Abstract
AT-hook motif nuclear-localized (AHL) genes play critical roles in chromatin remodeling and gene transcription regulation, profoundly influencing plant growth, development, and stress responses. While AHL genes have been extensively characterized in multiple plant species, their biological functions in pepper (Capsicum annuum L.) remain largely uncharacterized. In this study, we identified 45 CaAHL genes in the pepper genome through bioinformatics approaches. Comprehensive analyses were conducted to examine their chromosomal distribution, phylogenetic relationships, and the structural and functional features of their encoded proteins. Phylogenetic clustering classified the CaAHL proteins into six distinct subgroups. Transcriptome profiling revealed widespread expression of CaAHL genes across diverse tissues—including roots, stems, leaves, flowers, seeds, pericarp, placenta, and fruits—at various developmental stages. Quantitative real-time PCR further demonstrated that CaAHL1, CaAHL33, and CaAHL23 exhibited consistently high expression throughout flower bud development, whereas CaAHL36 showed preferential upregulation at early bud development stages. Expression profiling under hormone treatments and abiotic stresses indicated that CaAHL36 and CaAHL23 are auxin-inducible but are repressed by ABA, cold, heat, salt, and drought stress. Subcellular localization assays in Nicotiana benthamiana leaf epidermal cells showed that both CaAHL36 and CaAHL23 were predominantly localized in the nucleus, with faint expression also detected in the cytoplasm. Collectively, this study provides foundational insights into the CaAHL gene family, laying the groundwork for future functional investigations of these genes in pepper.
1. Introduction
Capsicum annuum L., commonly known as chili pepper, is a globally significant crop renowned for its distinctive pungency and rich nutritional profile. Its economic importance is underscored by its extensive cultivation and the diverse products derived from it, including fresh produce, spices, and pharmaceuticals. Advancements in genomic research, notably the sequencing of the pepper genome, have paved the way for in-depth studies into gene families that influence key agronomic traits, thereby enhancing breeding programs aimed at improving yield, disease resistance, and stress tolerance [1,2,3].
The AT-hook motif nuclear-localized (AHL) gene family is defined by the presence of an AT-hook motif, a small DNA-binding domain that specifically interacts with AT-rich DNA regions, as well as a conserved plant and prokaryote conserved (PPC) domain. Over the past decade, the role of AHLs in regulating various plant growth and developmental processes has been increasingly recognized. These processes include the elongation of hypocotyls [4,5,6], the formation of pollen walls and flower development [7,8], root growth [9,10], petiole elongation [11], and the modulation of phytohormone signaling [11]. Furthermore, AHL genes have been implicated in plant responses to pathogen infections, as well as to environmental stresses such as salt and drought [12,13,14,15]. Recent studies have also broadened our understanding of the AHL gene family in important crops like cotton, soybean, maize, and rice [16,17,18,19]. These findings emphasize the diverse roles of AHLs in plant development, offering new strategies for crop improvement.
In recent years, significant progress has been made in understanding the roles of specific AHL genes in plant flower development and male sterility. In Arabidopsis thaliana, AHL22 was identified as a critical regulator of flowering time by modulating FLOWERING LOCUS T (FT) chromatin state [20]. Similarly, AHL16 suppresses transposon activation and ensured timely flowering [21]. TRANSPOSABLE ELEMENT SILENCING VIA AT-HOOK (TEK), also known as AtAHL16, was found to be essential for male fertility. TEK regulates arabinogalactan-proteins (AGPs) in anthers critical for pollen wall formation. Mutations cause microspore development defects and sterility [22]. AHL15 promotes somatic embryogenesis, highlighting its biotechnological potential [23]. In rice, PERSISTENT TAPETAL CELL2 (PTC2) regulates tapetal programmed cell death and pollen wall patterning, with its loss leading to male sterility [8]. However, despite these advancements, comprehensive investigations into the AHL gene family in chili pepper remain scarce.
Despite these advances, AHL genes in chili pepper remain largely unexplored. Given the importance of pepper and the potential roles of AHLs in development and stress adaptation, a genome-wide analysis of the pepper AHL family is warranted. Here, we identify and characterize all CaAHL (Capsicum annuum AHL) genes in the pepper genome and analyzed their expression patterns across different tissues and developmental stages, as well as in response to exogenous hormone treatments and abiotic stress conditions. In particular, we examine the expression of several CaAHL genes during floral development, providing insights into their potential roles in reproductive processes. The comprehensive data generated in this study lay the foundation for future functional analyses and may inform molecular breeding strategies aimed at improving pepper cultivars through the manipulation of AHL genes.
2. Results
2.1. Identification and Basic Information on the CaAHL Gene Family
The Hidden Markov Model (HMM) profile of the PPC domain (PF03479) was used as a query sequence to identify AHL proteins in the Zhangshugang genome database [24] using the “Simple HMM Search” function in TBtools-II (v2.149). In total, 45 AHL genes containing the conserved PPC domain were identified. Only those showing consistent hits in both the “Sequence scores” and “Domain scores” outputs were retained to ensure domain completeness and sequence reliability. To facilitate subsequent studies, these pepper AHL genes were designated as CaAHL1 to CaAHL45 based on their positions across the 12 chromosomes in the Zhangshugang reference genome. Comprehensive details, including gene names, chromosomal locations, protein lengths, molecular weights, theoretical isoelectric points (pIs), instability indices, grand average of hydropathicity, and predicted subcellular localizations, are provided in Table 1.

Table 1.
Basic information on AHL genes identified in pepper.
The CaAHLs exhibited significant variation in protein length and physicochemical properties. The protein lengths ranged from 111 amino acids (CaAHL24) to 578 amino acids (CaAHL39). Correspondingly, the molecular weights (MWs) varied from 11.71 kDa (CaAHL40) to 60.51 kDa (CaAHL39). The theoretical isoelectric points (pIs) spanned from 4.44 (CaAHL34) to 11.25 (CaAHL40), while the instability indices ranged from 30.43 (CaAHL24) to 63.62 (CaAHL31). The grand average of hydropathicity (GRAVY) values ranged from −0.76 (CaAHL4) to 0.18 (CaAHL27). Subcellular localization predictions for the 45 CaAHL proteins revealed diverse distribution patterns: 20 localized to the nucleus, 17 to the chloroplast, 5 to the cytoplasm, and 1 each to the plasma membrane, vacuole membrane, and endoplasmic reticulum (ER) (Table 1).
2.2. Chromosomal Distribution of the CaAHL Gene Family
The 45 CaAHLs were unevenly distributed across 9 of the 12 chromosomes in the Zhangshugang genome (Figure 1a). Most CaAHLs were concentrated near the distal ends of the chromosomes. Only a few were located in central regions. Chromosome 1 had the highest number of CaAHLs, with a total of 21, followed by chromosomes 12 and 3, which contained 7 and 6 genes, respectively. In contrast, no CaAHLs were identified on chromosomes 10 and 11 (Figure 1b).
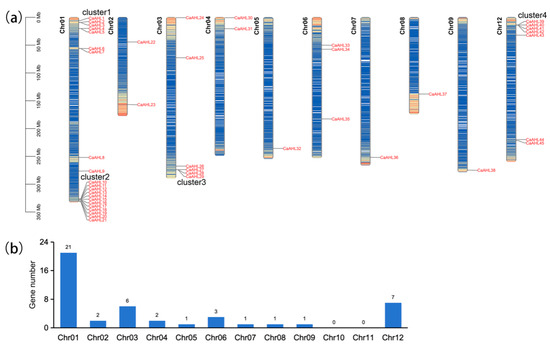
Figure 1.
Chromosome mapping and gene number of CaAHLs in Zhangshugang genome. (a) Chromosome mapping of CaAHLs. Chromosome numbers are represented on the top, and the scale is shown on the left. (b) Statistical analysis of CaAHLs on the 12 chromosomes.
Interestingly, CaAHLs were enriched in four specific regions of the genome: the proximal and distal ends of chromosome 1, the distal end of chromosome 3, and the proximal end of chromosome 12. These regions were designated as cluster 1, cluster 2, cluster 3, and cluster 4, respectively. Cluster 1 comprises five genes (CaAHL1 to CaAHL5), cluster 2 contains twelve genes (CaAHL10 to CaAHL21), cluster 3 includes three genes (CaAHL27 to CaAHL29), and cluster 4 consists of four genes (CaAHL39 to CaAHL42) (Figure 1a). The uneven distribution and clustering of CaAHLs suggest gene accumulation may have occurred through tandem duplication events in specific genomic regions.
2.3. Phylogenetic Analysis of the CaAHL Gene Family
To further delineate the evolutionary pathway of AHL genes across diverse species, a phylogenetic tree was constructed based on 45 pepper AHL amino acid sequences, along with those from tomato and Arabidopsis thaliana (Figure 2). The phylogenetic analysis revealed that the CaAHL gene family is divided into six main clades: Clade A, containing 2 genes; Clade B, containing 12 genes; Clade C, containing 6 genes; and Clade D, containing 6 genes; Clade E, containing 6 genes; Clade F, containing 13 genes. Interestingly, the number of AHL genes in pepper, tomato, and Arabidopsis thaliana shows only minor differences in Clade A (2, 2, and 2, respectively), Clade B (12, 11, and 13, respectively), Clade C (6, 4, and 5, respectively), and Clade D (6, 6, and 6, respectively) (Figure 2 and Table S1). However, more pronounced differences were observed in Clade E and Clade F. The number of AHL genes in Clade E is six, four, and one in pepper, tomato, and Arabidopsis thaliana, respectively. In Clade F, the corresponding values are 13, 10, and 2—indicating a notably higher number of AHL genes in pepper and tomato compared to Arabidopsis thaliana (Figure 2 and Table S1). Combined with chromosome localization analysis (Figure 1), we found that CaAHL11–CaAHL18 in gene cluster 2 and CaAHL27–CaAHL29 in gene cluster 3 are located in Clade E. Similarly, CaAHL39–CaAHL42 in gene cluster 4 are positioned in Clade F. These results suggest that during the evolutionary process in the Solanaceae family, the AHL genes in Clade E and Clade F may have undergone duplication events.
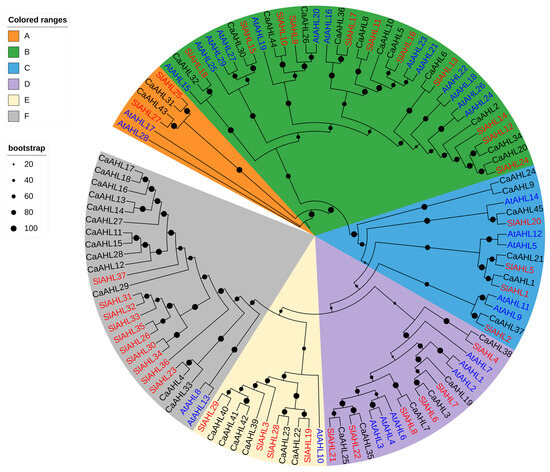
Figure 2.
Phylogenetic relationships among AHLs in Capsicum annuum, Solanum lycopersicum, and Arabidopsis thaliana. Clades are shaded in distinct colors to facilitate visual differentiation. Black dots indicate bootstrap support values, with only those greater than 20 displayed.
2.4. Analysis of Conserved CaAHL Motifs
We analyzed the motifs of the CaAHL genes and predicted their functions (Figure 3 and Table S1). The results indicated that most CaAHLs contained Motifs 1, 2, 3, and 4. Nearly all CaAHLs included Motif 1 and Motif 3, except for CaAHL41, which lacked Motif 1, and CaAHL17 and CaAHL18, which lacked Motif 3. Additionally, 82.22% and 73.33% of CaAHLs contained Motif 4 and Motif 2, respectively (Figure 3 and Table S2). Some CaAHLs contained multiple identical motifs. For example, CaAHL11, CaAHL12, and CaAHL15 each contained two Motif 4 elements. CaAHL31 and CaAHL4 contained two and three Motif 10 elements, respectively. We also found that CaAHL11 and CaAHL15; CaAHL13 and CaAHL14; as well as CaAHL23 and CaAHL24, exhibited highly similar motif structures, which is consistent with their clustering in the phylogenetic analysis (Figure 3 and Table S2). The four genes in cluster 4 exhibit significant differences in both motif number and protein length. This suggests that they are not tandem duplicates (Figure 3 and Table 1). However, the five genes in cluster 2 (CaAHL13, CaAHL14, CaAHL16, CaAHL17, and CaAHL18) exhibit high similarity in both gene structure and protein length. Notably, CaAHL16 contains Motif 3, Motif 7, and Motif 10, which are absent in CaAHL13 and CaAHL14, and possesses Motif 3, which is missing in CaAHL17 and CaAHL18. Overall, CaAHLs in the same subgroup in the phylogenetic tree tended to have similar structures and conserved motif distributions (Figure 3 and Table S2), indicating that CaAHLs contain highly conserved amino acid residues and that CaAHLs in the same cluster may have similar roles.
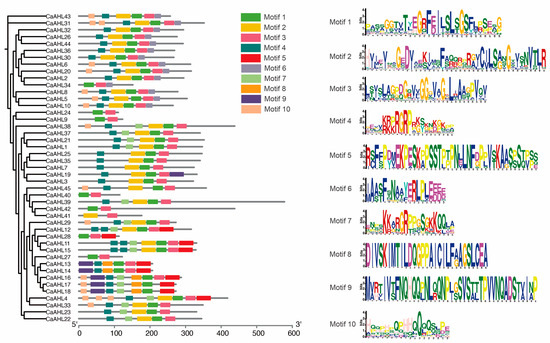
Figure 3.
Analysis of conserved CaAHL motifs. The evolutionary tree on the left is constructed based on the protein sequences of CaAHLs.
2.5. Cis-Regulatory Element Analysis of the CaAHL Promoters
Since CaAHLs play important roles in the response to plant growth, development, and stress responses, we utilized PlantCARE to analyze the cis-regulatory elements in the pre-2000 bp region of the CaAHLs promoter to explore their possible functions. We screened cis-regulatory elements associated with growth, development, stress, and hormone responses. These were identified in the promoter regions of CaAHLs and categorized into 18 distinct classes using TBtools (Figure 4 and Figure 5). The analysis revealed that the promoters of CaAHL22 and CaAHL24 are enriched with abscisic acid-responsive cis-regulatory elements, containing five and four elements, respectively. Promoters of CaAHL7, CaAHL1, and CaAHL34 exhibit a higher abundance of anaerobic-responsive elements, with five, four, and four elements, respectively. The promoter of CaAHL28 shows a significant presence of gibberellin-responsive elements. Similarly, the promoters of CaAHL23 and CaAHL4 are enriched with jasmonic acid-responsive elements, containing seven and five elements, respectively. CaAHL28 and CaAHL29 have higher numbers of MYB binding site-related elements, with six and five elements, respectively. The promoter of CaAHL21 contains four salicylic acid-responsive elements (Figure 4 and Figure 5). These distinct distributions of cis-regulatory elements suggest potential functional differentiation across the CaAHLs.
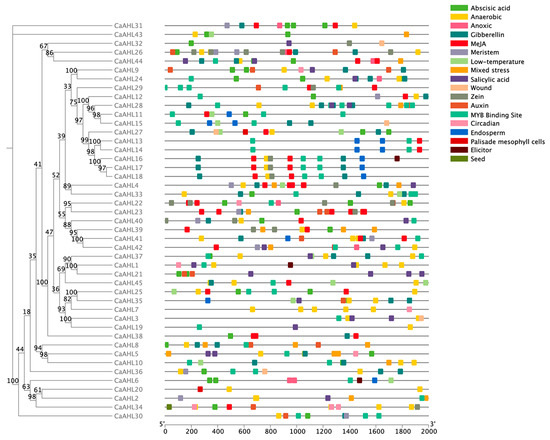
Figure 4.
The distribution of cis-regulatory elements predicted in the CaAHL promoters. Different colored boxes represent different cis-regulatory elements. The evolutionary tree on the left is constructed based on the protein sequences of CaAHLs.
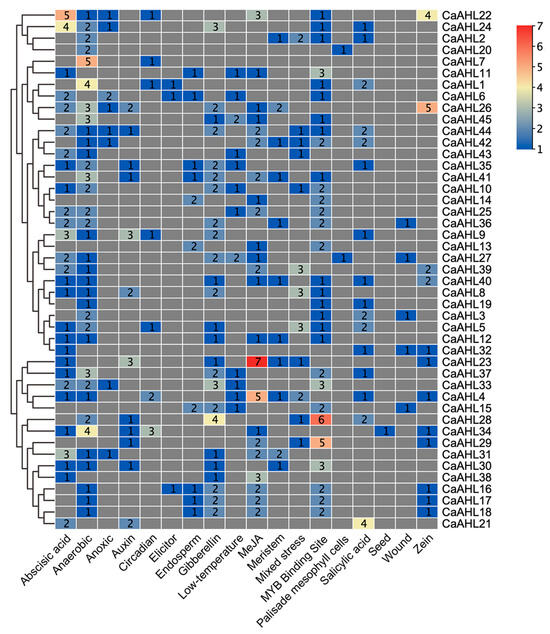
Figure 5.
Number of cis-regulatory elements in the CaAHL promoter regions. The evolutionary tree on the left is based on the analysis of the number and types of cis-regulatory elements.
2.6. Tissue-Specific Expression Profiles of CaAHLs in Pepper
To explore whether CaAHLs play a role in tissue development in pepper, the expression profiles of CaAHLs in the roots, stems, leaves, pericarp, and placenta were analyzed using RNA-seq data from the pepper line 6421 (Figure 6 and Table S3). The results revealed that several genes, including CaAHL1, CaAHL26, CaAHL5, CaAHL8, CaAHL10, CaAHL44, CaAHL2, and CaAHL20, exhibited root- and stem-specific expression (Figure 6). Certain genes, such as CaAHL9, CaAHL27, CaAHL28, and CaAHL11 to CaAHL18, were more highly expressed in seeds at 20 and 25 days after flowering (DAF). Some genes were specifically expressed in certain tissues or developmental stages. For instance, CaAHL29 displayed high expression in seeds at 60 DAF, suggesting a potential role in seed maturation. CaAHL36, CaAHL23, CaAHL12, and CaAHL16 to CaAHL18 were highly expressed during various stages of bud development, indicating their involvement in floral development. CaAHL21 showed specific expression in the placenta at 35 DAF, implying a role in placenta development (Figure 6). These findings suggest that most CaAHL genes are specifically expressed in roots, stems, and seeds at 20 and 25 DAF, highlighting their significant roles in root, stem, and seed development. The tissue-specific expression patterns of certain genes indicate functional differentiation across the CaAHL family members.
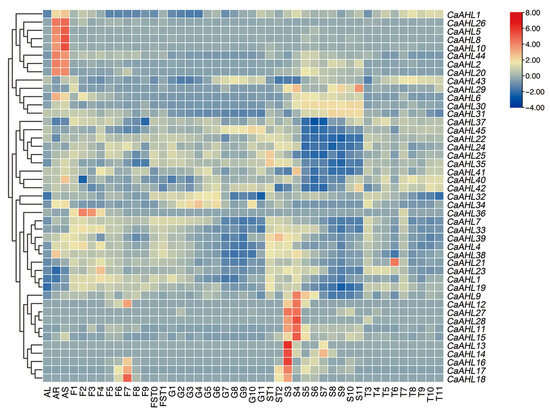
Figure 6.
Expression profile analysis of CaAHLs in various tissues and organs of pepper. Expression levels were determined in the following tissues and stages: leaf tissues were sampled 60 days after emergence and marked correspondingly as AL; stems and roots were marked AS and AR, respectively. Floral buds were sampled at 0.25, 0.35, 0.5, 0.7, 0.8, 1.0, 1.2, 1.45, and 1.7 cm and marked correspondingly as F1, F2, F3, F4, F5, F6, F7, F8, and F9; fruits were collected 3, 7, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, and 60 days after flowering (DAF) and marked correspondingly as FST0, FST1, G1, G2, G3, G4, G5, G6, G7, G8, G9, G10, G11; seed samples were collected 10, 15, 20, 25, 30, 35, 40, 45, 50, 55 and 60 DAF and marked correspondingly as ST1, ST2, S3, S4, S5, S6, S7, S8, S9, S10, S11; placenta samples were collected 20, 25, 30, 35, 40, 45, 50, 55 and 60 DAF and marked correspondingly as T3, T4, T5, T6, T7, T8, T9, T10, and T11. The FPKM values were log2-transformed, and a heatmap was generated using TBtools-II (v2.149) software. The evolutionary tree on the left is constructed based on gene expression levels. Expression values on the right are shown as a color gradient from low expression (blue) to high expression (red).
2.7. Expression Profiles of CaAHL Genes in Response to Exogenous Hormones and Abiotic Stresses
The transcriptome data used for analyzing hormone and abiotic stress responses were derived from a previously published dataset [25], where RNA-seq results were experimentally validated by qRT-PCR, confirming their reliability for downstream expression analysis.
In this study, heatmap visualization was utilized to analyze the expression profiles of all CaAHL genes in pepper roots under the treatments of plant hormones and abiotic stresses (Tables S4 and S5). Root tissues were subjected to five plant hormones—abscisic acid (ABA), gibberellin (GA), indole-3-acetic acid (IAA), jasmonic acid (JA), and salicylic acid (SA)—and five abiotic stresses, namely low-temperature stress, high-temperature stress, simulated drought stress (mannitol), salt stress, and oxidative stress (hydrogen peroxide). Notably, eight CaAHL genes (CaAHL12, CaAHL13, CaAHL14, CaAHL16, CaAHL17, CaAHL18, CaAHL27, and CaAHL28) were undetectable in root tissues before and after all hormone and abiotic stress treatments, suggesting that these genes are not expressed in roots. Following different hormone and abiotic stress treatments, the detectable CaAHL genes in roots exhibited varying degrees of upregulation or downregulation, indicating divergent functional roles among these genes (Figure 7).
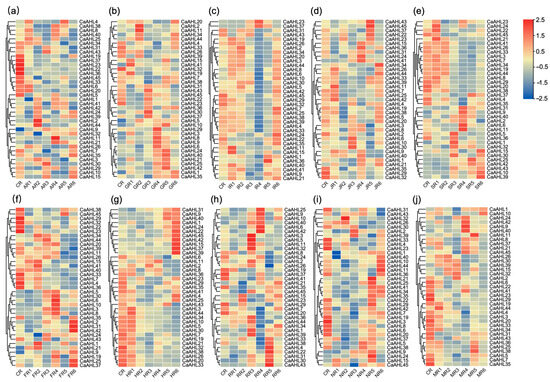
Figure 7.
Expression profiles of CaAHL genes under exogenous hormones and abiotic stresses. Red blocks denote genes with high expression levels, while blue blocks represent genes with low expression levels based on transcriptomic data. All samples were collected from roots, with three biological replicates per treatment. Each treatment group includes a corresponding untreated control sample (CR: control root). (a) AR: ABA-treated root; (b) GR: GA3-treated root; (c) IR: IAA-treated root; (d) JR: JA-treated root; (e) SR: SA-treated root; (f) FR: cold-treated root; (g) HR: heat-treated root; (h) RR: H2O2-treated root; (i) NR: NaCl-treated root; (j) MR: mannitol-treated root. Numbers 1 to 6 represent time points after treatment, 0.5 h, 1 h, 3 h, 6 h, 12 h, and 24 h, respectively.
Among the six genes that were highly expressed during floral bud development (Figure 6), only CaAHL23 and CaAHL36 were detectable in roots. Both genes were significantly downregulated by abscisic acid (ABA) treatment (Figure 7a). Under gibberellin treatment, CaAHL36 expression followed a dynamic pattern: it first decreased, then increased, then decreased again, and finally increased (Figure 7b). In contrast, CaAHL23 expression initially decreased, then increased, and subsequently decreased again (Figure 7b). Following IAA treatment, CaAHL23 and CaAHL36 transcript levels peaked at 6 and 12 h, respectively (Figure 7c). Interestingly, under JA treatment, the peak expression times of these genes were reversed: CaAHL23 peaked at 12 h and CaAHL36 at 6 h (Figure 7d). Under SA treatment, CaAHL23 expression showed fluctuations (increased, then decreased, then increased, and finally decreased), whereas CaAHL36 showed an initial decrease, followed by an increase and a subsequent decrease (Figure 7e). Under abiotic stresses (cold, heat, salt, and mannitol-induced drought), the expression of both genes generally decreased, consistent with the downregulation observed in response to ABA (Figure 7a,f,g,i,j). These results suggest that CaAHL23 and CaAHL36 may play critical roles in ABA-mediated stress responses. However, under oxidative stress, both genes displayed a transient expression pattern with an initial increase, followed by a decrease and a subsequent increase (Figure 7h).
2.8. Relative Expression Levels of Eight CaAHL Genes in Bud Development
Based on the transcriptomic data from different stages of flower bud development (Figure 6), we observed that eight CaAHL genes—CaAHL1, CaAHL4, CaAHL5, CaAHL23, CaAHL26, CaAHL33, CaAHL35, and CaAHL36—exhibited detectable expression at one or more stages. To gain a preliminary understanding of their potential involvement in floral development, we conducted qRT-PCR to examine their expression patterns during these stages (Figure 8). Expression levels were normalized using CaAHL5, which showed the lowest transcript abundance across all stages, as the reference. The qRT-PCR results revealed that CaAHL1, CaAHL4, CaAHL23, CaAHL33, and CaAHL35 consistently exhibited relatively high expression levels compared with CaAHL5 during all stages of floral development (Figure 8), suggesting their sustained roles throughout flower formation. Using CaAHL5 as the calibrator gene, we found that CaAHL26 consistently exhibited a slightly higher relative expression than CaAHL5 across most developmental stages, except at F2 and F3, where its relative expression was only marginally higher. Likewise, CaAHL36 showed relatively high expression compared to CaAHL5 at F1 and F2 (Figure 8). This suggests that CaAHL36 may be specifically involved in the early phase of floral bud initiation or organ differentiation. These findings indicate that while some CaAHL genes are broadly involved in flower development, others may function in a stage-specific manner, reflecting possible sub-functionalization within the gene family.
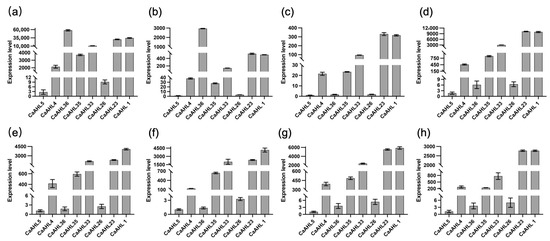
Figure 8.
Relative expression levels of eight CaAHL genes in bud development. Floral buds were sampled at 0.25, 0.35, 0.5, 0.7, 0.8, 1.0, 1.2, and 1.45 cm and marked correspondingly as F1 (a), F2 (b), F3 (c), F4 (d), F5 (e), F6 (f), F7 (g), and F8 (h).
2.9. Subcellular Localization of CaAHL23 and CaAHL36
To further investigate the cellular characteristics of CaAHL23 and CaAHL36, we conducted subcellular localization assays by fusing their coding sequences to GFP and transiently expressing them in Nicotiana benthamiana epidermal cells. The recombinant constructs (CaAHL23-GFP and CaAHL36-GFP) were driven by the CaMV 35S promoter, and infiltration was performed via Agrobacterium tumefaciens-mediated transformation. At 48 h post-infiltration, green fluorescence signals were detected using confocal laser scanning microscopy. The results showed that strong GFP fluorescence was predominantly localized in the nucleus for both CaAHL23 and CaAHL36 (Figure 9), consistent with their putative roles as transcription factors. Additionally, faint GFP signals were also observed in the cytoplasm, suggesting that a small proportion of the proteins may shuttle between the nucleus and cytoplasm or exhibit partial cytoplasmic distribution under these transient expression conditions. These findings provide preliminary evidence for the nuclear localization of CaAHL23 and CaAHL36 proteins, which is in line with their sequence-predicted function. However, further in vivo or stable expression studies would be necessary to confirm the dynamics and functional implications of their subcellular distribution.
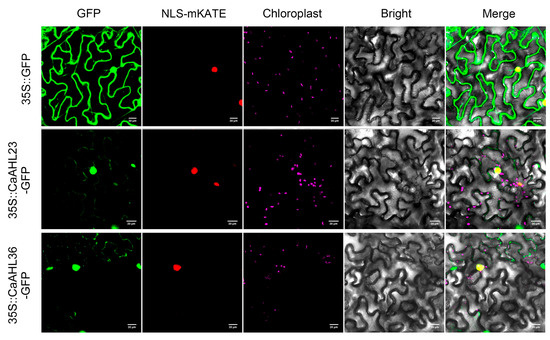
Figure 9.
Subcellular localization of CaAHL23 and CaAHL36. The vectors 35S::CaAHL23-GFP, 35S::CaAHL36-GFP, and 35S::GFP were separately co-transformed into Agrobacterium with the nuclear localization marker NLS-mKATE. GFP: green fluorescent protein fluorescence signal. Chloroplast: chlorophyll autofluorescence signal. Bright: field of bright light; Merged: overlay of the GFP and DAPI signals, nuclear localization signal, chlorophyll autofluorescence signal, and bright light field. Scale bar = 20 μm.
3. Discussion
Our genome-wide analysis identified 45 CaAHL genes in pepper, substantially more than the 29 AHLs in Arabidopsis or the 35–40 typically found in other diploid plants [17,18,19,26]. This expansion likely reflects a history of gene duplication. The uneven chromosomal distribution and presence of four CaAHL clusters suggest that tandem duplications contributed to family expansion. Indeed, phylogenetic mapping reveals that the tandem arrays on chromosomes 1, 3, and 12 correspond to lineage-specific expansions in Solanaceae. Comparative phylogeny (Figure 2) shows that Clades A–D are conserved in number across pepper, tomato, and Arabidopsis, indicating these subfamilies were present before the Solanaceae–Brassicaceae split and retained by purifying selection. In contrast, Clades E and F are greatly expanded in pepper (6 and 13 genes) relative to Arabidopsis (1–2 genes), implying pepper/tomato-specific duplications. Notably, genes in clusters 2 and 3 (CaAHL11–18 and CaAHL27–29) and cluster 4 (CaAHL39–42) all belong to Clades E/F, reinforcing that recent tandem duplication in pepper generated these extra copies. Tandem and proximal duplications are known to provide raw genetic material for the evolution of novel gene functions in plants [26,27], and CaAHL genes are no exception. Overall, the CaAHL family reflects a blend of ancient conserved regulators and newer, potentially neo-functionalized genes resulting from pepper-specific duplications.
Despite their diversification, CaAHL proteins maintain highly conserved structural features. Almost all members retain the canonical AT-hook and PPC domains (Motifs 1 and 3), underscoring their shared capacity for DNA binding and chromatin interaction [17,26]. The consistent motif architectures and intron–exon patterns within subgroups indicate that duplicated CaAHL genes often conserve core functions. At the same time, variable motif insertions or losses in some paralogs (e.g., CaAHL41 lacking Motif 1) hint at functional divergence. The promoters of CaAHL genes are rich in hormone- and stress-responsive elements, indicating tight regulation by environmental and developmental signals. For instance, multiple CaAHL promoters contain ABA, GA, JA, SA, and anaerobic response elements (Figure 4 and Figure 5). This cis-element diversity suggests that CaAHL transcription is modulated by hormonal cues. Consistent with this, related studies have shown AHL genes to be hormone-regulated: for example, Arabidopsis AHLs influence auxin and gibberellin signaling [11], and rice AHLs coregulate flowering and stress pathways [26]. In rice, OsAHL genes are markedly upregulated by drought and salinity and form co-expression networks with reproductive genes [26]. Similarly, pepper CaAHL promoters harbor stress-related elements, and our expression data show that many CaAHLs respond to drought, cold, salt, and oxidative stress. Thus, the conserved domains of AHL proteins are embedded in regulatory contexts that link development to environmental adaptation.
The diverse expression profiles of CaAHL genes imply extensive sub-functionalization. Many CaAHLs are expressed in vegetative tissues: for example, CaAHL1, 2, 5, 8, 10, 20, 26, 44 are root/stem-preferential (Figure 6), suggesting roles in root or stem development. Others are fruit/seed-enriched: notably, CaAHL9, 27, 28, 11–18 peak in seeds at 20–25 DAF, and CaAHL29 spikes only at 60 DAF (Figure 6), indicating possible roles in seed maturation. CaAHL21 is placenta-enriched (Figure 6), implying involvement in fruit tissue differentiation. In contrast, eight CaAHL genes (12, 13, 14, 16, 17, 18, 27, 28) were not expressed in roots even after stress/hormone treatments (Figure 7), aligning with their strictly reproductive expression. Such specialization parallels reported in other species: in rice, nearly all OsAHLs were expressed in at least some vegetative or reproductive tissue but with stage-specific peaks [26], while in Arabidopsis, different AHLs are known to govern distinct developmental processes [5,22]. These patterns suggest that, following duplication, pepper AHLs have partitioned developmental roles. Some paralogs, like CaAHL1 and CaAHL33, act broadly in organ development, while others (e.g., CaAHL29 or CaAHL36) are dedicated to specific stages of seed or flower formation.
To further investigate floral development-related candidates, we selected eight CaAHL genes that exhibited relatively high transcript levels during flower bud development and validated their expression patterns using qRT-PCR (Figure 8). In this analysis, CaAHL5 was used as the calibrator gene, and all expression levels of the other CaAHL genes were calculated relative to it at each developmental stage. On this basis, CaAHL4, CaAHL35, CaAHL33, CaAHL23, and CaAHL1 consistently exhibited higher expression levels than CaAHL5 across all stages. CaAHL26 also showed relatively high expression at most stages, except for F2 and F3. Notably, CaAHL36 displayed relatively strong expression at early stages (F1 and F2). These expression profiles suggest that certain CaAHLs are dynamically regulated during flower bud development and may play diverse roles in this process [8,22]. We also examined the subcellular localization of CaAHL23 and CaAHL36 to gain insight into their possible molecular functions. Both proteins were transiently expressed in Nicotiana benthamiana epidermal cells and fused with GFP for visualization. Strong fluorescence signals were observed in the nucleus, with weaker signals in the cytoplasm (Figure 9), indicating that these proteins are primarily nuclear-localized. This localization is consistent with their predicted roles as DNA-binding transcriptional regulators.
The identification of specific AHLs associated with flower and anther development provides candidate genes for hybrid breeding. For example, CaAHL36 could be targeted to create genic male-sterile or -fertile lines, facilitating hybrid seed production. Furthermore, the stress-responsive expression of many CaAHLs hints that they may affect stress resilience; breeding or engineering efforts might exploit AHL promoters or coding variants to improve tolerance. Future work should test these hypotheses by functional studies, such as CRISPR/Cas9 knockout or overexpression of selected CaAHLs to observe effects on fertility and stress tolerance. In conclusion, our genome-wide survey and expression analyses lay a foundation for understanding the roles of AHL proteins in pepper biology. The duplication-driven expansion of the CaAHL family has allowed pepper to evolve both conserved developmental regulators and novel genes adapted to its specific physiology and environment. This comprehensive resource will enable deeper investigations into how AHL transcription factors coordinate plant development and stress responses in Capsicum.
4. Materials and Methods
4.1. Plant Material and Growth Conditions
A highly elite breeding line of pepper (Capsicum annuum L.), designated as 6421, was employed in this study. Seeds of 6421 were surface-sterilized with 5% sodium hypochlorite for 15 min, rinsed thoroughly with sterile distilled water, and sown in 200-well seedling trays containing vermiculite. The trays were positioned atop a 30 L lightproof plastic container filled with Japanese garden test nutrient solution (pH 6.0). Seedlings were grown under controlled conditions: a day/night temperature of 25/18 °C, a 16/8 h light/dark cycle, 60–70% relative humidity, and a light intensity of 6000 Lux. Hormone and stress treatments were carried out as previously described [25].
4.2. Retrieval and Identification of AHL Genes in Pepper
In this study, the candidate AHL proteins were retrieved as follows: first, the protein, nucleotide, and genome sequences of Zhangshugang genomes were downloaded from the Pepper Genomics Database (http://ted.bti.cornell.edu/cgi-bin/pepper/search, accessed on 6 January 2025) [24]. Second, the HMM of AHL protein (PF03479) was downloaded from the Pfam database (http://pfam-legacy.xfam.org, accessed on 6 January 2025) [28]. Finally, the TBtools-II software (v2.149) was employed to identify putative AHL genes in the Zhangshugang genomes using the “Simple HMM Search” function, with the default cut-off parameters [29]. Specifically, the HMM profile of the AHL domain (PF03479) was used as a query, and candidate genes were selected based on the overlap of results in both the “Sequence scores” and “Domain scores” outputs.
4.3. Sequence Analysis and Structural Characteristics
The protein lengths, molecular weights, theoretical isoelectric points (pIs), instability indices, and grand average of hydropathicity of CaAHL proteins were analyzed using the TBtools-II (v2.149) software [29]. The supposed subcellular localizations of CaAHL proteins were predicted using the online tool WoLF PSORT (https://wolfpsort.hgc.jp, accessed on 7 January 2025) [30]. The protein sequences were submitted to the MEME program (version 5.5.7, https://meme-suite.org/meme/tools/meme, accessed on 7 January 2025) [31] to assess conserved motifs. The obtained conserved motifs were subjected to functional prediction using the Motif Comparison Tool Tomtom (version 5.5.7, https://meme-suite.org/meme/tools/tomtom, accessed on 8 January 2025) and predicted the potential functions of these motifs.
4.4. Chromosome Localization, Tandem Duplication, and Synteny Analysis
Chromosome locations and gene positions in pepper were obtained by searching the Sol Genomics Network. Chromosome mapping of the CaAHL gene family was visualized using MG2C (version 2.1, http://mg2c.iask.in/mg2c_v2.1, accessed on 11 January 2025) [32]. Tandem duplication events were further confirmed using the following criteria: (1) the alignment length had a coverage rate of more than 70% of the full length of the CaAHL genes; (2) the identity of the aligned region was over 70%; (3) an array of two or more genes was at less than 100 kb distance. TBtools-II (v2.149) software [29] was used to analyze the synteny of CaAHL genes across the three pepper genomes.
4.5. Phylogenetic Analysis
The phylogenetic tree was generated in the following three steps: first, the CaAHL protein sequences were imported into Clustal X to produce a multiple sequence alignment file. Second, the alignment result was used to build an unrooted tree using MEGA11 with a bootstrap of 1000 replicates and neighbor-joining (NJ) methods [33]. Third, the newly produced phylogenetic tree was visualized using the Interactive Tree of Life online website (version 7.2, https://itol.embl.de, accessed on 10 January 2025) [34].
4.6. RNA-Seq Analysis of CaAHL Genes
Transcriptome sequencing (RNA-seq) data for Capsicum line 6421 [25] (http://lifenglab.hzau.edu.cn/PepperHub/index.php, accessed on 5 January 2025) were utilized to investigate the expression profiles of the CaAHL gene family across various tissues and developmental stages. The treatment methods for all samples were based on those published by Liu et al. [25]. All data for the AHL genes were normalized (log2(FPKM+1)), and a heatmap was drawn using TBtools-II (version 2.149) software [29].
4.7. RNA Extraction and RT-qPCR Analysis
Total RNA was isolated from floral buds at various developmental stages (F1–F9) using the RNAprep Pure Plant Kit (DP421, TIANGEN, Beijing, China) according to the manufacturer’s instructions. The extracted RNA was then reverse-transcribed into cDNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Quantitative real-time PCR (RT-qPCR) was performed on a QuantStudio 3 Real-Time PCR System (Applied Biosystems/Thermo Fisher Scientific, Foster City, CA, USA) using 2×ChamQ Universal SYBR qPCR Master Mix (TransGen, Beijing, China) in accordance with the provided instructions. The primers were designed using Primer-BLAST, a tool available in NCBI (National Center for Biotechnology Information) for finding specific primers (https://www.ncbi.nlm.nih.gov/tools/primer-blast, accessed on 22 January 2025). The CaActin7 gene was used as the reference gene, with the forward primer sequence 5′-CTCGAGCAGTGTTTCCCAGT-3′ and the reverse primer sequence 5′-AGCTTCATCACCCACATAGGC-3′. Gene-specific primers were designed for CaAHLs as follows: CaAHL36 forward primer, 5′-CAACGTCATCAGGTCGATGT-3′, and reverse primer, 5′-CCAGGTGCCCATGAATAGAC-3′; CaAHL33 forward primer, 5′-GGAGTTGGCTTTACACCACA-3′, and reverse primer, 5′-TCCATGTGCAGAGAGAATGC-3′; CaAHL4 forward primer, 5′-TTTTCTTCACGTTGCCTTGTC-3′, and reverse primer, 5′- ATTGGCAGGCATGTTCGATT-3′; CaAHL23 forward primer, 5′-GAGTCTAGCGGTGGACCTAT-3′, and reverse primer, 5′-AGGTTCTCTCATCTCCAGGG-3′; CaAHL1 forward primer, 5′-GTCCTACTTCAGGGTCTGGT-3′, and reverse primer, 5′-CCAAATTGTGTCATCCACGC-3′; CaAHL26 forward primer, 5′-AGGGCTCATTCTAATTGGCG-3′, and reverse primer, 5′-GTGGGACTACTCCACCCTTA-3′; CaAHL5 forward primer, 5′-CTTCTGGGCTTCCGTTCTTT-3′, and reverse primer, 5′-AAAAGGTGGACGAAGAGCTG-3′; CaAHL35 forward primer, 5′-CTTGCTGTTTAAAGTTTTCAGTTCT-3′, and reverse primer, 5′-CCTAGAAAAGCCAAAACCCCTA-3′. qRT-PCR thermal cycling conditions were as follows: initial denaturation at 94 °C for 30 s, followed by 43 cycles of denaturation at 94 °C for 5 s and annealing/extension at 60 °C for 30 s. After amplification, a melt curve analysis was performed by heating to 95 °C, then cooling to 60 °C to verify the specificity of each amplicon. RT-qPCR experiments were conducted with three biological replicates and four technical replicates per sample. Relative gene expression levels were determined using the 2−ΔΔCt method.
4.8. Subcellular Localization
The full-length ORF sequences of CaAHL23 or CaAHL36 without the termination codon were cloned into the pCAMBIA1300-GFP vector and transformed into Agrobacterium tumefaciens GV3101. CaAHL23 or CaAHL36 fusion constructs were transformed into tobacco (Nicotiana benthamiana) leaves. After three days, fluorescence signals were observed and captured using a confocal laser scanning microscope (LSM 510 META, Carl Zeiss, Oberkochen, Germany).
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/ijms26136527/s1.
Author Contributions
Conceptualization, X.-Y.S. and Y.-L.L.; methodology, X.W.; software, X.-Y.S.; validation, Y.-L.L., Y.Z. and X.-Y.S.; formal analysis, Q.-Z.C. and Y.L.; investigation, B.-Q.T.; resources, F.L. and X.-X.Z.; data curation, X.-Y.S. and Y.-L.L.; writing—original draft preparation, X.-Y.S.; writing—review and editing, Y.-L.L., F.L. and X.-X.Z.; supervision, Y.-L.L.; project administration, F.L. and X.-X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 32372701) and China Agriculture Research System of MOF and MARA (CARS-24-A-15).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kim, S.; Park, M.; Yeom, S.-I.; Kim, Y.-M.; Lee, J.M.; Lee, H.-A.; Seo, E.; Choi, J.; Cheong, K.; Kim, K.-T.; et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 2014, 46, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, U.; Li, X.; Fan, Y.; Chang, W.; Niu, Y.; Li, J.; Qu, C.; Lu, K. Multi-omics revolution to promote plant breeding efficiency. Front. Plant Sci. 2022, 13, 1062952. [Google Scholar] [CrossRef]
- Islam, K.; Momo, J.; Rawoof, A.; Vijay, A.; Anusree, V.K.; Kumar, A.; Ramchiary, N. Integrated Use of Molecular and Omics Approaches for Breeding High Yield and Stress Resistance Chili Peppers. In Smart Plant Breeding for Vegetable Crops in Post-Genomics Era; Springer Nature: Singapore, 2023; pp. 279–335. [Google Scholar]
- Street, I.H.; Shah, P.K.; Smith, A.M.; Avery, N.; Neff, M.M. The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 2008, 54, 1–14. [Google Scholar] [CrossRef]
- Xiao, C.; Chen, F.; Yu, X.; Lin, C.; Fu, Y.-F. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 2009, 71, 39–50. [Google Scholar] [CrossRef]
- Xu, L.; Zheng, S.; Witzel, K.; Van De Slijke, E.; Baekelandt, A.; Mylle, E.; Van Damme, D.; Cheng, J.; De Jaeger, G.; Inzé, D.; et al. Chromatin attachment to the nuclear matrix represses hypocotyl elongation in Arabidopsis thaliana. Nat. Commun. 2024, 15, 1286. [Google Scholar] [CrossRef] [PubMed]
- Tayengwa, R.; Koirala, P.S.; Pierce, C.F.; Werner, B.E.; Neff, M.M. Overexpression of AtAHL20 causes delayed flowering in Arabidopsis via repression of FT expression. BMC Plant Biol. 2020, 20, 559. [Google Scholar] [CrossRef]
- Uzair, M.; Xu, D.; Schreiber, L.; Shi, J.; Liang, W.; Jung, K.-H.; Chen, M.; Luo, Z.; Zhang, Y.; Yu, J.; et al. PERSISTENT TAPETAL CELL2 Is Required for Normal Tapetal Programmed Cell Death and Pollen Wall Patterning. Plant Physiol. 2020, 182, 962–976. [Google Scholar] [CrossRef]
- Machaj, G.; Grzebelus, D. Characteristics of the AT-Hook Motif Containing Nuclear Localized (AHL) Genes in Carrot Provides Insight into Their Role in Plant Growth and Storage Root Development. Genes 2021, 12, 764. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Yang, H.; Sun, H.; Lu, P.; Yan, P.; Zhao, W.; Zeng, L.; Li, Z.; Zhang, L.; Hou, W. Characterization of AHL transcription factors and functional analysis of IbAHL10 in storage root development in sweetpotato. Sci. Hortic. 2024, 338, 113718. [Google Scholar] [CrossRef]
- Favero, D.S.; Kawamura, A.; Shibata, M.; Takebayashi, A.; Jung, J.-H.; Suzuki, T.; Jaeger, K.E.; Ishida, T.; Iwase, A.; Wigge, P.A.; et al. AT-Hook Transcription Factors Restrict Petiole Growth by Antagonizing PIFs. Curr. Biol. 2020, 30, 1454–1466.e6. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Z.; Liu, Y.; Kong, D.; Li, T.; Yu, S.; Mei, H.; Xu, X.; Liu, H.; Chen, L.; et al. A novel gene OsAHL1 improves both drought avoidance and drought tolerance in rice. Sci. Rep. 2016, 6, 30264. [Google Scholar] [CrossRef] [PubMed]
- Howden, A.J.M.; Stam, R.; Heredia, V.M.; Motion, G.B.; Have, S.T.; Hodge, K.; Amaro, T.M.M.M.; Huitema, E. Quantitative analysis of the tomato nuclear proteome during Phytophthora capsici infection unveils regulators of immunity. New Phytol. 2017, 215, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.-N.; Sun, H.-J.; Zuo, Z.-F.; Lee, D.H.; Song, P.-S.; Kang, H.-G.; Lee, H.-Y. Overexpression of ATHG1/AHL23 and ATPG3/AHL20, Arabidopsis AT-hook motif nuclear-localized genes, confers salt tolerance in transgenic Zoysia japonica. Plant Biotechnol. Rep. 2020, 14, 351–361. [Google Scholar] [CrossRef]
- Rayapuram, N.; Jarad, M.; Alhoraibi, H.M.; Bigeard, J.; Abulfaraj, A.A.; Völz, R.; Mariappan, K.G.; Almeida-Trapp, M.; Schlöffel, M.; Lastrucci, E.; et al. Chromatin phosphoproteomics unravels a function for AT-hook motif nuclear localized protein AHL13 in PAMP-triggered immunity. Proc. Natl. Acad. Sci. USA 2021, 118, e2004670118. [Google Scholar] [CrossRef]
- Bishop, E.H.; Kumar, R.; Luo, F.; Saski, C.; Sekhon, R.S. Genome-wide identification, expression profiling, and network analysis of AT-hook gene family in maize. Genomics 2020, 112, 1233–1244. [Google Scholar] [CrossRef]
- Zhao, L.; Lü, Y.; Chen, W.; Yao, J.; Li, Y.; Li, Q.; Pan, J.; Fang, S.; Sun, J.; Zhang, Y. Genome-wide identification and analyses of the AHL gene family in cotton (Gossypium). BMC Genom. 2020, 21, 69. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Zhou, W.; Xie, L.; Wang, L.; Zhang, Y.; Zhang, Q. Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genom. 2021, 22, 361. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Mishra, A. Genome-wide identification and analyses of the AHL gene family in rice (Oryza sativa). 3 Biotech 2023, 13, 248. [Google Scholar] [CrossRef]
- Yun, J.; Kim, Y.-S.; Jung, J.-H.; Seo, P.J.; Park, C.-M. The AT-hook Motif-containing Protein AHL22 Regulates Flowering Initiation by Modifying FLOWERING LOCUS T Chromatin in Arabidopsis. J. Biol. Chem. 2012, 287, 15307–15316. [Google Scholar] [CrossRef]
- Xu, Y.; Gan, E.-S.; Ito, T. The AT-hook/PPC domain protein TEK negatively regulates floral repressors including MAF4 and MAF5. Plant Signal. Behav. 2013, 8, e25006. [Google Scholar] [CrossRef]
- Lou, Y.; Xu, X.-F.; Zhu, J.; Gu, J.-N.; Blackmore, S.; Yang, Z.-N. The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat. Commun. 2014, 5, 3855. [Google Scholar] [CrossRef] [PubMed]
- Karami, O.; Rahimi, A.; Mak, P.; Horstman, A.; Boutilier, K.; Compier, M.; van der Zaal, B.; Offringa, R. An Arabidopsis AT-hook motif nuclear protein mediates somatic embryogenesis and coinciding genome duplication. Nat. Commun. 2021, 12, 2508. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhao, J.; Sun, H.; Xiong, C.; Sun, X.; Wang, X.; Wang, Z.; Jarret, R.; Wang, J.; Tang, B.; et al. Genomes of cultivated and wild Capsicum species provide insights into pepper domestication and population differentiation. Nat. Commun. 2023, 14, 5487. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S.; et al. PepperHub, an Informatics Hub for the Chili Pepper Research Community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef]
- Ambadas, D.A.; Singh, A.; Jha, R.K.; Chauhan, D.; Santhosh, B.; Sharma, V.K. Genome-wide dissection of AT-hook motif nuclear-localized gene family and their expression profiling for drought and salt stress in rice (Oryza sativa). Front. Plant Sci. 2023, 14, 1283555. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Intelligent Systems for Molecular Biology, Stanford University, Palo Alto, CA, USA, 14–17 August 1994; Volume 2, pp. 28–36. [Google Scholar]
- Chao, J.-T.; Kong, Y.-Z.; Wang, Q.; Sun, Y.; Gong, D.; Lv, J.; Liu, G. MapGene2Chrom, a tool to draw gene physical map based on Perl and SVG languages. Yi Chuan 2015, 37, 91–97. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).