a bacterial artificial chromosome library of Sorghum bicolor. Nucleic Acids Res. 1994, 22, 4922–4931. [Google Scholar] [CrossRef][Green Version]
- Fuchs, J.; Kloos, D.U.; Ganal, M.W.; Schubert, I. In situ localization of yeast artificial chromosome sequences on tomato and potato metaphase chromosomes. Chromosome Res. 1996, 4, 277–281. [Google Scholar] [CrossRef]
- Hagemann, S.; Scheer, B.; Schweizer, D. Repetitive sequences in the genome of Anemone blanda: Identification of tandem arrays and of dispersed repeats. Chromosoma 1993, 102, 312–324. [Google Scholar] [CrossRef]
- Moscone, E.A.; Matzke, M.A.; Matzke, A.J. The use of combined FISH/GISH in conjunction with DAPI counterstaining to identify chromosomes containing transgene inserts in amphidiploid tobacco. Chromosoma 1996, 105, 231–236. [Google Scholar] [CrossRef]
- Fuchs, J.; Strehl, S.; Brandes, A.; Schweizer, D.; Schubert, I. Molecular-cytogenetic characterization of the Vicia faba genome--heterochromatin differentiation, replication patterns and sequence localization. Chromosome Res. 1998, 6, 219–230. [Google Scholar] [CrossRef]
- Hasterok, R.; Jenkins, G.; Langdon, T.; Jones, R.N.; Maluszynska, J. Ribosomal DNA is an effective marker of Brassica chromosomes. Theor. Appl. Genet. 2001, 103, 486–490. [Google Scholar] [CrossRef]
- Moscone, E.A.; Klein, F.; Lambrou, M.; Fuchs, J.; Schweizer, D. Quantitative karyotyping and dual-color FISH mapping of 5S and 18S-25S rDNA probes in the cultivated Phaseolus species (Leguminosae). Genome 1999, 42, 1224–1233. [Google Scholar] [CrossRef]
- Snowdon, R.J.; Friedt, W.; Köhler, A.; Köhler, W. Molecular cytogenetic localization and characterization of 5S and 25S rDNA loci for chromosome identification in oilseed rape (Brassica napus L.). Ann. Bot. 2000, 86, 201–204. [Google Scholar] [CrossRef]
- Hasterok, R.; Dulawa, J.; Jenkins, G.; Leggett, M.; Langdon, T. Multi-substrate chromosome preparations for high throughput comparative FISH. BMC Biotechnol. 2006, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Hasterok, R.; Langdon, T.; Taylor, S.; Jenkins, G. Combinatorial labelling of DNA probes enables multicolour fluorescence in situ hybridisation in plants. Folia Histochem. Cytobiol. 2002, 40, 319–323. [Google Scholar] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Fransz, P.F.; Ali, H.B.; Schubert, I. Chromosome painting in Arabidopsis thaliana. Plant J. 2001, 28, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Pecinka, A.; Schubert, I. Recent progress in chromosome painting of Arabidopsis and related species. Chromosome Res. 2003, 11, 195–204. [Google Scholar] [CrossRef]
- Lysak, M.A.; Berr, A.; Pecinka, A.; Schmidt, R.; McBreen, K.; Schubert, I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 2006, 103, 5224–5229. [Google Scholar] [CrossRef]
- Pecinka, A.; Schubert, V.; Meister, A.; Kreth, G.; Klatte, M.; Lysak, M.A.; Fuchs, J.; Schubert, I. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 2004, 113, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Szinay, D.; Lang, C.; Ramanna, M.S.; van der Vossen, E.A.; Datema, E.; Lankhorst, R.K.; de Boer, J.; Peters, S.A.; Bachem, C.; et al. Cross-species bacterial artificial chromosome-fluorescence in situ hybridization painting of the tomato and potato chromosome 6 reveals undescribed chromosomal rearrangements. Genetics 2008, 180, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Idziak, D.; Betekhtin, A.; Wolny, E.; Lesniewska, K.; Wright, J.; Febrer, M.; Bevan, M.W.; Jenkins, G.; Hasterok, R. Painting the chromosomes of Brachypodium: Current status and future prospects. Chromosoma 2011, 120, 469–479. [Google Scholar] [CrossRef]
- Betekhtin, A.; Jenkins, G.; Hasterok, R. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS ONE 2014, 9, e115108. [Google Scholar] [CrossRef]
- Peterson, D.G.; Tomkins, J.P.; Frisch, D.A.; Wing, R.A.; Paterson, A.H. Construction of Plant Bacterial Artificial Chromosome (BAC) Libraries: An illustrated Guide. Available online: https://www.mgel.msstate.edu/pubs/bacman2.pdf (accessed on 15 July 2025).
- Beliveau, B.J.; Joyce, E.F.; Apostolopoulos, N.; Yilmaz, F.; Fonseka, C.Y.; McCole, R.B.; Chang, Y.; Li, J.B.; Senaratne, T.N.; Williams, B.R.; et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl. Acad. Sci. USA 2012, 109, 21301–21306. [Google Scholar] [CrossRef]
- Boyle, S.; Rodesch, M.J.; Halvensleben, H.A.; Jeddeloh, J.A.; Bickmore, W.A. Fluorescence in situ hybridization with high-complexity repeat-free oligonucleotide probes generated by massively parallel synthesis. Chromosome Res. 2011, 19, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Yamada, N.A.; Rector, L.S.; Tsang, P.; Carr, E.; Scheffer, A.; Sederberg, M.C.; Aston, M.E.; Ach, R.A.; Tsalenko, A.; Sampas, N.; et al. Visualization of fine-scale genomic structure by oligonucleotide-based high-resolution FISH. Cytogenet. Genome Res. 2011, 132, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, T.; Thammapichai, P.; Weng, Y.; Jiang, J. Chromosome-specific painting in Cucumis species using bulked oligonucleotides. Genetics 2015, 200, 771–779. [Google Scholar] [CrossRef]
- Liu, G.; Zhang, T. Single copy oligonucleotide fluorescence in situ hybridization probe design platforms: Development, application and evaluation. Int. J. Mol. Sci. 2021, 22, 7124. [Google Scholar] [CrossRef]
- Harun, A.; Liu, H.; Song, S.; Asghar, S.; Wen, X.; Fang, Z.; Chen, C. Oligonucleotide fluorescence in situ hybridization: An efficient chromosome painting method in plants. Plants 2023, 12, 2816. [Google Scholar] [CrossRef]
- Schwacke, R.; Bolger, M.; Usadel, B. PubPlant—A continuously updated online resource for sequenced and published plant genomes. Front. Plant Sci. 2025, 16, 1603547. [Google Scholar] [CrossRef]
- Schwacke, R.; Bolger, M.; Usadel, B. PubPlant—A Continuously Updated Resource for Sequenced and Published Plant Genomes. Available online: https://www.plabipd.de/pubplant_timeline.html (accessed on 15 July 2025).
- Albert, P.S.; Zhang, T.; Semrau, K.; Rouillard, J.M.; Kao, Y.H.; Wang, C.R.; Danilova, T.V.; Jiang, J.; Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc. Natl. Acad. Sci. USA 2019, 116, 1679–1685. [Google Scholar] [CrossRef]
- Li, G.R.; Zhang, T.; Yu, Z.H.; Wang, H.J.; Yang, E.N.; Yang, Z.J. An efficient Oligo-FISH painting system for revealing chromosome rearrangements and polyploidization in Triticeae. Plant J. 2021, 105, 978–993. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Sun, S.; Wu, Y.; Zhou, Y.; Gu, S.; Yu, H.; Yi, C.; Gu, M.; Jiang, J.; Liu, B.; et al. Dual-color oligo-FISH can reveal chromosomal variations and evolution in Oryza species. Plant J. 2020, 101, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, J.; Demidov, D.; Houben, A.; Schubert, I. Chromosomal histone modification patterns—from conservation to diversity. Trends Plant Sci. 2006, 11, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Wolny, E.; Braszewska-Zalewska, A.; Hasterok, R. Spatial distribution of epigenetic modifications in Brachypodium distachyon embryos during seed maturation and germination. PLoS ONE 2014, 9, e101246. [Google Scholar] [CrossRef] [PubMed]
- Wolny, E.; Braszewska-Zalewska, A.; Kroczek, D.; Hasterok, R. Histone H3 and H4 acetylation patterns are more dynamic than those of DNA methylation in Brachypodium distachyon embryos during seed maturation and germination. Protoplasma 2017, 254, 2045–2052. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Hasterok, R. Epigenetics of the preferential silencing of Brachypodium stacei-originated 35S rDNA loci in the allotetraploid grass Brachypodium hybridum. Sci. Rep. 2017, 7, 5260. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Nibau, C.; Phillips, D.W.; Jenkins, G.; Armstrong, S.J.; Doonan, J.H. CDKG1 protein kinase is essential for synapsis and male meiosis at high ambient temperature in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2014, 111, 2182–2187. [Google Scholar] [CrossRef]
- Choi, K.; Zhao, X.; Tock, A.J.; Lambing, C.; Underwood, C.J.; Hardcastle, T.J.; Serra, H.; Kim, J.; Cho, H.S.; Kim, J.; et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 2018, 28, 532–546. [Google Scholar] [CrossRef]
- Bara, A.W.; Braszewska, A.; Kwasniewska, J. DNA nethylation-an epigenetic mark in mutagen-treated Brachypodium distachyon cells. Plants 2021, 10, 1408. [Google Scholar] [CrossRef] [PubMed]
- Bara-Halama, A.W.; Idziak-Helmcke, D.; Kwasniewska, J. Unraveling the DNA methylation in the rDNA foci in mutagen-induced Brachypodium distachyon micronuclei. Int. J. Mol. Sci. 2022, 23, 6797. [Google Scholar] [CrossRef] [PubMed]
- Wolny, E.; Skalska, A.; Braszewska, A.; Mur, L.A.J.; Hasterok, R. Defining the cell wall, cell cycle and chromatin landmarks in the responses of Brachypodium distachyon to salinity. Int. J. Mol. Sci. 2021, 22, 949. [Google Scholar] [CrossRef] [PubMed]
- Ohmido, N.; Polosoro, A. Chromatin immunostaining of plant nuclei. In Plant Cytogenetics and Cytogenomics. Methods in Molecular Biology; Heitkam, T., Garcia, S., Eds.; Humana: New York, NY, USA, 2023; Volume 2672, pp. 233–244. [Google Scholar]
- Heintzmann, R.; Huser, T. Super-resolution structured illumination microscopy. Chem. Rev. 2017, 117, 13890–13908. [Google Scholar] [CrossRef] [PubMed]
- Hickey, S.M.; Ung, B.; Bader, C.; Brooks, R.; Lazniewska, J.; Johnson, I.R.D.; Sorvina, A.; Logan, J.; Martini, C.; Moore, C.R.; et al. Fluorescence microscopy-an outline of hardware, biological handling, and fluorophore considerations. Cells 2021, 11, 35. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhong, S.; Hou, Y.; Cao, R.; Wang, W.; Li, D.; Dai, Q.; Kim, D.; Xi, P. Superresolution structured illumination microscopy reconstruction algorithms: A review. Light Sci. Appl. 2023, 12, 172. [Google Scholar] [CrossRef]
- Imaris AI Microscopy Image Analysis Software. Available online: https://imaris.oxinst.com/ (accessed on 15 July 2025).
- ImageJ Image Processing and Analysis in Java. Available online: https://imagej.net/ij/ (accessed on 15 July 2025).
- Febrer, M.; Goicoechea, J.L.; Wright, J.; McKenzie, N.; Song, X.; Lin, J.; Collura, K.; Wissotski, M.; Yu, Y.; Ammiraju, J.S.; et al. An integrated physical, genetic and cytogenetic map of Brachypodium distachyon, a model system for grass research. PLoS ONE 2010, 5, e13461. [Google Scholar] [CrossRef]
- The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Xiong, Y.; Yuan, S.; Xiong, Y.; Li, L.; Peng, J.; Zhang, J.; Fan, X.; Jiang, C.; Sha, L.N.; Wang, Z.; et al. Analysis of allohexaploid wheatgrass genome reveals its Y haplome origin in Triticeae and high-altitude adaptation. Nat. Commun. 2025, 16, 3104. [Google Scholar] [CrossRef]
- Ten Thousand Plant Genome Project. Available online: https://en.genomics.cn/en-project-dzwyjs-6184.html (accessed on 15 July 2025).
- Jenkins, G.; Hasterok, R. BAC ‘landing’ on chromosomes of Brachypodium distachyon for comparative genome alignment. Nat. Protoc. 2007, 2, 88–98. [Google Scholar] [CrossRef]
- Dolezalova, A.; Sladekova, L.; Simonikova, D.; Holusova, K.; Karafiatova, M.; Varshney, R.K.; Dolezel, J.; Hribova, E. Karyotype differentiation in cultivated chickpea revealed by oligopainting fluorescence in situ hybridization. Front. Plant Sci. 2021, 12, 791303. [Google Scholar] [CrossRef]
- Farhat, P.; Mandakova, T.; Divisek, J.; Kudoh, H.; German, D.A.; Lysak, M.A. The evolution of the hypotetraploid Catolobus pendulus genome—The poorly known sister species of Capsella. Front. Plant Sci. 2023, 14, 1165140. [Google Scholar] [CrossRef] [PubMed]
- Berankova, D.; Cizkova, J.; Majzlikova, G.; Dolezalova, A.; Mduma, H.; Brown, A.; Swennen, R.; Hribova, E. Striking variation in chromosome structure within Musa acuminata subspecies, diploid cultivars, and F1 diploid hybrids. Front. Plant Sci. 2024, 15, 1387055. [Google Scholar] [CrossRef] [PubMed]
- Dolezalova, A.; Berankova, D.; Kolackova, V.; Hribova, E. Insight into chromatin compaction and spatial organization in rice interphase nuclei. Front. Plant Sci. 2024, 15, 1358760. [Google Scholar] [CrossRef] [PubMed]
- Municio, C.; Antosz, W.; Grasser, K.D.; Kornobis, E.; Van Bel, M.; Eguinoa, I.; Coppens, F.; Brautigam, A.; Lermontova, I.; Bruckmann, A.; et al. The Arabidopsis condensin CAP-D subunits arrange interphase chromatin. New Phytol. 2021, 230, 972–987. [Google Scholar] [CrossRef]
- Wang, N.; Wang, Z.; Tzourtzou, S.; Wang, X.; Bi, X.; Leimeister, J.; Xu, L.; Sakamoto, T.; Matsunaga, S.; Schaller, A.; et al. The plant nuclear lamina disassembles to regulate genome folding in stress conditions. Nat. Plants 2023, 9, 1081–1093. [Google Scholar] [CrossRef]
- Macas, J.; Avila Robledillo, L.; Kreplak, J.; Novak, P.; Koblizkova, A.; Vrbova, I.; Burstin, J.; Neumann, P. Assembly of the 81.6 Mb centromere of pea chromosome 6 elucidates the structure and evolution of metapolycentric chromosomes. PLoS Genet. 2023, 19, e1010633. [Google Scholar] [CrossRef]
- Baez, M.; Kuo, Y.T.; Dias, Y.; Souza, T.; Boudichevskaia, A.; Fuchs, J.; Schubert, V.; Vanzela, A.L.L.; Pedrosa-Harand, A.; Houben, A. Analysis of the small chromosomal Prionium serratum (Cyperid) demonstrates the importance of reliable methods to differentiate between mono- and holocentricity. Chromosoma 2020, 129, 285–297. [Google Scholar] [CrossRef]
- Randall, R.S.; Jourdain, C.; Nowicka, A.; Kaduchova, K.; Kubova, M.; Ayoub, M.A.; Schubert, V.; Tatout, C.; Colas, I.; Kalyanikrishna; et al. Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes. Nucleus 2022, 13, 277–299. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, Z.; Han, J.L.; Khurshid, H.; Esh, A.; Hasterok, R.; Wang, K. Characterization of meiotic chromosome behavior in the autopolyploid Saccharum spontaneum reveals preferential chromosome pairing without distinct DNA sequence variation. Crop J. 2023, 11, 1550–1558. [Google Scholar] [CrossRef]
- Han, B.H.; Wang, X.; Sun, Y.Y.; Kang, X.L.; Zhang, M.; Luo, J.W.; Han, H.M.; Zhou, S.H.; Lu, Y.Q.; Liu, W.H.; et al. Pre-breeding of spontaneous Robertsonian translocations for density planting architecture by transferring Agropyron cristatum chromosome 1P into wheat. Theor. Appl. Genet. 2024, 137, 110. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Xu, M.; Wang, Y.; Cheng, X.; Huang, C.; Zhang, H.; Li, T.; Wang, C.; Chen, C.; Wang, Y.; et al. Development and molecular cytogenetic identification of two wheat-Aegilops geniculata Roth 7Mg chromosome substitution lines with resistance to Fusarium head blight, powdery mildew and stripe rust. Int. J. Mol. Sci. 2022, 23, 7056. [Google Scholar] [CrossRef] [PubMed]
- Hasterok, R.; Wang, K.; Jenkins, G. Progressive refinement of the karyotyping of Brachypodium genomes. New Phytol. 2020, 227, 1668–1675. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, H.; Khurshid, H.; Esh, A.; Wu, C.W.; Wang, Q.N.; Piperidis, N. Past and recent advances in sugarcane cytogenetics. Crop J. 2023, 11, 1–8. [Google Scholar] [CrossRef]
- Bayat, S.; Lysak, M.A.; Mandakova, T. Genome structure and evolution in the cruciferous tribe Thlaspideae (Brassicaceae). Plant J. 2021, 108, 1768–1785. [Google Scholar] [CrossRef]
- Gordon, S.P.; Contreras-Moreira, B.; Levy, J.J.; Djamei, A.; Czedik-Eysenberg, A.; Tartaglio, V.S.; Session, A.; Martin, J.; Cartwright, A.; Katz, A.; et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020, 11, 3670. [Google Scholar] [CrossRef] [PubMed]
- Sancho, R.; Inda, L.A.; Diaz-Perez, A.; Des Marais, D.L.; Gordon, S.; Vogel, J.P.; Lusinska, J.; Hasterok, R.; Contreras-Moreira, B.; Catalan, P. Tracking the ancestry of known and ‘ghost’ homeologous subgenomes in model grass Brachypodium polyploids. Plant J. 2022, 109, 1535–1558. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.; Martins, L.D.V.; Bustamante, F.O.; Munoz-Amatriain, M.; Close, T.; da Costa, A.F.; Benko-Iseppon, A.M.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Breaks of macrosynteny and collinearity among moth bean (Vigna aconitifolia), cowpea (V. unguiculata), and common bean (Phaseolus vulgaris). Chromosome Res. 2020, 28, 293–306. [Google Scholar] [CrossRef]
- Ferraz, M.E.; Fonseca, A.; Pedrosa-Harand, A. Multiple and independent rearrangements revealed by comparative cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosome Res. 2020, 28, 395–405. [Google Scholar] [CrossRef]
- de Barros, D.; Montenegro, C.; Gomes, M.; Ferraz, M.E.; Miotto, S.T.S.; Pedrosa-Harand, A. Cytogenetic characterization and karyotype evolution in six Macroptilium species (Leguminosae). Genome 2023, 66, 165–174. [Google Scholar] [CrossRef]
- Dias, Y.; Sader, M.A.; Vieira, M.L.C.; Pedrosa-Harand, A. Comparative cytogenetic maps of Passiflora alata and P. watsoniana (Passifloraceae) using BAC-FISH. Plant Syst. Evol. 2020, 306, 51. [Google Scholar] [CrossRef]
- Do Vale Martins, L.; de Oliveira Bustamante, F.; da Silva Oliveira, A.R.; da Costa, A.F.; de Lima Feitoza, L.; Liang, Q.; Zhao, H.; Benko-Iseppon, A.M.; Munoz-Amatriain, M.; Pedrosa-Harand, A.; et al. BAC- and oligo-FISH mapping reveals chromosome evolution among Vigna angularis, V. unguiculata, and Phaseolus vulgaris. Chromosoma 2021, 130, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Bielski, W.; Ksiazkiewicz, M.; Simonikova, D.; Hribova, E.; Susek, K.; Naganowska, B. The puzzling fate of a lupin chromosome revealed by reciprocal oligo-FISH and BAC-FISH mapping. Genes 2020, 11, 1489. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.; de Oliveira Bustamante, F.; do Vale Martins, L.; da Costa, V.A.; Montenegro, C.; Oliveira, A.; de Lima, G.S.; Braz, G.T.; Jiang, J.; da Costa, A.F.; et al. Translocations and inversions: Major chromosomal rearrangements during Vigna (Leguminosae) evolution. Theor. Appl. Genet. 2024, 137, 29. [Google Scholar] [CrossRef]
- Serafim, L.; Silva, J.H.; Dias, S.; Oliveira, A.; Nunes, M.C.; da Costa, A.F.; Benko-Iseppon, A.M.; Jiang, J.; do Vale Martins, L.; Brasileiro-Vidal, A.C. “End-to-end chromosome fusion” as the main driver of descending dysploidy in Vigna lasiocarpa (Mart. ex Benth.) Verdc. (Leguminosae Juss.). Plants 2025, 14, 1872. [Google Scholar] [CrossRef]
- Chen, C.; Han, Y.; Xiao, H.; Zou, B.; Wu, D.; Sha, L.; Yang, C.; Liu, S.; Cheng, Y.; Wang, Y.; et al. Chromosome-specific painting in Thinopyrum species using bulked oligonucleotides. Theor. Appl. Genet. 2023, 136, 177. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, X.; Li, Y.; Zou, B.; Xiao, H.; Han, Y.; Yang, X.; Wu, D.; Sha, L.; Yang, C.; et al. Chromosome-specific painting reveals the Y genome origin and chromosome rearrangements of the St genome in Triticeae. Plant Physiol. 2024, 196, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Liu, X.; Yang, Z.; Li, G. Chromosome rearrangement in Elymus dahuricus revealed by ND-FISH and oligo-FISH painting. Plants 2023, 12, 3268. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Jiang, C.; Yuan, W.; Zhang, M.; Fang, Z.; Li, Y.; Li, G.; Jia, J.; Yang, Z. A universal karyotypic system for hexaploid and diploid Avena species brings oat cytogenetics into the genomics era. BMC Plant Biol. 2021, 21, 213. [Google Scholar] [CrossRef]
- Mata-Sucre, Y.; Parteka, L.M.; Ritz, C.M.; Gatica-Arias, A.; Felix, L.P.; Thomas, W.W.; Souza, G.; Vanzela, A.L.L.; Pedrosa-Harand, A.; Marques, A. Oligo-barcode illuminates holocentric karyotype evolution in Rhynchospora (Cyperaceae). Front. Plant Sci. 2024, 15, 1330927. [Google Scholar] [CrossRef]
- Shi, P.; Sun, H.; Liu, G.; Zhang, X.; Zhou, J.; Song, R.; Xiao, J.; Yuan, C.; Sun, L.; Wang, Z.; et al. Chromosome painting reveals inter-chromosomal rearrangements and evolution of subgenome D of wheat. Plant J. 2022, 112, 55–67. [Google Scholar] [CrossRef]
- Simonikova, D.; Nemeckova, A.; Cizkova, J.; Brown, A.; Swennen, R.; Dolezel, J.; Hribova, E. Chromosome painting in cultivated bananas and their wild relatives (Musa spp.) reveals differences in chromosome structure. Int. J. Mol. Sci. 2020, 21, 7915. [Google Scholar] [CrossRef]
- Yu, F.; Zhao, X.; Chai, J.; Ding, X.; Li, X.; Huang, Y.; Wang, X.; Wu, J.; Zhang, M.; Yang, Q.; et al. Chromosome-specific painting unveils chromosomal fusions and distinct allopolyploid species in the Saccharum complex. New Phytol. 2022, 233, 1953–1965. [Google Scholar] [CrossRef]
- Zaki, N.M.; Schwarzacher, T.; Singh, R.; Madon, M.; Wischmeyer, C.; Hanim Mohd Nor, N.; Zulkifli, M.A.; Heslop-Harrison, J.S.P. Chromosome identification in oil palm (Elaeis guineensis) using in situ hybridization with massive pools of single copy oligonucleotides and transferability across Arecaceae species. Chromosome Res. 2021, 29, 373–390. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhao, H.; He, J.; Yang, Z.; Guan, B.; Chen, K.; Hong, Q.; Wang, J.; Liu, J.; Jiang, J. Extraordinarily conserved chromosomal synteny of Citrus species revealed by chromosome-specific painting. Plant J. 2020, 103, 2225–2235. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Zhang, L.; Li, K.; Sun, J.; Li, Z.; Han, Y. Karyotypic stability of Fragaria (strawberry) species revealed by cross-species chromosome painting. Chromosome Res. 2021, 29, 285–300. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhang, Q.; Xu, M.; Yan, M.; Liu, X.; Sun, J.; Cao, Q.; Wang, H.; Yang, J.; Li, Z.; et al. Comparative karyotype analysis provides cytogenetic evidence for the origin of sweetpotato. Chromosome Res. 2024, 32, 14. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Zheng, Q.; Shi, S.; Wang, W.; Wang, F.; Xie, Q.; Chen, X.; Shen, H.; Xiao, G.; Li, H. Whole-chromosome oligo-painting in licorice unveils interspecific chromosomal evolutionary relationships and possible origin of triploid genome species. Plant J. 2024, 120, 2089–2100. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Guo, H.; Wang, Y.; Zhou, B. Identification of chromosomes by fluorescence in situ hybridization in Gossypium hirsutum via developing oligonucleotide probes. Genome 2024, 67, 64–77. [Google Scholar] [CrossRef]
- Bi, Y.; Zhao, Q.; Yan, W.; Li, M.; Liu, Y.; Cheng, C.; Zhang, L.; Yu, X.; Li, J.; Qian, C.; et al. Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J. 2020, 102, 178–186. [Google Scholar] [CrossRef]
- Zhao, Q.; Meng, Y.; Wang, P.; Qin, X.; Cheng, C.; Zhou, J.; Yu, X.; Li, J.; Lou, Q.; Jahn, M.; et al. Reconstruction of ancestral karyotype illuminates chromosome evolution in the genus Cucumis. Plant J. 2021, 107, 1243–1259. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Chen, L.; Sun, J.; Liu, Z.; Han, Y. Cross-species chromosome painting offers new insights into the phylogenetic relationships among 16 representative species of Ipomoeeae. Front. Plant Sci. 2025, 16, 1610698. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Chai, J.; Li, X.; Yu, Z.; Yang, R.; Ding, X.; Wang, Q.; Wu, J.; Yang, X.; Deng, Z. Chromosomal characterization of Tripidium arundinaceum revealed by oligo-FISH. Int. J. Mol. Sci. 2021, 22, 8539. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, T.; Pedrosa-Harand, A. High rates of structural rearrangements have shaped the chromosome evolution in dysploid Phaseolus beans. Theor. Appl. Genet. 2023, 136, 215. [Google Scholar] [CrossRef]
- Bacovsky, V.; Cegan, R.; Simonikova, D.; Hribova, E.; Hobza, R. The formation of sex chromosomes in Silene latifolia and S. dioica was accompanied by multiple chromosomal rearrangements. Front. Plant Sci. 2020, 11, 205. [Google Scholar] [CrossRef]
- Montenegro, C.; do Vale Martins, L.; Bustamante, F.O.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Comparative cytogenomics reveals genome reshuffling and centromere repositioning in the legume tribe Phaseoleae. Chromosome Res. 2022, 30, 477–492. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Bustamante, F.; do Nascimento, T.H.; Montenegro, C.; Dias, S.; do Vale Martins, L.; Braz, G.T.; Benko-Iseppon, A.M.; Jiang, J.; Pedrosa-Harand, A.; Brasileiro-Vidal, A.C. Oligo-FISH barcode in beans: A new chromosome identification system. Theor. Appl. Genet. 2021, 134, 3675–3686. [Google Scholar] [CrossRef] [PubMed]
- Kobrlova, L.; Cizkova, J.; Zoulova, V.; Vejvodova, K.; Hribova, E. First insight into the genomes of the Pulmonaria officinalis group (Boraginaceae) provided by repeatome analysis and comparative karyotyping. BMC Plant Biol. 2024, 24, 859. [Google Scholar] [CrossRef] [PubMed]
- Mahelka, V.; Krak, K.; Fehrer, J.; Caklova, P.; Nagy Nejedla, M.; Cegan, R.; Kopecky, D.; Safar, J. A Panicum-derived chromosomal segment captured by Hordeum a few million years ago preserves a set of stress-related genes. Plant J. 2021, 105, 1141–1164. [Google Scholar] [CrossRef]
- Yucel, G.; Betekhtin, A.; Cabi, E.; Tuna, M.; Hasterok, R.; Kolano, B. The chromosome number and rDNA loci evolution in Onobrychis (Fabaceae). Int. J. Mol. Sci. 2022, 23, 11033. [Google Scholar] [CrossRef]
- Kumar, S.; Kaur, S.; Seem, K.; Kumar, S.; Mohapatra, T. Understanding 3D genome organization and its effect on transcriptional gene regulation under environmental stress in plant: A chromatin perspective. Front. Cell Dev. Biol. 2021, 9, 774719. [Google Scholar] [CrossRef] [PubMed]
- Dogan, E.S.; Liu, C. Three-dimensional chromatin packing and positioning of plant genomes. Nat. Plants 2018, 4, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Pei, L.; Li, G.; Lindsey, K.; Zhang, X.; Wang, M. Plant 3D genomics: The exploration and application of chromatin organization. New Phytol. 2021, 230, 1772–1786. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Xiong, D.; Li, G.; Li, X. Unraveling the 3D genome architecture in plants: Present and future. Mol. Plant 2020, 13, 1676–1693. [Google Scholar] [CrossRef] [PubMed]
- Johann To Berens, P.; Schivre, G.; Theune, M.; Peter, J.; Sall, S.O.; Mutterer, J.; Barneche, F.; Bourbousse, C.; Molinier, J. Advanced image analysis methods for automated segmentation of subnuclear chromatin domains. Epigenomes 2022, 6, 34. [Google Scholar] [CrossRef]
- Ning, Y.; Shang, D.; Xin, H.; Ni, R.; Wang, Z.; Zhen, Y.; Liu, G.; Xi, M. Establishing of 3D-FISH on frozen section and its applying in chromosome territories analysis in Populus trichocarpa. Plant Cell Rep. 2024, 43, 255. [Google Scholar] [CrossRef]
- Hu, B.; Wang, N.; Bi, X.; Karaaslan, E.S.; Weber, A.L.; Zhu, W.; Berendzen, K.W.; Liu, C. Plant lamin-like proteins mediate chromatin tethering at the nuclear periphery. Genome Biol. 2019, 20, 87. [Google Scholar] [CrossRef]
- Schubert, V.; Lermontova, I.; Schubert, I. The Arabidopsis CAP-D proteins are required for correct chromatin organisation, growth and fertility. Chromosoma 2013, 122, 517–533. [Google Scholar] [CrossRef]
- Shan, W.; Kubova, M.; Mandakova, T.; Lysak, M.A. Nuclear organization in crucifer genomes: Nucleolus-associated telomere clustering is not a universal interphase configuration in Brassicaceae. Plant J. 2021, 108, 528–540. [Google Scholar] [CrossRef]
- Nemeckova, A.; Kolackova, V.; Vrana, J.; Dolezel, J.; Hribova, E. DNA replication and chromosome positioning throughout the interphase in three-dimensional space of plant nuclei. J. Exp. Bot. 2020, 71, 6262–6272. [Google Scholar] [CrossRef]
- Zuo, S.; Mandakova, T.; Kubova, M.; Lysak, M.A. Genomes, repeatomes and interphase chromosome organization in the meadowfoam family (Limnanthaceae, Brassicales). Plant J. 2022, 110, 1462–1475. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Jiang, J. Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 1998, 6, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Robaszkiewicz, E.; Idziak-Helmcke, D.; Tkacz, M.A.; Chrominski, K.; Hasterok, R. The arrangement of Brachypodium distachyon chromosomes in interphase nuclei. J. Exp. Bot. 2016, 67, 5571–5583. [Google Scholar] [CrossRef]
- Nowicka, A.; Ferkova, L.; Said, M.; Kovacik, M.; Zwyrtkova, J.; Baroux, C.; Pecinka, A. Non-Rabl chromosome organization in endoreduplicated nuclei of barley embryo and endosperm tissues. J. Exp. Bot. 2023, 74, 2527–2541. [Google Scholar] [CrossRef]
- Kubalova, I.; Camara, A.S.; Capal, P.; Beseda, T.; Rouillard, J.M.; Krause, G.M.; Holusova, K.; Toegelova, H.; Himmelbach, A.; Stein, N.; et al. Helical coiling of metaphase chromatids. Nucleic Acids Res. 2023, 51, 2641–2654. [Google Scholar] [CrossRef]
- Hayashida, M.; Sartsanga, C.; Phengchat, R.; Malac, M.; Harada, K.; Akashi, T.; Fukui, K.; Ohmido, N. Higher-order structure of barley chromosomes observed by electron tomography. Micron 2022, 160, 103328. [Google Scholar] [CrossRef]
- Siregar, A.Y.; Sartsanga, C.; Arifudin, F.S.; Phengchat, R.; Salamah, A.; Ohmido, N.; Fukui, K.; Dwiranti, A. Calcium ion significance on the maintenance of barley (Hordeum vulgare) chromosome compaction. Micron 2021, 145, 103046. [Google Scholar] [CrossRef]
- Kubalova, I.; Nemeckova, A.; Weisshart, K.; Hribova, E.; Schubert, V. Comparing super-resolution microscopy techniques to analyze chromosomes. Int. J. Mol. Sci. 2021, 22, 1903. [Google Scholar] [CrossRef]
- Kubalova, I.; Weisshart, K.; Houben, A.; Schubert, V. Super-resolution microscopy reveals the number and distribution of topoisomerase IIalpha and CENH3 molecules within barley metaphase chromosomes. Chromosoma 2023, 132, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Sartsanga, C.; Phengchat, R.; Wako, T.; Fukui, K.; Ohmido, N. Localization and quantitative distribution of a chromatin structural protein Topoisomerase II on plant chromosome using HVTEM and UHVTEM. Micron 2024, 179, 103596. [Google Scholar] [CrossRef] [PubMed]
- Ohmido, N.; Sartsanga, C.; Dwiranti, A. Structure and compaction of plant chromosomes: Studies using advanced electron microscopy. Micron 2025, 196–197, 103860. [Google Scholar] [CrossRef]
- Kaduchova, K.; Marchetti, C.; Ovecka, M.; Galuszka, P.; Bergougnoux, V.; Samaj, J.; Pecinka, A. Spatial organization and dynamics of chromosomes and microtubules during barley mitosis. Plant J. 2023, 115, 602–613. [Google Scholar] [CrossRef]
- Kuo, Y.T.; Schubert, V.; Marques, A.; Schubert, I.; Houben, A. Centromere diversity: How different repeat-based holocentromeres may have evolved. Bioessays 2024, 46, e2400013. [Google Scholar] [CrossRef] [PubMed]
- Naish, M.; Henderson, I.R. The structure, function, and evolution of plant centromeres. Genome Res. 2024, 34, 161–178. [Google Scholar] [CrossRef] [PubMed]
- Schubert, V.; Neumann, P.; Marques, A.; Heckmann, S.; Macas, J.; Pedrosa-Harand, A.; Schubert, I.; Jang, T.S.; Houben, A. Super-resolution microscopy reveals diversity of plant centromere architecture. Int. J. Mol. Sci. 2020, 21, 3488. [Google Scholar] [CrossRef] [PubMed]
- Plackova, K.; Zedek, F.; Schubert, V.; Houben, A.; Bures, P. Kinetochore size scales with chromosome size in bimodal karyotypes of Agavoideae. Ann. Bot. 2022, 130, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.T.; Camara, A.S.; Schubert, V.; Neumann, P.; Macas, J.; Melzer, M.; Chen, J.; Fuchs, J.; Abel, S.; Klocke, E.; et al. Holocentromeres can consist of merely a few megabase-sized satellite arrays. Nat. Commun. 2023, 14, 3502. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Oliveira, L.; Jang, T.S.; Novak, P.; Koblizkova, A.; Schubert, V.; Houben, A.; Macas, J. Disruption of the standard kinetochore in holocentric Cuscuta species. Proc. Natl. Acad. Sci. USA 2023, 120, e2300877120. [Google Scholar] [CrossRef]
- Ding, W.; Zhu, Y.; Han, J.; Zhang, H.; Xu, Z.; Khurshid, H.; Liu, F.; Hasterok, R.; Shen, X.; Wang, K. Characterization of centromeric DNA of Gossypium anomalum reveals sequence-independent enrichment dynamics of centromeric repeats. Chromosome Res. 2023, 31, 12. [Google Scholar] [CrossRef]
- Oliveira, L.; Neumann, P.; Mata-Sucre, Y.; Kuo, Y.T.; Marques, A.; Schubert, V.; Macas, J. KNL1 and NDC80 represent new universal markers for the detection of functional centromeres in plants. Chromosome Res. 2024, 32, 3. [Google Scholar] [CrossRef]
- Melters, D.P.; Paliulis, L.V.; Korf, I.F.; Chan, S.W. Holocentric chromosomes: Convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012, 20, 579–593. [Google Scholar] [CrossRef]
- Mata-Sucre, Y.; Kratka, M.; Oliveira, L.; Neumann, P.; Macas, J.; Schubert, V.; Huettel, B.; Kejnovsky, E.; Houben, A.; Pedrosa-Harand, A.; et al. Repeat-based holocentromeres of the woodrush Luzula sylvatica reveal insights into the evolutionary transition to holocentricity. Nat. Commun. 2024, 15, 9565. [Google Scholar] [CrossRef] [PubMed]
- Alisawi, O.; Richert-Poggeler, K.R.; Heslop-Harrison, J.S.P.; Schwarzacher, T. The nature and organization of satellite DNAs in Petunia hybrida, related, and ancestral genomes. Front. Plant Sci. 2023, 14, 1232588. [Google Scholar] [CrossRef] [PubMed]
- Vozarova, R.; Herklotz, V.; Kovarik, A.; Tynkevich, Y.O.; Volkov, R.A.; Ritz, C.M.; Lunerova, J. Ancient origin of two 5S rDNA families dominating in the genus Rosa and their behavior in the Canina-type meiosis. Front. Plant Sci. 2021, 12, 643548. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; He, Z.; Liu, J.; Wu, H.; Gong, X. FISH mapping of telomeric and non-telomeric (AG3T3)3 reveal the chromosome numbers and chromosome rearrangements of 41 woody plants. Genes 2022, 13, 1239. [Google Scholar] [CrossRef]
- Liu, Y.; Liaw, Y.M.; Teo, C.H.; Capal, P.; Wada, N.; Fukui, K.; Dolezel, J.; Ohmido, N. Molecular organization of recombinant human-Arabidopsis chromosomes in hybrid cell lines. Sci. Rep. 2021, 11, 7160. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bartos, J.; Boudichevskaia, A.; Voigt, A.; Rabanus-Wallace, M.T.; Dreissig, S.; Tulpova, Z.; Simkova, H.; Macas, J.; Kim, G.; et al. The genetic mechanism of B chromosome drive in rye illuminated by chromosome-scale assembly. Nat. Commun. 2024, 15, 9686. [Google Scholar] [CrossRef]
- Karafiatova, M.; Bojdova, T.; Stejskalova, M.; Harnadkova, N.; Kumar, V.; Houben, A.; Chen, J.; Dolezalova, A.; Honys, D.; Bartos, J. Unravelling the unusual: Chromosome elimination, nondisjunction and extra pollen mitosis characterize the B chromosome in wild sorghum. New Phytol. 2024, 243, 1840–1854. [Google Scholar] [CrossRef]
- Shams, I.; Raskina, O. Supernumerary B chromosomes and plant genome changes: A snapshot of wild populations of Aegilops speltoides Tausch (Poaceae, Triticeae). Int. J. Mol. Sci. 2020, 21, 3768. [Google Scholar] [CrossRef]
- Chen, J.; Birchler, J.A.; Houben, A. The non-Mendelian behavior of plant B chromosomes. Chromosome Res. 2022, 30, 229–239. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, Y.; Yi, C.; Gao, Z.; Zhang, Z.; Zhu, C.; Birchler, J.A.; Han, F. Genome assembly of the maize B chromosome provides insight into its epigenetic characteristics and effects on the host genome. Genome Biol. 2025, 26, 47. [Google Scholar] [CrossRef]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Salse, J. Deciphering the evolutionary interplay between subgenomes following polyploidy: A paleogenomics approach in grasses. Am. J. Bot. 2016, 103, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J.S.P.; Schwarzacher, T.; Liu, Q. Polyploidy: Its consequences and enabling role in plant diversification and evolution. Ann. Bot. 2023, 131, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chester, M.; Leitch, A.R.; Soltis, P.S.; Soltis, D.E. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridisation). Genes 2010, 1, 166–192. [Google Scholar] [CrossRef]
- Han, J.; Zhou, B.; Shan, W.; Yu, L.; Wu, W.; Wang, K. A and D genomes spatial separation at somatic metaphase in tetraploid cotton: Evidence for genomic disposition in a polyploid plant. Plant J. 2015, 84, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Hasterok, R.; Draper, J.; Jenkins, G. Laying the cytotaxonomic foundations of a new model grass, Brachypodium distachyon (L.) Beauv. Chromosome Res. 2004, 12, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Chrtek, J.; Mraz, P.; Belyayev, A.; Pastova, L.; Mrazova, V.; Caklova, P.; Josefiova, J.; Zagorski, D.; Hartmann, M.; Jandova, M.; et al. Evolutionary history and genetic diversity of apomictic allopolyploids in Hieracium s.str.: Morphological versus genomic features. Am. J. Bot. 2020, 107, 66–90. [Google Scholar] [CrossRef] [PubMed]
- Mandáková, T.; Lysak, M.A. The identification of the missing maternal genome of the allohexaploid camelina (Camelina sativa). Plant J. 2022, 112, 622–629. [Google Scholar] [CrossRef]
- Köhler, M.; Oakley, L.J.; Font, F.; Peñas, M.L.L.; Majure, L.C. On the continuum of evolution: A putative new hybrid speciation event in Opuntia (Cactaceae) between a native and an introduced species in southern South America. Syst. Biodivers. 2021, 19, 1026–1039. [Google Scholar] [CrossRef]
- Shimomai, H.; Taichi, N.; Katsuhara, K.R.; Kato, S.; Ushimaru, A.; Ohmido, N. Allopolyploidy enhances survival advantages for urban environments in the native plant genus Commelina. Ann. Bot. 2024, 134, 1055–1066. [Google Scholar] [CrossRef]
- Amosova, A.V.; Zemtsova, L.V.; Yurkevich, O.Y.; Zhidkova, E.N.; Ksiazczyk, T.; Shostak, N.G.; Muravlev, A.A.; Artemyeva, A.M.; Samatadze, T.E.; Zoshchuk, S.A.; et al. Genomic changes in generations of synthetic rapeseed-like allopolyploid grown under selection. Euphytica 2017, 213, 217. [Google Scholar] [CrossRef]
- Kruppa, K.; Turkosi, E.; Mayer, M.; Toth, V.; Vida, G.; Szakacs, E.; Molnar-Lang, M. McGISH identification and phenotypic description of leaf rust and yellow rust resistant partial amphiploids originating from a wheat x Thinopyrum synthetic hybrid cross. J. Appl. Genet. 2016, 57, 427–437. [Google Scholar] [CrossRef]
- Song, Z.P.; Zuo, Y.Y.; Li, W.J.; Dai, S.F.; Liu, G.; Pu, Z.J.; Yan, Z.H. Chromosome stability of synthetic Triticum turgidum–Aegilops umbellulata hybrids. BMC Plant Biol. 2024, 24, 391. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Shi, S.D.; Shen, H.T.; Xie, Q.L.; Li, H.B. Haplotype-specific chromosome painting provides insights into the chromosomal characteristics in self-duplicating autotetraploid sugarcane. Ind. Crops Prod. 2023, 202, 117085. [Google Scholar] [CrossRef]
- Zhang, J.; Fan, C.; Liu, Y.; Shi, Q.; Sun, Y.; Huang, Y.; Yuan, J.; Han, F. Cytological analysis of the diploid-like inheritance of newly synthesized allotetraploid wheat. Chromosome Res. 2023, 32, 1. [Google Scholar] [CrossRef] [PubMed]
- Borowska-Zuchowska, N.; Robaszkiewicz, E.; Mykhailyk, S.; Wartini, J.; Pinski, A.; Kovarik, A.; Hasterok, R. To be or not to be expressed: The first evidence of a nucleolar dominance tissue-specificity in Brachypodium hybridum. Front. Plant Sci. 2021, 12, 768347. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Kovarik, A.; Robaszkiewicz, E.; Tuna, M.; Tuna, G.S.; Gordon, S.; Vogel, J.P.; Hasterok, R. The fate of 35S rRNA genes in the allotetraploid grass Brachypodium hybridum. Plant J. 2020, 103, 1810–1825. [Google Scholar] [CrossRef]
- Trunova, D.; Borowska-Zuchowska, N.; Mykhailyk, S.; Xia, K.; Zhu, Y.; Sancho, R.; Rojek-Jelonek, M.; Garcia, S.; Wang, K.; Catalan, P.; et al. Does time matter? Intraspecific diversity of ribosomal RNA genes in lineages of the allopolyploid model grass Brachypodium hybridum with different evolutionary ages. BMC Plant Biol. 2024, 24, 981. [Google Scholar] [CrossRef]
- Mahelka, V.; Kopecky, D.; Majka, J.; Krak, K. Uniparental expression of ribosomal RNA in xFestulolium grasses: A link between the genome and nucleolar dominance. Front. Plant Sci. 2023, 14, 1276252. [Google Scholar] [CrossRef]
- Matyasek, R.; Kalfusova, R.; Kuderova, A.; Rehurkova, K.; Sochorova, J.; Kovarik, A. Transcriptional silencing of 35S rDNA in Tragopogon porrifolius correlates with cytosine methylation in sequence-specific manner. Int. J. Mol. Sci. 2024, 25, 7540. [Google Scholar] [CrossRef]
- Borowska-Zuchowska, N.; Mykhailyk, S.; Robaszkiewicz, E.; Matysiak, N.; Mielanczyk, L.; Wojnicz, R.; Kovarik, A.; Hasterok, R. Switch them off or not: Selective rRNA gene repression in grasses. Trends Plant Sci. 2023, 28, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Idziak, D.; Hasterok, R. Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 2008, 51, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Mandakova, T.; Krumpolcova, A.; Matyasek, R.; Volkov, R.; Lysak, M.A.; Kovarik, A. Uniparental silencing of 5S rRNA genes in plant allopolyploids—Insights from Cardamine (Brassicaceae). Plant J. 2024, 119, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Gramazio, P.; Prohens, J.; Toppino, L.; Plazas, M. Editorial: Introgression breeding in cultivated plants. Front. Plant Sci. 2021, 12, 764533. [Google Scholar] [CrossRef]
- Kashyap, A.; Garg, P.; Tanwar, K.; Sharma, J.; Gupta, N.C.; Ha, P.T.T.; Bhattacharya, R.C.; Mason, A.S.; Rao, M. Strategies for utilization of crop wild relatives in plant breeding programs. Theor. Appl. Genet. 2022, 135, 4151–4167. [Google Scholar] [CrossRef] [PubMed]
- Kwiatek, M.T.; Nawracala, J. Chromosome manipulations for progress of triticale (×Triticosecale) breeding. Plant Breed. 2018, 137, 823–831. [Google Scholar] [CrossRef]
- Rustgi, S. Plant cytogenetics blurring disciplinary boundaries to sustain global food security. Nucleus 2023, 66, 239–243. [Google Scholar] [CrossRef]
- Kumar, R.; Das, S.P.; Choudhury, B.U.; Kumar, A.; Prakash, N.R.; Verma, R.; Chakraborti, M.; Devi, A.G.; Bhattacharjee, B.; Das, R.; et al. Advances in genomic tools for plant breeding: Harnessing DNA molecular markers, genomic selection, and genome editing. Biol. Res. 2024, 57, 80. [Google Scholar] [CrossRef]
- Salina, E.A.; Adonina, I.G. Cytogenetics in the dtudy of chromosomal rearrangement during wheat evolution and breeding. In Cytogenetics—Past, Present and Further Perspectives; Larramendy, M.L., Soloneski, S., Eds.; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar]
- Kwiatek, M.T.; Belter, J.; Ulaszewski, W.; Skowronska, R.; Noweiska, A.; Wisniewska, H. Molecular identification of triticale introgression lines carrying leaf rust resistance genes transferred from Aegilops kotschyi Boiss. and Ae. tauschii Coss. J. Appl. Genet. 2021, 62, 431–439. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, Q.; Fan, T.; Zhao, L.; Ren, Z.; Tan, F.; Luo, P.; Ren, T. Molecular cytogenetic and physiological characterization of a novel wheat-rye T1RS.1BL translocation line from Secale cereal L. Weining with resistance to stripe rust and functional “Stay Green” trait. Int. J. Mol. Sci. 2022, 23, 4626. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ren, Z.; Tan, F.; Luo, P.; Ren, T. Molecular cytogenetic characterization of novel 1RS.1BL translocation and complex chromosome translocation lines with stripe rust resistance. Int. J. Mol. Sci. 2022, 23, 2731. [Google Scholar] [CrossRef] [PubMed]
- Jamwal, N.S.; Badiyal, A.; Chaudhary, H.K.; Schwarzacher, T.; Heslop-Harrison, J.S. Molecular cytogenetic analysis of newly developed progenies from triticale x wheat crosses for yield and stress tolerance. Cereal Res. Commun. 2024, 52, 859–865. [Google Scholar] [CrossRef]
- Li, Q.F.; Lu, Y.Q.; Pan, C.L.; Wang, Z.J.; Liu, F.L.; Zhang, J.P.; Yang, X.M.; Li, X.Q.; Liu, W.H.; Li, L.H. Characterization, identification and evaluation of a novel wheat-Agropyron cristatum (L.) Gaertn. disomic addition line II-30-5. Genet. Resour. Crop Evol. 2020, 67, 2213–2223. [Google Scholar] [CrossRef]
- Song, L.; Zhao, H.; Zhang, Z.; Zhang, S.; Liu, J.; Zhang, W.; Zhang, N.; Ji, J.; Li, L.; Li, J. Molecular cytogenetic identification of wheat-Aegilops biuncialis 5Mb disomic addition line with tenacious and black glumes. Int. J. Mol. Sci. 2020, 21, 4053. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Yang, X.; Wang, C.; Zhang, H.; Deng, P.; Liu, X.; Chen, C.; Ji, W.; Wang, Y. Molecular cytogenetics for a wheat-Aegilops geniculata 3Mg alien addition line with resistance to stripe rust and powdery mildew. BMC Plant Biol. 2021, 21, 575. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, J.; Xiao, Y.; Feng, X.; Zhang, H.; Chen, C.; Ji, W.; Wang, Y. Genetic analysis of resistance to powdery mildew on 7Mg chromosome of wheat-Aegilops geniculata, development and utilization of specific molecular markers. BMC Plant Biol. 2022, 22, 564. [Google Scholar] [CrossRef]
- Motsnyi, I.I.; Halaiev, O.V.; Alieksieieva, T.G.; Chebotar, G.O.; Chebotar, S.V.; Betekhtin, A.; Hasterok, R.; Armoniene, R.; Rahmatov, M. Cytogenetic and molecular identification of novel wheat-Elymus sibiricus addition lines with resistance to leaf rust and the presence of leaf pubescence trait. Front. Plant Sci. 2024, 15, 1482211. [Google Scholar] [CrossRef]
- Feng, X.; Du, X.; Wang, S.; Deng, P.; Wang, Y.; Shang, L.; Tian, Z.; Wang, C.; Chen, C.; Zhao, J.; et al. Identification and DNA marker development for a wheat-Leymus mollis 2Ns (2D) disomic chromosome substitution. Int. J. Mol. Sci. 2022, 23, 2676. [Google Scholar] [CrossRef]
- Hou, C.; Han, J.; Zhang, L.; Geng, Q.; Zhao, L.; Liu, S.; Yang, Q.; Chen, X.; Wu, J. Identification of resistance to Fusarium head blight and molecular cytogenetics of interspecific derivatives between wheat and Psathyrostachys huashanica. Breed. Sci. 2022, 72, 213–221. [Google Scholar] [CrossRef]
- Pang, J.; Huang, C.; Wang, Y.; Wen, X.; Deng, P.; Li, T.; Wang, C.; Liu, X.; Chen, C.; Zhao, J.; et al. Molecular cytological analysis and specific marker development in wheat-Psathyrostachys huashanica Keng 3Ns additional line with elongated glume. Int. J. Mol. Sci. 2023, 24, 6726. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, S.; Shi, Y.; Zhang, S.; Yan, W.; Song, W.; Yang, X.; Song, Q.; Jang, B.; Qi, X.; et al. Molecular cytogenetic characterization and fusarium head blight resistance of five wheat-Thinopyrum intermedium partial amphiploids. Mol. Cytogenet. 2021, 14, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, C.; Feng, X.; Zhao, J.; Deng, P.; Wang, Y.; Zhang, H.; Liu, X.; Li, T.; Chen, C.; et al. Molecular cytogenetics and development of St-chromosome-specific molecular markers of novel stripe rust resistant wheat-Thinopyrum intermedium and wheat-Thinopyrum ponticum substitution lines. BMC Plant Biol. 2022, 22, 111. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Gupta, M.; Atri, C.; Akhatar, J.; Kumar, S.; Heslop-Harrison, P.; Banga, S.S. Anchoring alien chromosome segment substitutions bearing gene(s) for resistance to mustard aphid in Brassica juncea-B. fruticulosa introgression lines and their possible disruption through gamma irradiation. Theor. Appl. Genet. 2021, 134, 3209–3224. [Google Scholar] [CrossRef] [PubMed]
- Ankrah, N.A.; El-nagish, A.; Breitenbach, S.; Tetteh, A.Y.; Heitkam, T. Comparative cytogenetics of kenaf (Hibiscus cannabinus L.) breeding lines reveal chromosomal variability and instability. Genet. Resour. Crop Evol. 2024, 72, 3423–3436. [Google Scholar] [CrossRef]
- Savas-Tuna, G.; Yücel, G.; Asçiogul, T.K.; Ates, D.; Esiyok, D.; Tanyolaç, M.B.; Tuna, M. Molecular cytogenetic characterization of common bean (Phaseolus vulgaris L.) accessions. Turk. J. Agric. For. 2020, 44, 612–630. [Google Scholar] [CrossRef]
- Kwiatek, M.T.; Drozdowska, Z.; Kurasiak-Popowska, D.; Noweiska, A.; Nawracala, J. Cytomolecular analysis of mutants, breeding lines, and varieties of camelina (Camelina sativa L. Crantz). J. Appl. Genet. 2021, 62, 199–205. [Google Scholar] [CrossRef]
- Park, H.W.; Sevilleno, S.S.; Ha, M.; Cabahug-Braza, R.A.; Yi, J.H.; Lim, K.B.; Cho, W.; Hwang, Y.J. The application of fluorescence in situ hybridization in the prescreening of Veronica hybrids. Plants 2024, 13, 1264. [Google Scholar] [CrossRef]
- Tomiczak, K. Molecular and cytogenetic description of somatic hybrids between Gentiana cruciata L. and G. tibetica King. J. Appl. Genet. 2020, 61, 13–24. [Google Scholar] [CrossRef]
- Zhou, M.; Yong, X.; Zhu, J.; Xu, Q.; Liu, X.; Zhang, L.; Mou, L.; Zeng, L.; Wu, M.; Jiang, B.; et al. Chromosomal analysis of progenies between Lilium intersectional hybrids and wild species using ND-FISH and GISH. Front. Plant Sci. 2024, 15, 1461798. [Google Scholar] [CrossRef]
- Xu, Z.; Chen, J.; Meng, S.; Xu, P.; Zhai, C.; Huang, F.; Guo, Q.; Zhao, L.; Quan, Y.; Shangguan, Y.; et al. Genome sequence of Gossypium anomalum facilitates interspecific introgression breeding. Plant Commun. 2022, 3, 100350. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhu, W.; Zhou, J.; Li, H.; Xu, X.; Zhang, B.; Gao, X. Repetitive DNA sequence detection and its role in the human genome. Commun. Biol. 2023, 6, 954. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef]
- Leitch, I.J.; Johnston, E.; Pellicer, J.; Hidalgo, O.; Bennett, M.D. Plant DNA C-Values Database (Release 7.1, Apr 2019). Available online: https://cvalues.science.kew.org/ (accessed on 15 July 2025).
- Belyayev, A.; Josefiova, J.; Jandova, M.; Kalendar, R.; Krak, K.; Mandak, B. Natural history of a aatellite DNA family: From the ancestral genome component to species-specific sequences, concerted and non-concerted evolution. Int. J. Mol. Sci. 2019, 20, 1201. [Google Scholar] [CrossRef]
- Novak, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef]
- Novak, P.; Neumann, P.; Macas, J. Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat. Protoc. 2020, 15, 3745–3776. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Avila Robledillo, L.; Koblizkova, A.; Vrbova, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Rouard, M.; Biswas, M.K.; Droc, G.; Cui, D.; Roux, N.; Baurens, F.C.; Ge, X.J.; Schwarzacher, T.; Heslop-Harrison, P.J.S.; et al. A chromosome-level reference genome of Ensete glaucum gives insight into diversity and chromosomal and repetitive sequence evolution in the Musaceae. Gigascience 2022, 11, giac027. [Google Scholar] [CrossRef] [PubMed]
- Dias, Y.; Mata-Sucre, Y.; Thangavel, G.; Costa, L.; Baez, M.; Houben, A.; Marques, A.; Pedrosa-Harand, A. How diverse a monocentric chromosome can be? Repeatome and centromeric organization of Juncus effusus (Juncaceae). Plant J. 2024, 118, 1832–1847. [Google Scholar] [CrossRef]
- Rathore, P.; Schwarzacher, T.; Heslop-Harrison, J.S.; Bhat, V.; Tomaszewska, P. The repetitive DNA sequence landscape and DNA methylation in chromosomes of an apomictic tropical forage grass, Cenchrus ciliaris. Front. Plant Sci. 2022, 13, 952968. [Google Scholar] [CrossRef]
- Ribeiro, T.; Vasconcelos, E.; Dos Santos, K.G.B.; Vaio, M.; Brasileiro-Vidal, A.C.; Pedrosa-Harand, A. Diversity of repetitive sequences within compact genomes of Phaseolus L. beans and allied genera Cajanus L. and Vigna Savi. Chromosome Res. 2020, 28, 139–153. [Google Scholar] [CrossRef]
- Ibiapino, A.; Baez, M.; Garcia, M.A.; Costea, M.; Stefanovic, S.; Pedrosa-Harand, A. Karyotype asymmetry in Cuscuta L. subgenus Pachystigma reflects its repeat DNA composition. Chromosome Res. 2022, 30, 91–107. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, S.; Taniguchi, S.; Schmidt, N.; Jost, M.; Wanke, S.; Heitkam, T.; Ohmido, N. Repeatome landscapes and cytogenetics of hortensias provide a framework to trace Hydrangea evolution and domestication. Ann. Bot. 2025, 135, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Mata-Sucre, Y.; Sader, M.; Van-Lume, B.; Gagnon, E.; Pedrosa-Harand, A.; Leitch, I.J.; Lewis, G.P.; Souza, G. How diverse is heterochromatin in the Caesalpinia group? Cytogenomic characterization of Erythrostemon hughesii Gagnon & G.P. Lewis (Leguminosae: Caesalpinioideae). Planta 2020, 252, 49. [Google Scholar] [CrossRef] [PubMed]
- Ferraz, M.E.; Ribeiro, T.; Sader, M.; Nascimento, T.; Pedrosa-Harand, A. Comparative analysis of repetitive DNA in dysploid and non-dysploid Phaseolus beans. Chromosome Res. 2023, 31, 30. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.T.; Ishii, T.; Fuchs, J.; Hsieh, W.H.; Houben, A.; Lin, Y.R. The evolutionary dynamics of repetitive DNA and its impact on the genome diversification in the genus Sorghum. Front. Plant Sci. 2021, 12, 729734. [Google Scholar] [CrossRef]
- Tomaszewska, P.; Vorontsova, M.S.; Renvoize, S.A.; Ficinski, S.Z.; Tohme, J.; Schwarzacher, T.; Castiblanco, V.; de Vega, J.J.; Mitchell, R.A.C.; Heslop-Harrison, J.S.P. Complex polyploid and hybrid species in an apomictic and sexual tropical forage grass group: Genomic composition and evolution in Urochloa (Brachiaria) species. Ann. Bot. 2023, 131, 87–108. [Google Scholar] [CrossRef]
- Tomaszewska, P.; Schwarzacher, T.; Heslop-Harrison, J.S.P. Oat chromosome and genome evolution defined by widespread terminal intergenomic translocations in polyploids. Front. Plant Sci. 2022, 13, 1026364. [Google Scholar] [CrossRef]
- Huang, Y.; Ding, W.; Zhang, M.; Han, J.; Jing, Y.; Yao, W.; Hasterok, R.; Wang, Z.; Wang, K. The formation and evolution of centromeric satellite repeats in Saccharum species. Plant J. 2021, 106, 616–629. [Google Scholar] [CrossRef]
- Xin, H.; Wang, Y.; Zhang, W.; Bao, Y.; Neumann, P.; Ning, Y.; Zhang, T.; Wu, Y.; Jiang, N.; Jiang, J.; et al. Celine, a long interspersed nuclear element retrotransposon, colonizes in the centromeres of poplar chromosomes. Plant Physiol. 2024, 195, 2787–2798. [Google Scholar] [CrossRef]
- Galvez-Galvan, A.; Garrido-Ramos, M.A.; Prieto, P. Bread wheat satellitome: A complex scenario in a huge genome. Plant Mol. Biol. 2024, 114, 8. [Google Scholar] [CrossRef] [PubMed]
- Kwasniewska, J.; Bara, A.W. Plant cytogenetics in the micronuclei investigation-the past, current status, and perspectives. Int. J. Mol. Sci. 2022, 23, 1306. [Google Scholar] [CrossRef] [PubMed]
- Juchimiuk-Kwasniewska, J.; Brodziak, L.; Maluszynska, J. FISH in analysis of gamma ray-induced micronuclei formation in barley. J. Appl. Genet. 2011, 52, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Kus, A.; Kwasniewska, J.; Hasterok, R. Brachypodium distachyon—A useful model in the qualification of mutagen-induced micronuclei using multicolor FISH. PLoS ONE 2017, 12, e0170618. [Google Scholar] [CrossRef][Green Version]
- Tostes, N.V.; Ferreira, M.V.R.; Soares, F.A.F.; Silva, J.C.; Bhering, L.L.; Clarindo, W.R. DNA content, repeatome composition and origin of the Zea mays micronuclei. Sci. Rep. 2025, 15, 14997. [Google Scholar] [CrossRef]
- Kus, A.; Kwasniewska, J.; Szymanowska-Pulka, J.; Hasterok, R. Dissecting the chromosomal composition of mutagen-induced micronuclei in Brachypodium distachyon using multicolour FISH. Ann. Bot. 2018, 122, 1161–1171. [Google Scholar] [CrossRef]
- Kus, A.; Szymanowska-Pulka, J.; Kwasniewska, J.; Hasterok, R. Detecting Brachypodium distachyon chromosomes Bd4 and Bd5 in MH- and X-ray-induced micronuclei using mcFISH. Int. J. Mol. Sci. 2019, 20, 2848. [Google Scholar] [CrossRef]
- You, H.; Tang, D.; Liu, H.; Zhou, Y.; Li, Y.; Shen, Y.; Gong, Z.; Yu, H.; Gu, M.; Jiang, J.; et al. Chromosome ends initiate homologous chromosome pairing during rice meiosis. Plant Physiol. 2024, 195, 2617–2634. [Google Scholar] [CrossRef]
- Braz, G.T.; Yu, F.; Zhao, H.; Deng, Z.; Birchler, J.A.; Jiang, J. Preferential meiotic chromosome pairing among homologous chromosomes with cryptic sequence variation in tetraploid maize. New Phytol. 2021, 229, 3294–3302. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V.; Kimber, G. Genetic control of chromosome pairing in intergeneric hybrids with wheat. Nature 1959, 183, 1244–1246. [Google Scholar] [CrossRef]
- Higgins, E.E.; Howell, E.C.; Armstrong, S.J.; Parkin, I.A.P. A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus. New Phytol. 2021, 229, 3281–3293. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, A.R.; Dluzewska, J.; Szymanska-Lejman, M.; Desjardins, S.; Tock, A.J.; Kbiri, N.; Lambing, C.; Lawrence, E.J.; Bieluszewski, T.; Rowan, B.; et al. MSH2 shapes the meiotic crossover landscape in relation to interhomolog polymorphism in Arabidopsis. EMBO J. 2020, 39, e104858. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Fernandez-Jimenez, N.; Szymanska-Lejman, M.; Pele, A.; Underwood, C.J.; Serra, H.; Lambing, C.; Dluzewska, J.; Bieluszewski, T.; Pradillo, M.; et al. Natural variation identifies SNI1, the SMC5/6 component, as a modifier of meiotic crossover in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2021970118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Dluzewska, J.; Fernandez-Jimenez, N.; Ranjan, R.; Pele, A.; Dziegielewski, W.; Szymanska-Lejman, M.; Hus, K.; Gorna, J.; Pradillo, M.; et al. The kinase ATR controls meiotic crossover distribution at the genome scale in Arabidopsis. Plant Cell 2024, 37, koae292. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, E.; Doudna, J.A. Biotechnology: Rewriting a genome. Nature 2013, 495, 50–51. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Khosravi, S.; Ishii, T.; Dreissig, S.; Houben, A. Application and prospects of CRISPR/Cas9-based methods to trace defined genomic sequences in living and fixed plant cells. Chromosome Res. 2020, 28, 7–17. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.W.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Anton, T.; Bultmann, S.; Leonhardt, H.; Markaki, Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus 2014, 5, 163–172. [Google Scholar] [CrossRef]
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Gladilin, E.; Mette, M.F.; Puchta, H.; Houben, A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017, 91, 565–573. [Google Scholar] [CrossRef]
- Fujimoto, S.; Matsunaga, S. Visualization of chromatin loci with transiently expressed CRISPR/Cas9 in plants. Cytologia 2017, 82, 559–562. [Google Scholar] [CrossRef]
- Ishii, T.; Schubert, V.; Khosravi, S.; Dreissig, S.; Metje-Sprink, J.; Sprink, T.; Fuchs, J.; Meister, A.; Houben, A. RNA-guided endonuclease—In situ labelling (RGEN-ISL): A fast CRISPR/Cas9-based method to label genomic sequences in various species. New Phytol. 2019, 222, 1652–1661. [Google Scholar] [CrossRef] [PubMed]
- Beying, N.; Schmidt, C.; Pacher, M.; Houben, A.; Puchta, H. CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 2020, 6, 638–645. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Fransz, P.; Ronspies, M.; Dreissig, S.; Fuchs, J.; Heckmann, S.; Houben, A.; Puchta, H. Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat. Commun. 2020, 11, 4418. [Google Scholar] [CrossRef] [PubMed]
- Ronspies, M.; Schmidt, C.; Schindele, P.; Lieberman-Lazarovich, M.; Houben, A.; Puchta, H. Massive crossover suppression by CRISPR-Cas-mediated plant chromosome engineering. Nat. Plants 2022, 8, 1153–1159. [Google Scholar] [CrossRef]
- Puchta, H.; Houben, A. Plant chromosome engineering—Past, present and future. New Phytol. 2024, 241, 541–552. [Google Scholar] [CrossRef]
- Helia, O.; Matusova, B.; Havlova, K.; Hyskova, A.; Lycka, M.; Beying, N.; Puchta, H.; Fajkus, J.; Fojtova, M. Chromosome engineering points to the cis-acting mechanism of chromosome arm-specific telomere length setting and robustness of plant phenotype, chromatin structure and gene expression. Plant J. 2025, 121, e70024. [Google Scholar] [CrossRef]
- Khosravi, S.; Hinrichs, R.; Ronspies, M.; Haghi, R.; Puchta, H.; Houben, A. Epigenetic state and gene expression remain stable after CRISPR/Cas-mediated chromosomal inversions. New Phytol. 2025, 245, 2527–2539. [Google Scholar] [CrossRef]
- Revolutionizing Cytogenetics with AI. Available online: https://www.hnl.com/resources/blog/revolutionizing-cytogenetics-with-ai (accessed on 15 July 2025).
- Rosenblum, L.S.; Holmes, J.; Taghiyev, A.F. The emergence of artificial intelligence-guided karyotyping: A review and reflection. Genes 2025, 16, 685. [Google Scholar] [CrossRef]
- Nakayama, S.; Fukui, K. Quantitative chromosome mapping of small plant chromosomes by improved imaging on CHIAS II. Genes Genet. Syst. 1997, 72, 35–40. [Google Scholar] [CrossRef]
- Kato, S.; Fukui, K. Condensation pattern (CP) analysis of plant chromosomes by an improved chromosome image analysing system, CHIAS III. Chromosome Res. 1998, 6, 473–479. [Google Scholar] [CrossRef]
- Kato, S.; Ohmido, N.; Hara, M.; Kataoka, R.; Fukui, K. Image analysis of small plant chromosomes by using an improved system, CHIAS IV. Chromosome Sci. 2009, 12, 43–50. [Google Scholar]
- Fukui, K. Standardization of karyotyping plant chromosomes by a newly developed chromosome image analyzing system (CHIAS). Theor. Appl. Genet. 1986, 72, 27–32. [Google Scholar] [CrossRef]
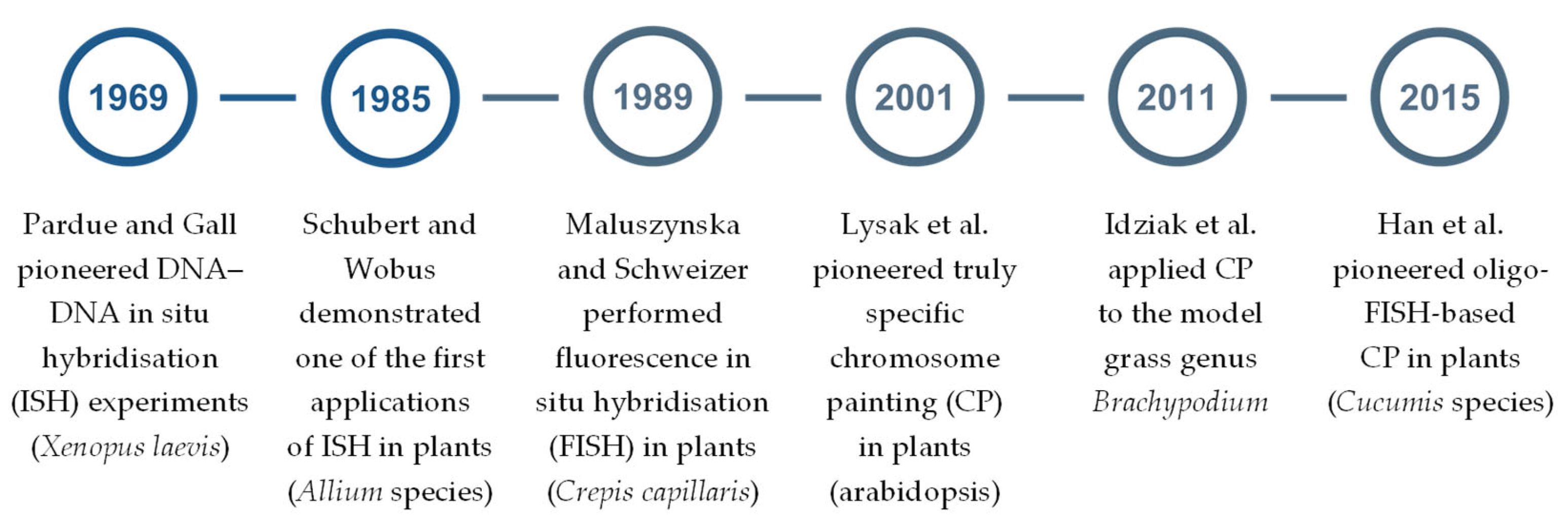

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wolny, E.; Mur, L.A.J.; Ohmido, N.; Yin, Z.; Wang, K.; Hasterok, R. Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century. Int. J. Mol. Sci. 2025, 26, 7013. https://doi.org/10.3390/ijms26147013
Wolny E, Mur LAJ, Ohmido N, Yin Z, Wang K, Hasterok R. Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century. International Journal of Molecular Sciences. 2025; 26(14):7013. https://doi.org/10.3390/ijms26147013
Chicago/Turabian StyleWolny, Elzbieta, Luis A. J. Mur, Nobuko Ohmido, Zujun Yin, Kai Wang, and Robert Hasterok. 2025. "Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century" International Journal of Molecular Sciences 26, no. 14: 7013. https://doi.org/10.3390/ijms26147013
APA StyleWolny, E., Mur, L. A. J., Ohmido, N., Yin, Z., Wang, K., & Hasterok, R. (2025). Thriving or Withering? Plant Molecular Cytogenetics in the First Quarter of the 21st Century. International Journal of Molecular Sciences, 26(14), 7013. https://doi.org/10.3390/ijms26147013








