Impact of Aerobic Training on Transcriptomic Changes in Skeletal Muscle of Rats with Cardiac Cachexia
Abstract
1. Introduction
2. Results
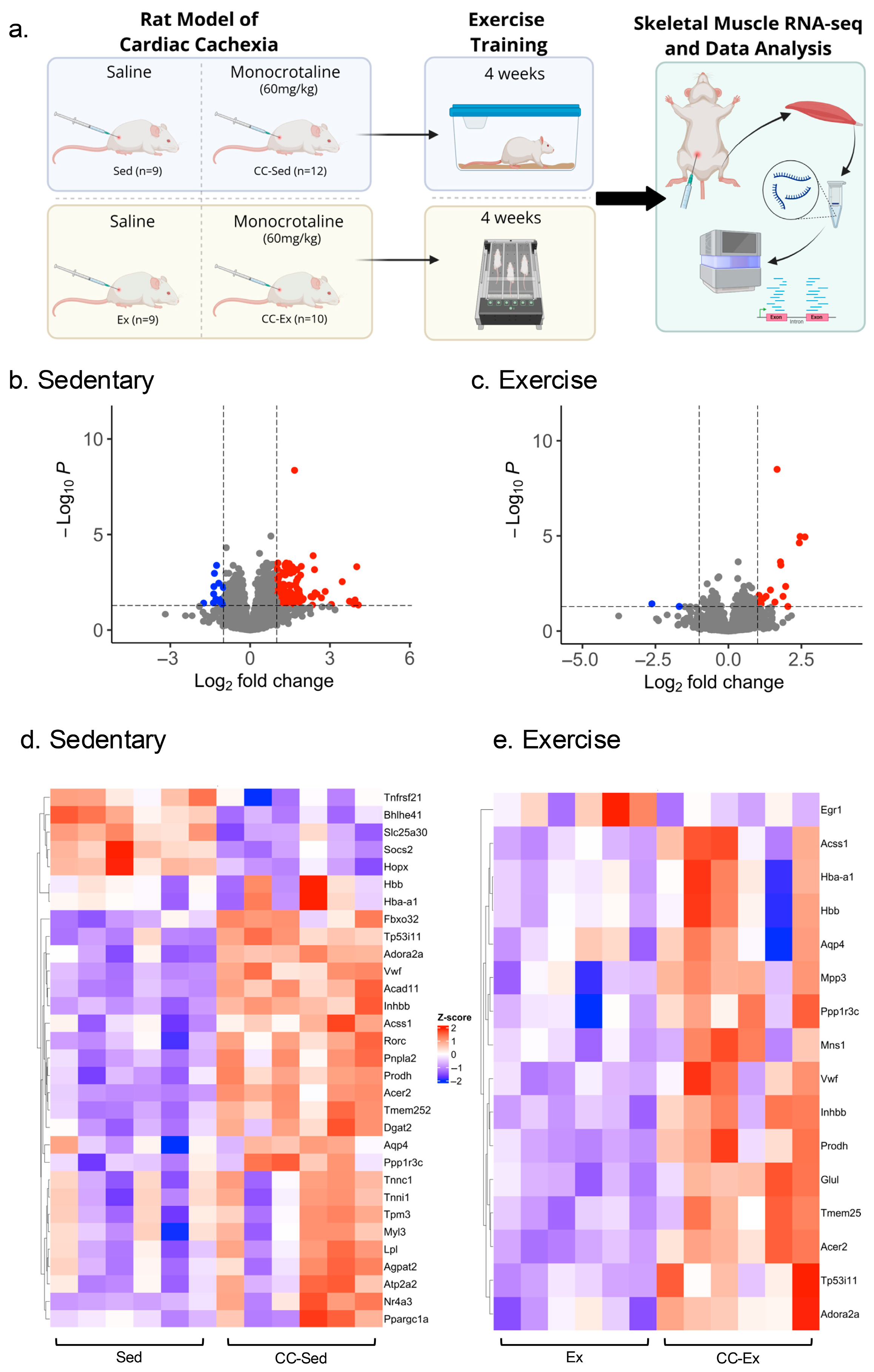
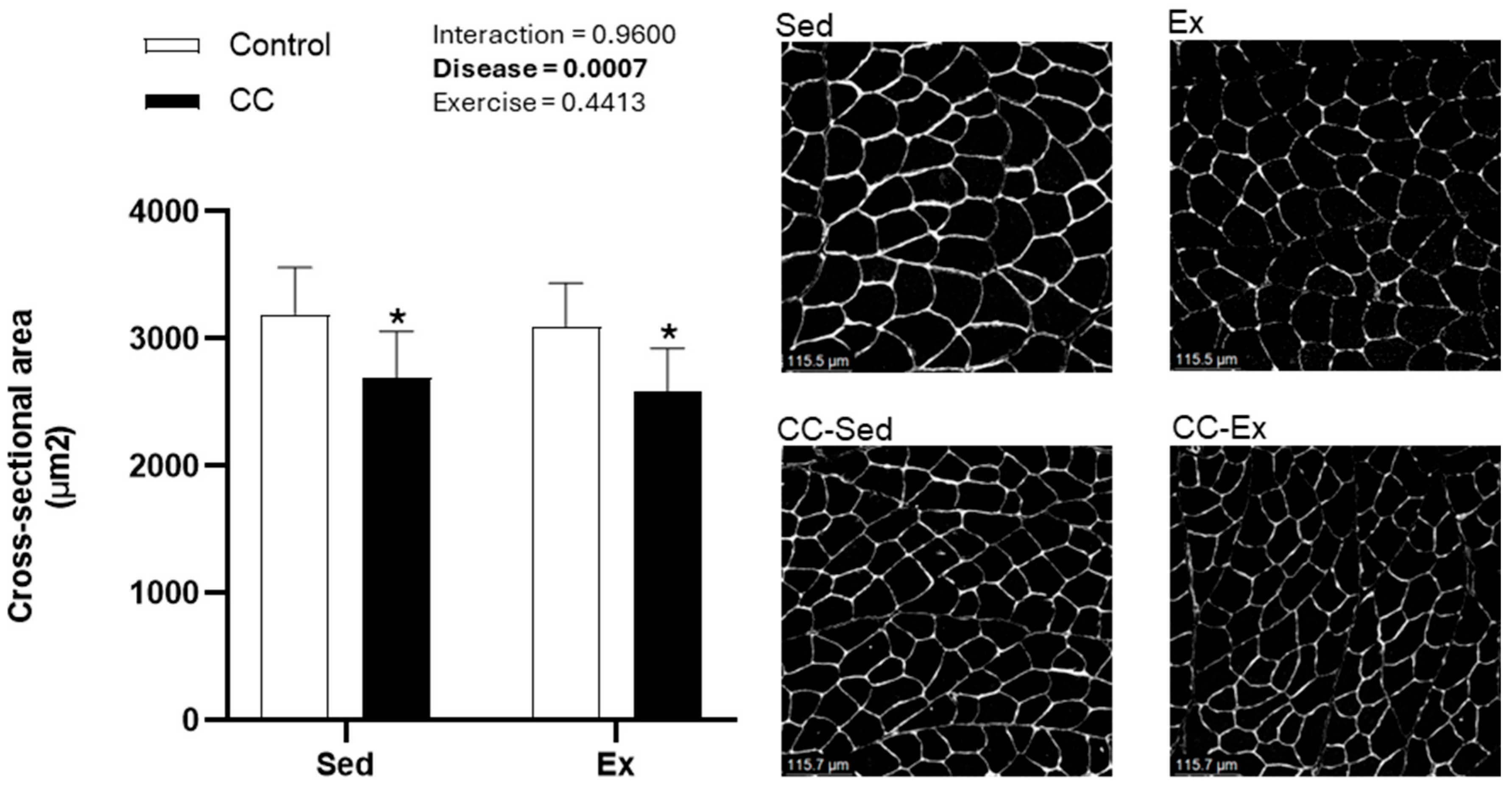
2.1. Characterization of Right Ventricle Remodeling and Cardiac Cachexia Indicators
2.2. Differential Gene Expression Analysis Identified a Larger Transcriptome Shift in the Sedentary CC Group
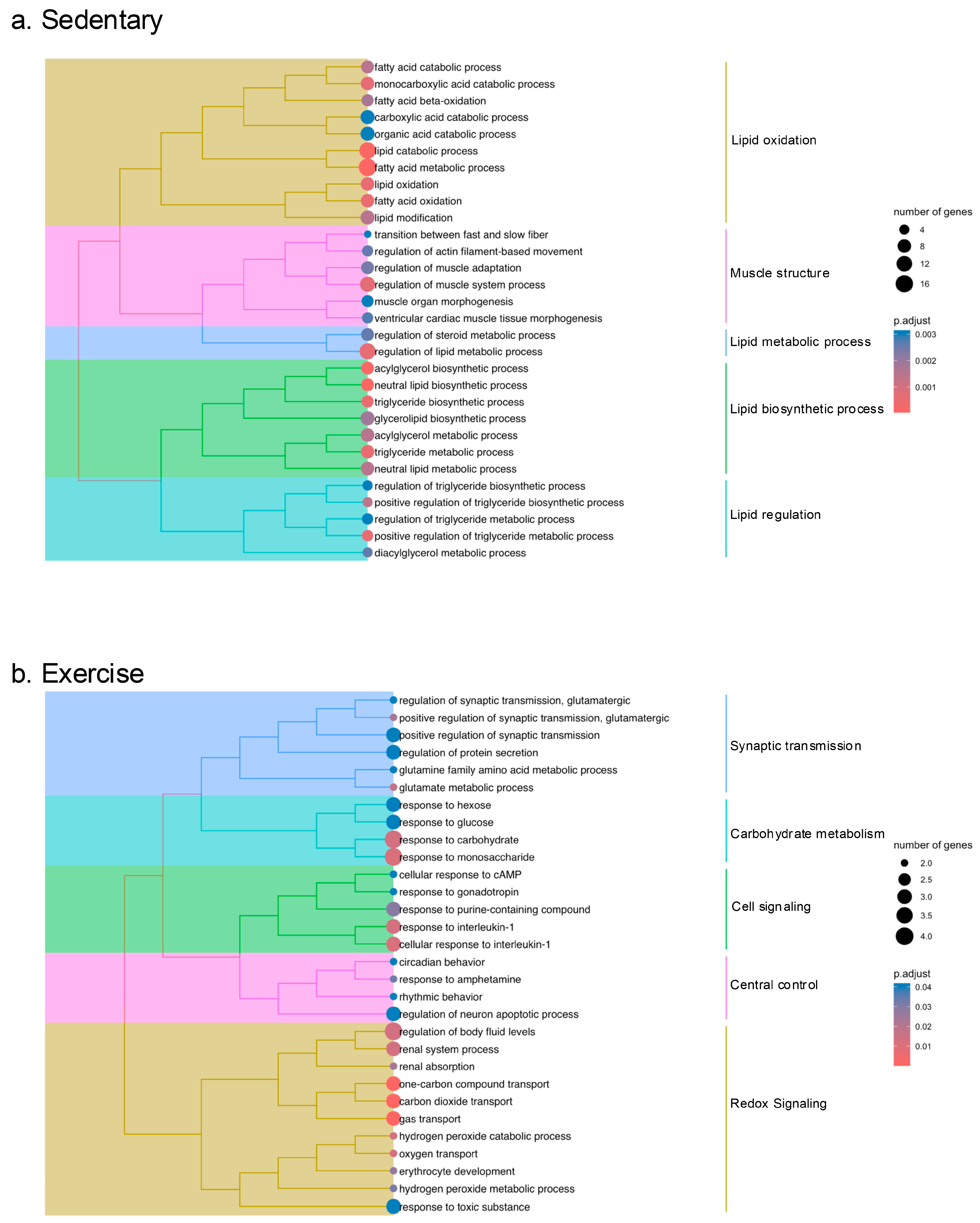
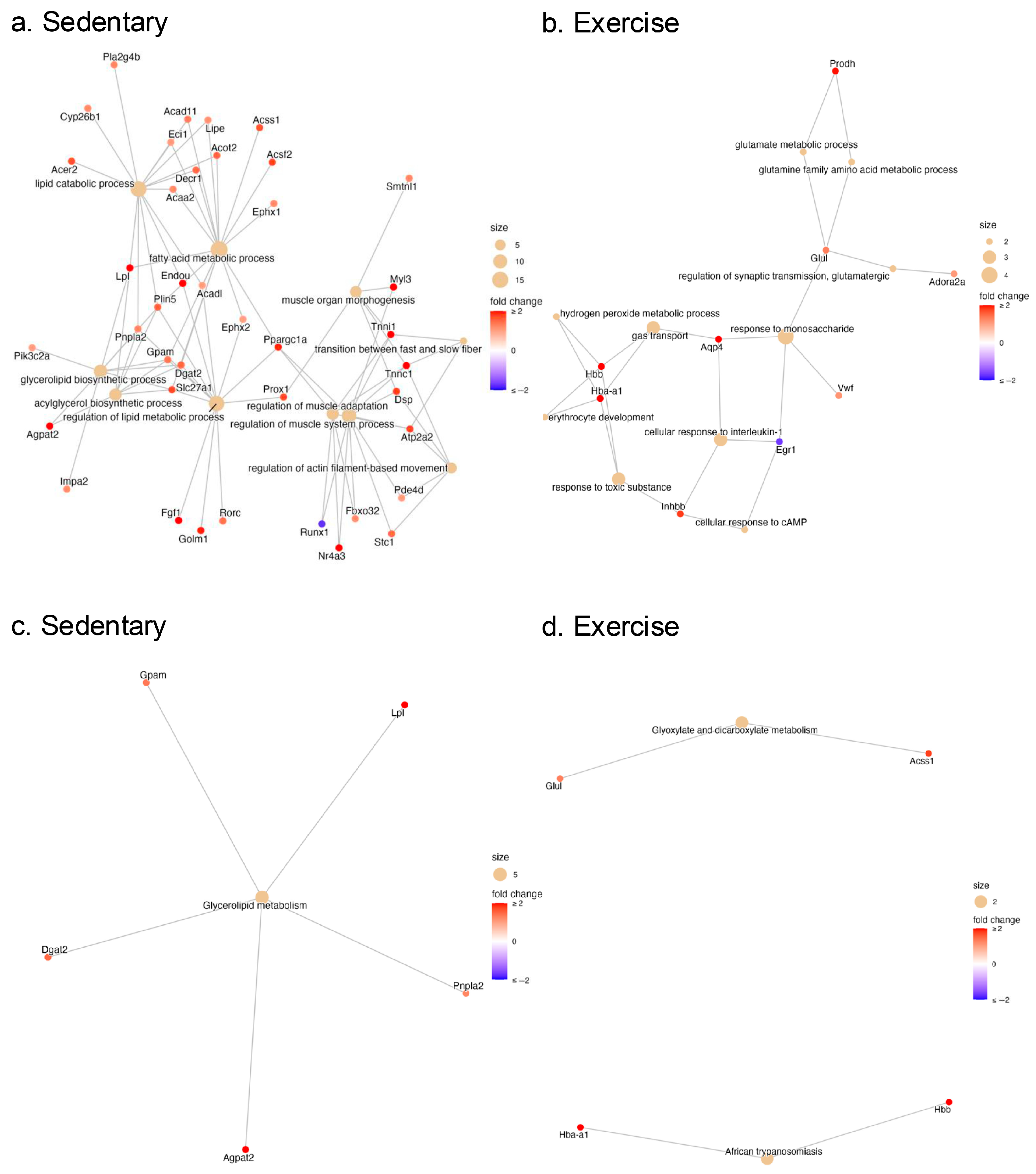
2.3. Pathway Enrichment Analysis of the DEGs in the Gastrocnemius Muscle of Sedentary and Exercised CC Rats
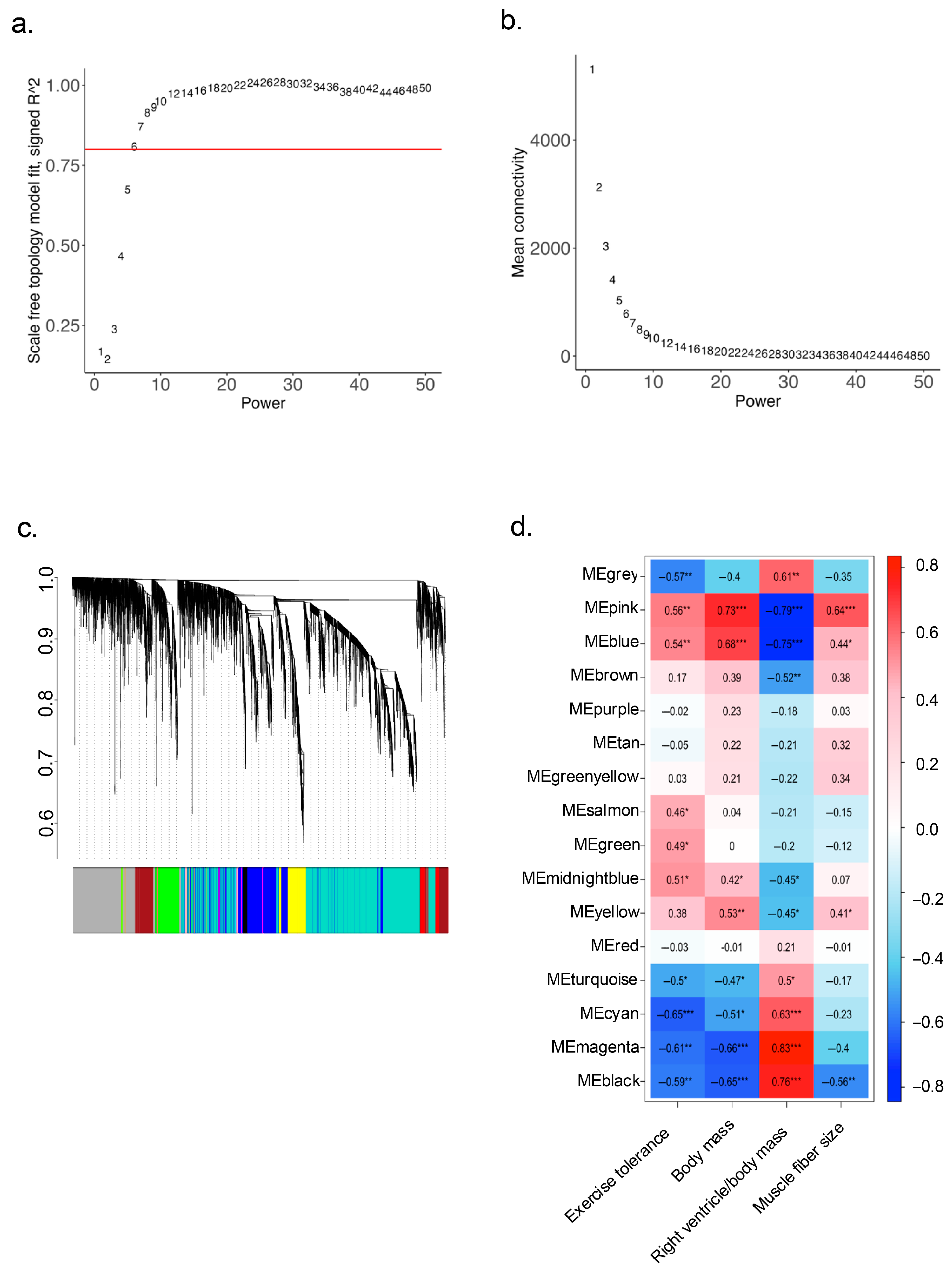
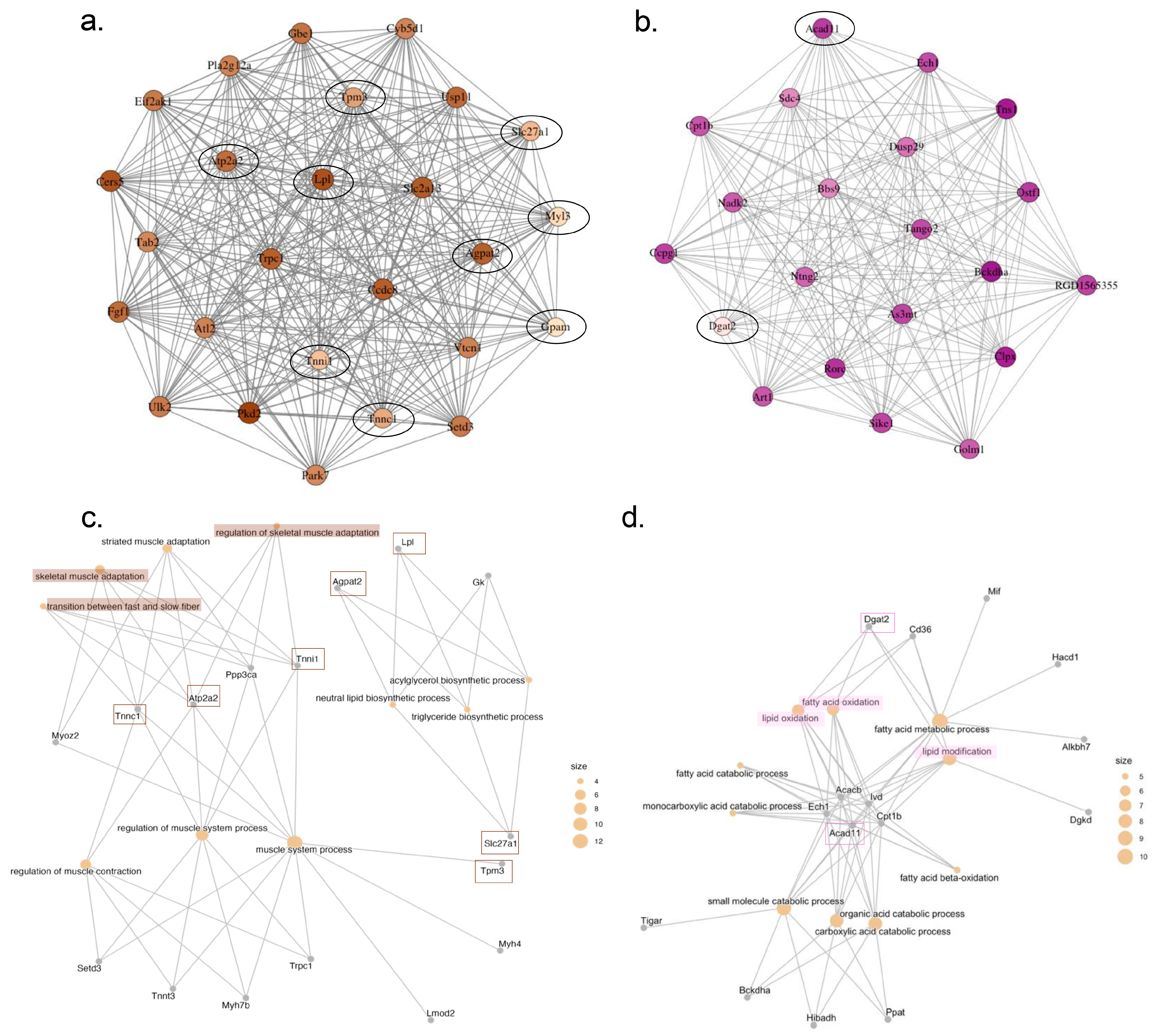
2.4. Identification of Key Regulatory Genes for Cardiac Cachexia by Weighted Gene Co-Expression Network Analysis (WGCNA)
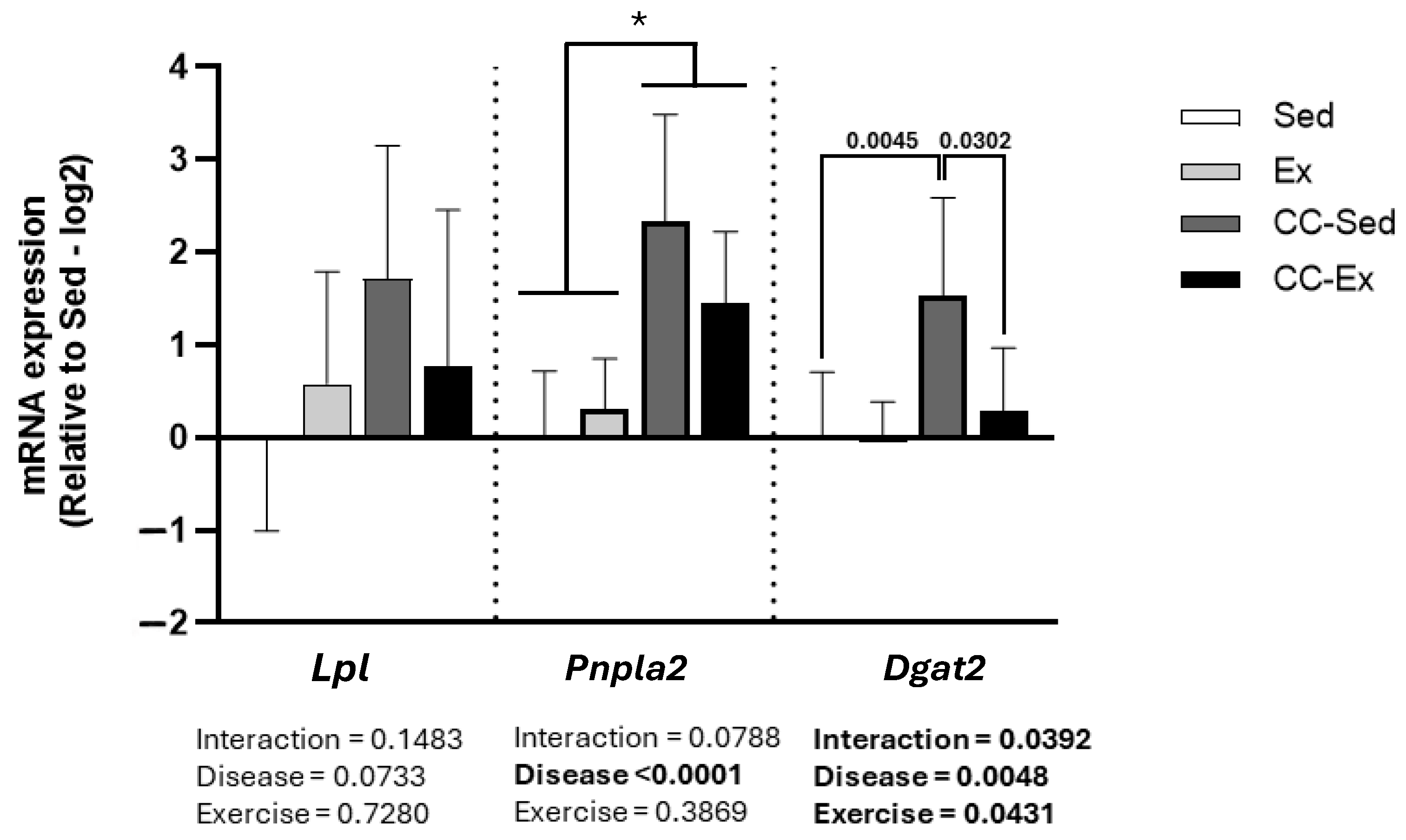
2.5. Validation of DEGs Related to Lipid Metabolism
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Treadmill Familiarization, Exercise Tolerance Capacity Test, and Aerobic Training
4.3. Euthanasia and Tissue Collection
4.4. Skeletal Muscle Cross-Sectional Area
4.5. RNA Isolation and RNA-Seq Library Preparation
4.6. RNA-Seq Data Processing and Differential Expression Analysis
4.7. WGCNA
4.8. Real Time Quantitative Polymerase Chain Reaction (RT-qPCR)
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BPs | Biological Processes |
| CC | Cardiac cachexia |
| CC-Ex | Cardiac cachexia exercise group |
| CC-Sed | Cardiac cachexia sedentary group |
| DEG | Differentially expressed genes |
| Ex | Control exercise group |
| GO | Gene ontology |
| HF | Heart failure |
| KEGG | Kyoto Encyclopedia of Gene and Genomes |
| ME | Module eigengene |
| RV | Right ventricle |
| Sed | Control sedentary group |
| RIN | RNA integrity numbers |
| RNA-seq | RNA sequence |
| WGCNA | Weighted gene co-expression network analysis |
References
- von Haehling, S.; Anker, S.D. Prevalence, incidence and clinical impact of cachexia: Facts and numbers—Update 2014. J. Cachexia Sarcopenia Muscle 2014, 5, 261–263. [Google Scholar] [CrossRef]
- von Haehling, S.; Anker, M.S.; Anker, S.D. Prevalence and clinical impact of cachexia in chronic illness in Europe, USA, and Japan: Facts and numbers update 2016. J. Cachexia Sarcopenia Muscle 2016, 7, 507–509. [Google Scholar] [CrossRef]
- Groenewegen, A.; Rutten, F.H.; Mosterd, A.; Hoes, A.W. Epidemiology of heart failure. Eur. J. Heart Fail. 2020, 22, 1342–1356. [Google Scholar] [CrossRef]
- Bozkurt, B.; Ahmad, T.; Alexander, K.; Baker, W.L.; Bosak, K.; Breathett, K.; Carter, S.; Drazner, M.H.; Dunlay, S.M.; Fonarow, G.C.; et al. HF STATS 2024: Heart Failure Epidemiology and Outcomes Statistics An Updated 2024 Report from the Heart Failure Society of America. J. Card. Fail. 2025, 31, 66–116. [Google Scholar] [CrossRef]
- Marra, A.M.; Arcopinto, M.; Bossone, E.; Ehlken, N.; Cittadini, A.; Grunig, E. Pulmonary arterial hypertension-related myopathy: An overview of current data and future perspectives. Nutr. Metab. Cardiovasc. Dis. 2015, 25, 131–139. [Google Scholar] [CrossRef]
- Menezes, T.C.F.; Lee, M.H.; Fonseca Balladares, D.C.; Nolan, K.; Sharma, S.; Kumar, R.; Ferreira, E.V.M.; Graham, B.B.; Oliveira, R.K.F. Skeletal Muscle Pathology in Pulmonary Arterial Hypertension and Its Contribution to Exercise Intolerance. J. Am. Heart Assoc. 2025, 14, e036952. [Google Scholar] [CrossRef]
- McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Bohm, M.; Burri, H.; Butler, J.; Celutkiene, J.; Chioncel, O.; et al. Corrigendum to: 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2021, 42, 4901. [Google Scholar] [CrossRef]
- Sachdev, V.; Sharma, K.; Keteyian, S.J.; Alcain, C.F.; Desvigne-Nickens, P.; Fleg, J.L.; Florea, V.G.; Franklin, B.A.; Guglin, M.; Halle, M.; et al. Supervised Exercise Training for Chronic Heart Failure with Preserved Ejection Fraction: A Scientific Statement from the American Heart Association and American College of Cardiology. J. Am. Coll. Cardiol. 2023, 81, 1524–1542. [Google Scholar] [CrossRef]
- Alves, C.R.R.; da Cunha, T.F.; da Paixao, N.A.; Brum, P.C. Aerobic exercise training as therapy for cardiac and cancer cachexia. Life Sci. 2015, 125, 9–14. [Google Scholar] [CrossRef]
- Mehra, M.R. Fat, cachexia, and the right ventricle in heart failure: A web of complicity. J. Am. Coll. Cardiol. 2013, 62, 1671–1673. [Google Scholar] [CrossRef]
- Yegorova, S.; Yegorov, O.; Ferreira, L.F. RNA-sequencing reveals transcriptional signature of pathological remodeling in the diaphragm of rats after myocardial infarction. Gene 2021, 770, 145356. [Google Scholar] [CrossRef]
- Moraes, L.N.; Fernandez, G.J.; Vechetti, I.J.r.; Freire, P.P.; Souza, R.W.A.; Villacis, R.A.R.; Rogatto, S.R.; Reis, P.P.; Dal-Pai-Silva, M.; Carvalho, R.F. Integration of miRNA and mRNA expression profiles reveals microRNA-regulated networks during muscle wasting in cardiac cachexia. Sci. Rep. 2017, 7, 6998. [Google Scholar] [CrossRef]
- Gomes, M.J.; Pagan, L.U.; Lima, A.R.R.; Reyes, D.R.A.; Martinez, P.F.; Damatto, F.C.; Pontes, T.H.D.; Rodrigues, E.A.; Souza, L.M.; Tosta, I.F.; et al. Effects of aerobic and resistance exercise on cardiac remodelling and skeletal muscle oxidative stress of infarcted rats. J. Cell. Mol. Med. 2020, 24, 5352–5362. [Google Scholar] [CrossRef]
- Wang, Y.; Pessin, J.E. Mechanisms for fiber-type specificity of skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 243–250. [Google Scholar] [CrossRef]
- Geng, T.; Li, P.; Yin, X.; Yan, Z. PGC-1alpha promotes nitric oxide antioxidant defenses and inhibits FOXO signaling against cardiac cachexia in mice. Am. J. Pathol. 2011, 178, 1738–1748. [Google Scholar] [CrossRef]
- Gene Ontology, C.; Aleksander, S.A.; Balhoff, J.; Carbon, S.; Cherry, J.M.; Drabkin, H.J.; Ebert, D.; Feuermann, M.; Gaudet, P.; Harris, N.L.; et al. The Gene Ontology knowledgebase in 2023. Genetics 2023, 224, iyad031. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Furumichi, M.; Tanabe, M.; Hirakawa, M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010, 38, D355–D360. [Google Scholar] [CrossRef]
- Molinari, F.; Malara, N.; Mollace, V.; Rosano, G.; Ferraro, E. Animal models of cardiac cachexia. Int. J. Cardiol. 2016, 219, 105–110. [Google Scholar] [CrossRef]
- Hassoun, P.M. Pulmonary Arterial Hypertension. N. Engl. J. Med. 2021, 385, 2361–2376. [Google Scholar] [CrossRef]
- Knapp, F.; Niemann, B.; Li, L.; Molenda, N.; Kracht, M.; Schulz, R.; Rohrbach, S. Differential effects of right and left heart failure on skeletal muscle in rats. J. Cachexia Sarcopenia Muscle 2020, 11, 1830–1849. [Google Scholar] [CrossRef]
- Friedberg, M.K.; Redington, A.N. Right Versus Left Ventricular Failure Differences, Similarities, and Interactions. Circulation 2014, 129, 1033–1044. [Google Scholar] [CrossRef]
- Batt, J.; Ahmed, S.S.; Correa, J.; Bain, A.; Granton, J. Skeletal muscle dysfunction in idiopathic pulmonary arterial hypertension. Am. J. Respir. Cell Mol. Biol. 2014, 50, 74–86. [Google Scholar] [CrossRef]
- Dalla Libera, L.; Zennaro, R.; Sandri, M.; Ambrosio, G.B.; Vescovo, G. Apoptosis and atrophy in rat slow skeletal muscles in chronic heart failure. Am. J. Physiol.-Cell Physiol. 1999, 277, C982–C986. [Google Scholar] [CrossRef]
- Moreira-Goncalves, D.; Padrao, A.I.; Ferreira, R.; Justino, J.; Nogueira-Ferreira, R.; Neuparth, M.J.; Vitorino, R.; Fonseca, H.; Silva, A.F.; Duarte, J.A.; et al. Signaling pathways underlying skeletal muscle wasting in experimental pulmonary arterial hypertension. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 2722–2731. [Google Scholar] [CrossRef]
- Xiang, G.L.; Ying, K.L.; Jiang, P.; Jia, M.P.; Sun, Y.P.; Li, S.Q.; Wu, X.D.; Hao, S.Y. Growth differentiation factor 11 induces skeletal muscle atrophy via a STAT3-dependent mechanism in pulmonary arterial hypertension. Skelet. Muscle 2022, 12, 10. [Google Scholar] [CrossRef]
- De Moraes, W.; de Souza, P.R.M.; da Paixao, N.A.; de Sousa, L.G.O.; Ribeiro, D.A.; Marshall, A.G.; Prestes, J.; Irigoyen, M.C.; Brum, P.C.; Medeiros, A. Aerobic exercise training rescues protein quality control disruption on white skeletal muscle induced by chronic kidney disease in rats. J. Cell. Mol. Med. 2018, 22, 1452–1463. [Google Scholar] [CrossRef]
- Kosmas, K.; Michael, Z.; Papathanasiou, A.E.; Spyropoulos, F.; Adib, E.; Jasuja, R.; Christou, H. Skeletal Muscle Dysfunction in Experimental Pulmonary Hypertension. Int. J. Mol. Sci. 2022, 23, 10912. [Google Scholar] [CrossRef]
- Johnston, J.R.; Chase, P.B.; Pinto, J.R. Troponin through the looking-glass: Emerging roles beyond regulation of striated muscle contraction. Oncotarget 2018, 9, 1461–1482. [Google Scholar] [CrossRef]
- Yuen, M.; Cooper, S.T.; Marston, S.B.; Nowak, K.J.; McNamara, E.; Mokbel, N.; Ilkovski, B.; Ravenscroft, G.; Rendu, J.; de Winter, J.M.; et al. Muscle weakness in TPM3-myopathy is due to reduced Ca.-sensitivity and impaired acto-myosin cross-bridge cycling in slow fibres. Hum. Mol. Genet. 2015, 24, 6278–6292. [Google Scholar] [CrossRef]
- Cheung, W.W.; Zheng, R.; Hao, S.; Wang, Z.; Gonzalez, A.; Zhou, P.; Hoffman, H.M.; Mak, R.H. The role of IL-1 in adipose browning and muscle wasting in CKD-associated cachexia. Sci. Rep. 2021, 11, 15141. [Google Scholar] [CrossRef]
- Gleeson, M.; Bishop, N.C.; Stensel, D.J.; Lindley, M.R.; Mastana, S.S.; Nimmo, M.A. The anti-inflammatory effects of exercise: Mechanisms and implications for the prevention and treatment of disease. Nat. Rev. Immunol. 2011, 11, 607–615. [Google Scholar] [CrossRef]
- Damluji, A.A.; Alfaraidhy, M.; AlHajri, N.; Rohant, N.N.; Kumar, M.; Al Malouf, C.; Bahrainy, S.; Kwak, M.J.; Batchelor, W.B.; Forman, D.E.; et al. Sarcopenia and Cardiovascular Diseases. Circulation 2023, 147, 1534–1553. [Google Scholar] [CrossRef]
- Hirabayashi, K.; Kinugawa, S.; Yokota, T.; Takada, S.; Fukushima, A.; Suga, T.; Takahashi, M.; Ono, T.; Morita, N.; Omokawa, M.; et al. Intramyocellular lipid is increased in the skeletal muscle of patients with dilated cardiomyopathy with lowered exercise capacity. Int. J. Cardiol. 2014, 176, 1110–1112. [Google Scholar] [CrossRef]
- Li, X.T.; Bi, X.K. Integrated Control of Fatty Acid Metabolism in Heart Failure. Metabolites 2023, 13, 615. [Google Scholar] [CrossRef]
- Zadoorian, A.; Du, X.M.; Yang, H.Y. Lipid droplet biogenesis and functions in health and disease. Nat. Rev. Endocrinol. 2023, 19, 443–459. [Google Scholar] [CrossRef]
- Emmens, J.E.; Jia, C.; Ng, L.L.; van Veldhuisen, D.J.; Dickstein, K.; Anker, S.D.; Lang, C.C.; Filippatos, G.; Cleland, J.G.F.; Metra, M.; et al. Impaired High-Density Lipoprotein Function in Patients with Heart Failure. J. Am. Heart Assoc. 2021, 10, e019123. [Google Scholar] [CrossRef]
- Pradhan, A.; Bhandari, M.; Vishwakarma, P.; Gualtieri, P.; Di Renzo, L.; Iellamo, F.; Sethi, R.; Perrone, M.A. Dyslipidemia and heart failure: Current evidence and perspectives of use of statins. Eur. Rev. Med. Pharmacol. Sci. 2024, 28, 2860–2877. [Google Scholar] [CrossRef]
- Das, S.K.; Hoefler, G. The role of triglyceride lipases in cancer associated cachexia. Trends Mol. Med. 2013, 19, 292–301. [Google Scholar] [CrossRef]
- He, M.A.; Pei, Z.T.; Mohsen, A.W.; Watkins, P.; Murdoch, G.; Van Veldhoven, P.P.; Ensenauer, R.; Vockley, J. Identification and characterization of new long chain Acyl-CoA dehydrogenases. Mol. Genet. Metab. 2011, 102, 418–429. [Google Scholar] [CrossRef]
- Manz, X.D.; Bogaard, H.J.; Aman, J. Regulation of VWF (Von Willebrand Factor) in Inflammatory Thrombosis. Arterioscler. Thromb. Vas. 2022, 42, 1307–1320. [Google Scholar] [CrossRef]
- Moreira, J.B.N.; Wohlwend, M.; Fenk, S.; Åmellem, I.; Flatberg, A.; Kraljevic, J.; Marinovic, J.; Ljubkovic, M.; Bjorkoy, G.; Wisloff, U. Exercise Reveals Proline Dehydrogenase as a Potential Target in Heart Failure. Prog. Cardiovasc. Dis. 2019, 62, 193–202. [Google Scholar] [CrossRef]
- Wang, J.C.; Xue, Z.M.; Hua, C.T.; Lin, J.; Shen, Z.D.; Song, Y.J.; Ying, H.Y.; Lv, Q.B.; Wang, M.H.; Zhou, B.Q. Metabolomic Analysis of the Ameliorative Effect of Enhanced Proline Metabolism on Hypoxia-Induced Injury in Cardiomyocytes. Oxid. Med. Cell. Longev. 2020, 2020, 1–15. [Google Scholar] [CrossRef]
- Lv, Q.B.; Li, D.B.; Zhao, L.D.; Yu, P.C.; Tao, Y.C.; Zhu, Q.J.; Wang, Y.; Wang, M.H.; Fu, G.S.; Shang, M.; et al. Proline metabolic reprogramming modulates cardiac remodeling induced by pressure overload in the heart. Sci. Adv. 2024, 10, eadl3549. [Google Scholar] [CrossRef]
- Lee, J.M.; Hammarén, H.M.; Savitski, M.M.; Baek, S.H. Control of protein stability by post-translational modifications. Nat. Commun. 2023, 14, 201. [Google Scholar] [CrossRef]
- Krueger, F. TrimGalore. 2021. [Software]. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 12 September 2024).
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
| Sed n = 9 | Ex n = 8 | CC-Sed n = 7 | CC-Ex n = 6 | p-Value | ||
|---|---|---|---|---|---|---|
| Body mass (g) | 410.3 ± 29.5 | 392.6 ± 26.1 | 331.5 ± 31.3 | 338.7 ± 39.8 | Interaction | p = 0.2918 |
| Exercise | p = 0.6538 | |||||
| Disease | p < 0.0001 | |||||
| Relative RV mass (mg/g) | 0.514 ± 0.07 | 0.491 ± 0.03 | 1.175 ± 0.20 | 1.188 ± 0.20 | Interaction | p = 0.7402 |
| Exercise | p = 0.9233 | |||||
| Disease | p < 0.0001 | |||||
| Exercise Tolerance (min) | 16.97 ± 3.4 | 23.54 ± 2.9 * | 11.40 ± 2.6 *# | 10.26 ± 3.8 *# | Interaction | p = 0.0036 |
| Exercise | p = 0.0320 | |||||
| Disease | p < 0.0001 |
| Sedentary | Exercise | Description | |||
|---|---|---|---|---|---|
| Symbol | Log2 FC | p-Adjusted | Log2 FC | p-Adjusted | |
| Acer2 | 1.66966 | 4.30 × 10−9 | 1.66688 | 3.14 × 10−9 | alkaline ceramidase 2 |
| Acss1 | 1.63399 | 0.00060 | 1.78190 | 0.00023 | acyl-CoA synthetase short-chain family member 1 |
| Adora2a | 1.04996 | 0.00374 | 1.04944 | 0.01295 | adenosine A2a receptor |
| Aqp4 | 1.99764 | 0.02221 | 2.03651 | 0.04934 | aquaporin 4 |
| Hba-a1 | 1.71166 | 0.00391 | 2.45445 | 1.08 × 10−5 | hemoglobin alpha, adult chain 1 |
| Hbb | 1.76343 | 0.00642 | 2.62157 | 1.13 × 10−5 | hemoglobin subunit beta |
| Inhbb | 1.62991 | 0.00091 | 1.79615 | 0.00034 | inhibin subunit beta B |
| Ppp1r3c | 1.25889 | 0.00389 | 1.18808 | 0.01937 | protein phosphatase 1, regulatory subunit 3C |
| Prodh | 2.36806 | 0.00013 | 1.96116 | 0.00442 | proline dehydrogenase |
| Tmem252 | 1.44794 | 0.00091 | 1.12137 | 0.03606 | transmembrane protein 252 |
| Tp53i11 | 1.18541 | 0.01490 | 1.43972 | 0.00680 | tumor protein p53 inducible protein 11 |
| Vwf | 1.35910 | 0.00091 | 1.10128 | 0.02476 | von Willebrand factor |
| Symbol | Log2FC | Name | ||
|---|---|---|---|---|
| Sedentary | Upregulated | Tnnc1 | 4.06665 | troponin C1, slow skeletal and cardiac type |
| Nr4a3 | 4.00686 | nuclear receptor subfamily 4, group A, member 3 | ||
| Tpm3 | 3.94351 | tropomyosin 3 | ||
| Tnni1 | 3.89482 | troponin T1, slow skeletal type | ||
| Myl3 | 3.73428 | myosin light chain 3 | ||
| Downregulated | Runx1 | −1.74698 | RUNX family transcription factor 1 | |
| Bhlhe41 | −1.37910 | basic helix-loop-helix family, member e41 | ||
| Slc25a30 | −1.36981 | solute carrier family 25, member 30 | ||
| Tnfrsf21 | −1.36134 | TNF receptor superfamily member 21 | ||
| Map3k14 | −1.34111 | mitogen-activated protein kinase kinase kinase 14 | ||
| Exercise | Upregulated | Hbb | 2.62157 | hemoglobin subunit beta |
| Hba-a1 | 2.45445 | hemoglobin alpha, adult chain 1 | ||
| Hba-a3 | 2.42841 | hemoglobin alpha, adult chain 3 | ||
| Aqp4 | 2.03651 | aquaporin 4 | ||
| Prodh | 1.96115 | proline dehydrogenase | ||
| Downregulated | Nrip3 | −2.62039 | nuclear receptor interacting protein 3 | |
| Egr1 | −1.68517 | early growth response 1 |
| Sedentary | Exercise | ||||||
|---|---|---|---|---|---|---|---|
| GO ID | Cellular Component | p-Adj | GeneID | GO ID | Cellular Component | p-Adj | GeneID |
| 0005833 | Hemoglobin complex | 0.025 | Hba-a1/Hbb/Hba-a3 | 0005833 | Hemoglobin complex | 0.000 | Hba-a1/Hbb/Hba-a3 |
| 0097386 | Glial cell projection | 0.000 | Aqp4/Glul | ||||
| GO ID | Molecular Function | p-Adj | GeneID | GO ID | Molecular Function | p-Adj | GeneID |
| 0015645 | fatty acid ligase activity | 0.007 | Acsf2/Acss3/Acss1/Slc27a1 | 0005344 | oxygen carrier activity | 0.000 | Hba-a1/Hbb/Hba-a3 |
| 0016405 | CoA-ligase activity | 0.007 | Acsf2/Acss3/Acss1/Slc27a1 | 0019825 | oxygen binding | 0.000 | Hba-a1/Hbb/Hba-a3 |
| 0016878 | acid-thiol ligase activity | 0.007 | Acsf2/Acss3/Acss1/Slc27a1 | 0004601 | peroxidase activity | 0.000 | Hba-a1/Hbb/Hba-a3 |
| 0016877 | ligase activity, forming carbon-sulfur bonds | 0.019 | Acsf2/Acss3/Acss1/Slc27a1 | 0016684 | oxidoreductase activity, acting on peroxide as acceptor | 0.000 | Hba-a1/Hbb/Hba-a3 |
| 0004806 | triglyceride lipase activity | 0.029 | Lpl/Lipe/Pnpla2 | 0016209 | antioxidant activity | 0.001 | Hba-a1/Hbb/Hba-a3 |
| 0016411 | acylglycerol O-acyltransferase activity | 0.030 | Dgat2/Agpat2/Pnpla2 | 0140104 | molecular carrier activity | 0.001 | Hba-a1/Hbb/Hba-a3 |
| 0009975 | cyclase activity | 0.030 | Dglucy/Gucy2g/Npr2 | 0020037 | heme binding | 0.006 | Hba-a1/Hbb/Hba-a3 |
| 0008374 | O-acyltransferase activity | 0.030 | Gpam/Dgat2/Agpat2/Pnpla2 | 0046906 | tetrapyrrole binding | 0.006 | Hba-a1/Hbb/Hba-a3 |
| 0019825 | oxygen binding | 0.033 | Hba-a1/Hbb/Hba-a3 | 0043177 | organic acid binding | 0.000 | Prodh/Hba-a1/Hbb/Hba-a3/Glul |
| 0005344 | oxygen carrier activity | 0.019 | Hba-a1/Hbb/Hba-a3 | 0001664 | G protein-coupled receptor binding | 0.020 | Adora2a/Hba-a1/Hba-a3 |
| 0016597 | amino acid binding | 0.015 | Prodh/Glul | ||||
| Sedentary Groups Comparison (S vs. CC-S) | |||
|---|---|---|---|
| GO ID | Biological Process (BP) | p-Adj | GeneID |
| GO:0006631 | fatty acid metabolic process | 0.000 | Acsf2/Ephx1/Ppargc1a/Nr4a3/Acss1/Decr1/Eci1/Acot2/Acad11/Lpl/Acadl/ Acaa2/Gpam/Dgat2/Ephx2/Slc27a1/Lipe/Plin5 |
| GO:0044242 | cellular lipid catabolic process | 0.000 | Pla2g4b/Acer2/Decr1/Eci1/Acad11/Lpl/Acadl/Acaa2/Cyp26b1/Lipe/Plin5/Pnpla2 |
| GO:0019395 | fatty acid oxidation | 0.000 | Ppargc1a/Nr4a3/Decr1/Eci1/Acad11/Acadl/Acaa2/Dgat2/Plin5 |
| GO:0016042 | lipid catabolic process | 0.000 | Pla2g4b/Acer2/Decr1/Eci1/Acot2/Acad11/Lpl/Acadl/Acaa2/Cyp26b1/Lipe/Plin5/Endou/Pnpla2 |
| GO:0046460 | neutral lipid biosynthetic process | 0.000 | Lpl/Gpam/Dgat2/Slc27a1/Agpat2/Plin5/Pnpla2 |
| GO:0046463 | acylglycerol biosynthetic process | 0.000 | Lpl/Gpam/Dgat2/Slc27a1/Agpat2/Plin5/Pnpla2 |
| GO:0034440 | lipid oxidation | 0.000 | Ppargc1a/Nr4a3/Decr1/Eci1/Acad11/Acadl/Acaa2/Dgat2/Plin5 |
| GO:0019216 | regulation of lipid metabolic process | 0.000 | Prox1/Ppargc1a/Nr4a3/Acadl/Fgf1/Gpam/Dgat2/Ephx2/Slc27a1/Golm1/Rorc/Plin5/Endou/Pnpla2 |
| GO:0019432 | triglyceride biosynthetic process | 0.000 | Lpl/Gpam/Dgat2/Slc27a1/Agpat2/Plin5 |
| GO:0090208 | positive regulation of triglyceride metabolic process | 0.001 | Gpam/Dgat2/Slc27a1/Plin5/Pnpla2 |
| Exercise Groups Comparison (Ex vs. CC-Ex) | |||
| GO ID | Biological Process (BP) | p-Adj | GeneID |
| GO:0015669 | gas transport | 0.000 | Aqp4/Hba-a1/Hbb/Hba-a3 |
| GO:0015670 | carbon dioxide transport | 0.000 | Aqp4/Hba-a1/Hbb |
| GO:0015671 | oxygen transport | 0.000 | Hba-a1/Hbb/Hba-a3 |
| GO:0019755 | one-carbon compound transport | 0.000 | Aqp4/Hba-a1/Hbb |
| GO:0042744 | hydrogen peroxide catabolic process | 0.000 | Hba-a1/Hbb/Hba-a3 |
| GO:0042743 | hydrogen peroxide metabolic process | 0.001 | Hba-a1/Hbb/Hba-a3 |
| GO:0098869 | cellular oxidant detoxification | 0.005 | Hba-a1/Hbb/Hba-a3 |
| GO:0009636 | response to toxic substance | 0.008 | Hba-a1/Hbb/Hba-a3/Inhbb |
| GO:1990748 | cellular detoxification | 0.008 | Hba-a1/Hbb/Hba-a3 |
| GO:0034284 | response to monosaccharide | 0.009 | Aqp4/Egr1/Vwf/Glul |
| ME | Biological Process | p-Adj |
|---|---|---|
| MEgrey | regulation of mitochondrial membrane potential | 0.000788 |
| glial cell activation | 0.010507 | |
| negative regulation of amyloid fibril formation | 0.014577 | |
| MEpink | mitochondrial membrane organization | 0.039761 |
| inner mitochondrial membrane organization | 0.043771 | |
| MEblue | purine ribonucleotide catabolic process | 0.008092 |
| ribonucleotide catabolic process | 0.008092 | |
| purine nucleotide catabolic process | 0.008092 | |
| MEbrown | transition between fast and slow fiber | 0.000257 |
| skeletal muscle adaptation | 0.000402 | |
| regulation of skeletal muscle adaptation | 0.000402 | |
| MEpurple | cell proliferation in external granule layer | 0.007065 |
| cerebellar granule cell precursor proliferation | 0.007065 | |
| cell proliferation in hindbrain | 0.007065 | |
| MEgreenyellow | microautophagy | 0.013788 |
| protein refolding | 0.013788 | |
| snRNA processing | 0.015794 | |
| MEsalmon | peripheral nervous system development | 0.000392 |
| glial cell development | 0.002722 | |
| myelin assembly | 0.002722 | |
| MEgreen | positive regulation of cell adhesion | 1.51 × 10−9 |
| antigen processing and presentation of exogenous peptide antigen via MHC class II | 9.66 × 10−7 | |
| extracellular matrix organization | 9.66 × 10−7 | |
| MEmidnightblue | defense response to virus | 1.67 × 10−25 |
| response to virus | 6.84 × 10−24 | |
| antiviral innate immune response | 8.36 × 10−16 | |
| MEyellow | aerobic respiration | 2.17 × 10−73 |
| cellular respiration | 3.31 × 10−70 | |
| energy derivation by oxidation of organic compounds | 2.21 × 10−61 | |
| MEturquoise | ribonucleoprotein complex biogenesis | 2.24 × 10−11 |
| ribosome biogenesis | 7.77 × 10−8 | |
| rRNA metabolic process | 1.41 × 10−6 | |
| MEcyan | regulation of circadian rhythm | 0.000112 |
| rhythmic process | 0.000112 | |
| circadian rhythm | 0.000133 | |
| MEmagenta | fatty acid oxidation | 0.001356 |
| lipid oxidation | 0.001356 | |
| lipid modification | 0.002489 | |
| MEblack | ribosome biogenesis | 1.09 × 10−33 |
| ribonucleoprotein complex biogenesis | 3.39 × 10−33 | |
| ribosomal small subunit biogenesis | 4.85 × 10−33 |
| ME | Groups | Overlapping DEGs |
|---|---|---|
| MEpink | Sedentary | Aldoc/Nrbp2/Slc25a30/Vwf/Prodh |
| Exercise | Vwf/Prodh | |
| MEblue | Sedentary | Spsb1/Maff/Pnpla2/Plim3/Galnt15 |
| Exercise | - | |
| MEbrown | Sedentary | Lpl/Slc2a13/Ccdc8/Agpat2/Fgf1/Atp2a2/Smtnl1/Prox1/Ldhb/Acsf2/Rps6ka5/Dixdc1/Tpm3/Npr2/Tpd52l1/Tnnc1/Adamtsl4/Klhl34/Slc27a1/Kremen1/Tnni1/Nnt/Myl3/Gpam/Ift81/Decr1/Rerg/Eci1/Fgfrl1 |
| Exercise | - | |
| MEyellow | Sedentary | Vegfa |
| Exercise | - | |
| MEturquoise | Sedentary | Ptpn3/Cep85l/Fbxo32 |
| Exercise | - | |
| MEmagenta | Sedentary | Tns1/Rorc/Acad11/Golm1/Sdc4/Dgat2 |
| Exercise | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inoue, D.S.; Pigg, Q.W.; Harris, D.R.; Zhang, D.; Boland, D.J.; Janini Gomes, M. Impact of Aerobic Training on Transcriptomic Changes in Skeletal Muscle of Rats with Cardiac Cachexia. Int. J. Mol. Sci. 2025, 26, 6525. https://doi.org/10.3390/ijms26136525
Inoue DS, Pigg QW, Harris DR, Zhang D, Boland DJ, Janini Gomes M. Impact of Aerobic Training on Transcriptomic Changes in Skeletal Muscle of Rats with Cardiac Cachexia. International Journal of Molecular Sciences. 2025; 26(13):6525. https://doi.org/10.3390/ijms26136525
Chicago/Turabian StyleInoue, Daniela Sayuri, Quinten W. Pigg, Dillon R. Harris, Dongmei Zhang, Devon J. Boland, and Mariana Janini Gomes. 2025. "Impact of Aerobic Training on Transcriptomic Changes in Skeletal Muscle of Rats with Cardiac Cachexia" International Journal of Molecular Sciences 26, no. 13: 6525. https://doi.org/10.3390/ijms26136525
APA StyleInoue, D. S., Pigg, Q. W., Harris, D. R., Zhang, D., Boland, D. J., & Janini Gomes, M. (2025). Impact of Aerobic Training on Transcriptomic Changes in Skeletal Muscle of Rats with Cardiac Cachexia. International Journal of Molecular Sciences, 26(13), 6525. https://doi.org/10.3390/ijms26136525







