The Potential of Extracellular Vesicle-Mediated Spread of Self-Amplifying RNA and a Way to Mitigate It
Abstract
1. Introduction
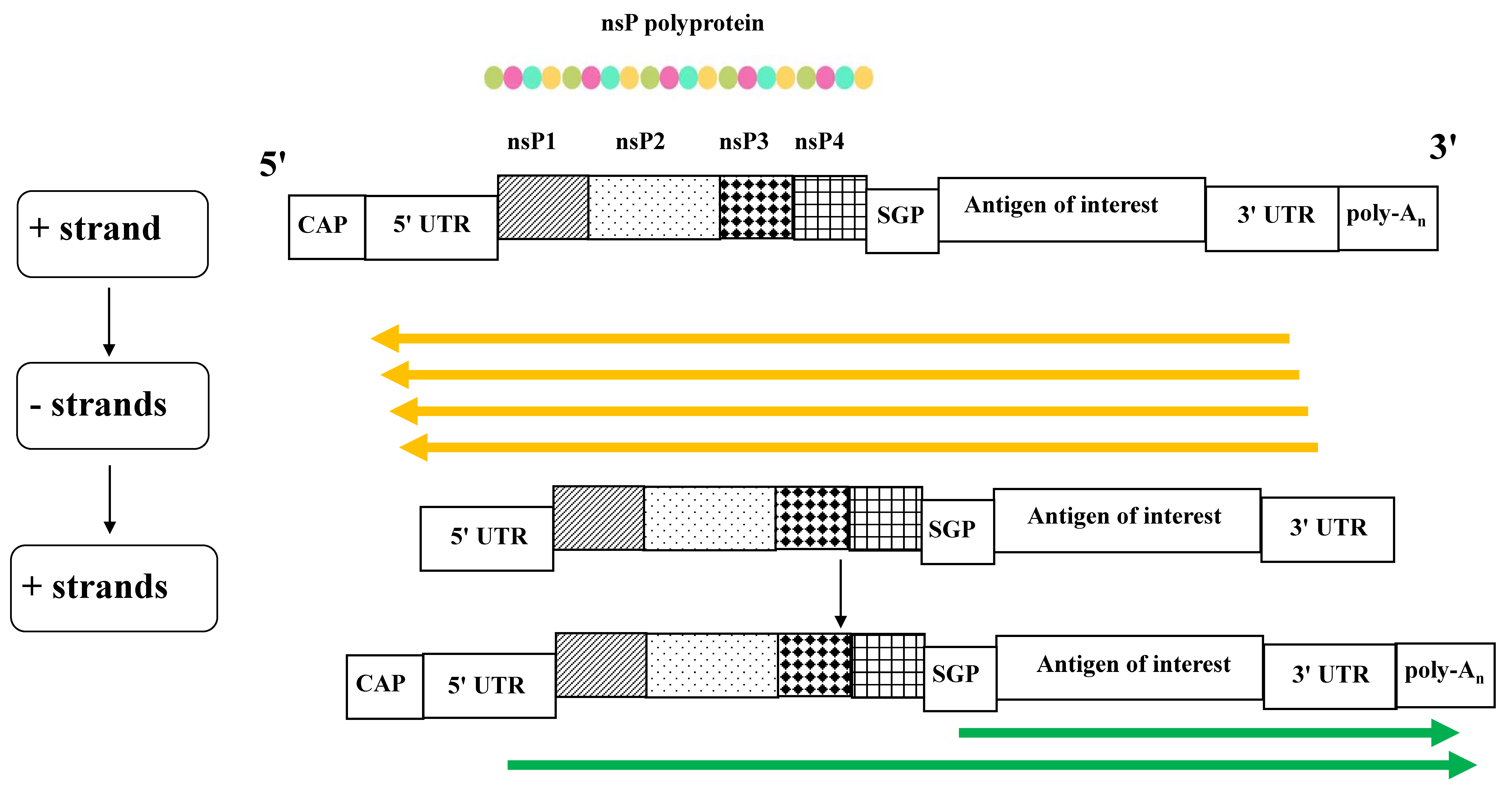
2. The saRNA Replication Cycle
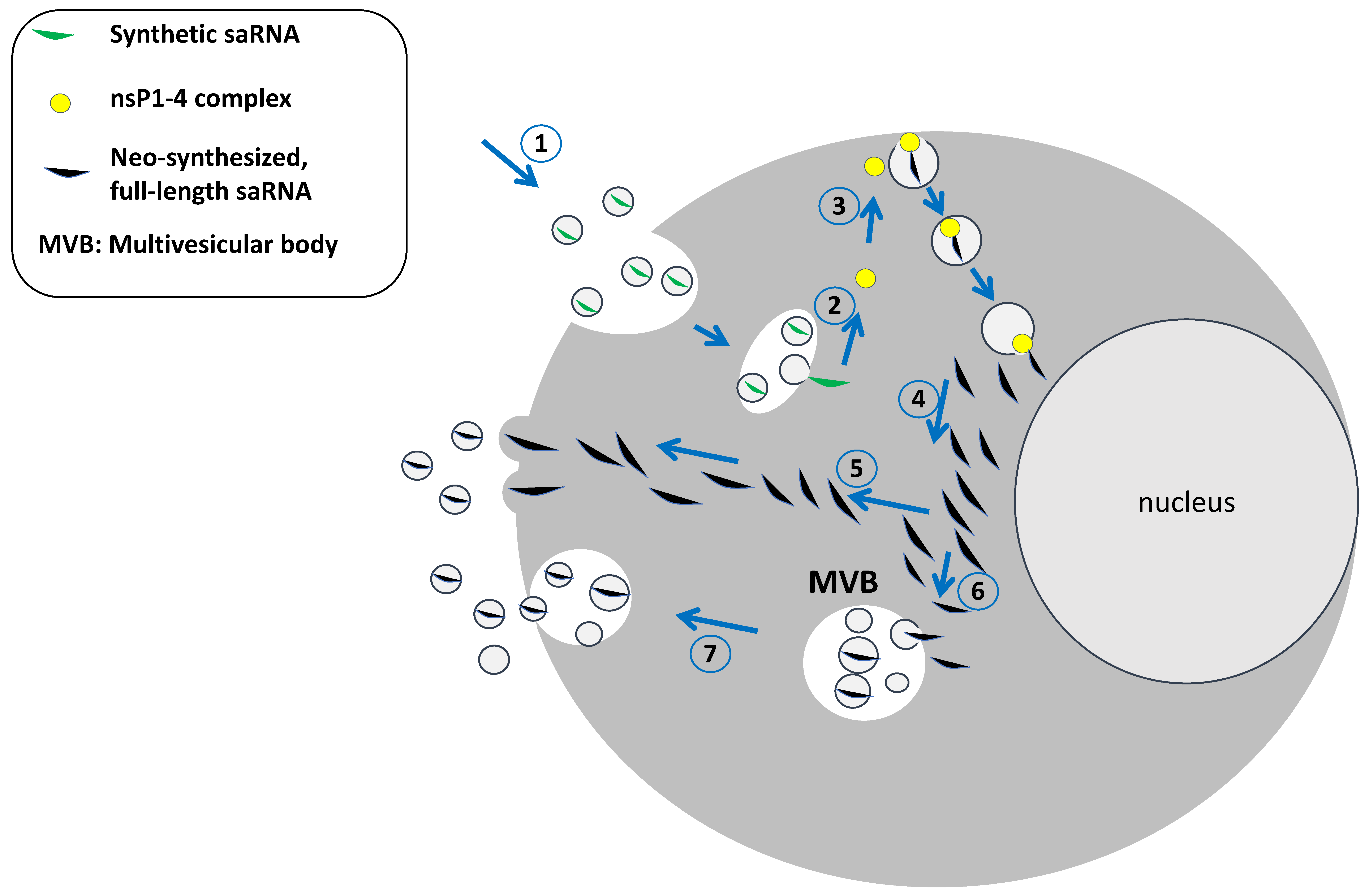
3. The Intracellular Fate of saRNA and Its Loading into Extracellular Vesicles
4. EVs as Vehicles of Propagation of the Alphavirus Genome: The Potential of EV-Associated saRNA Spread
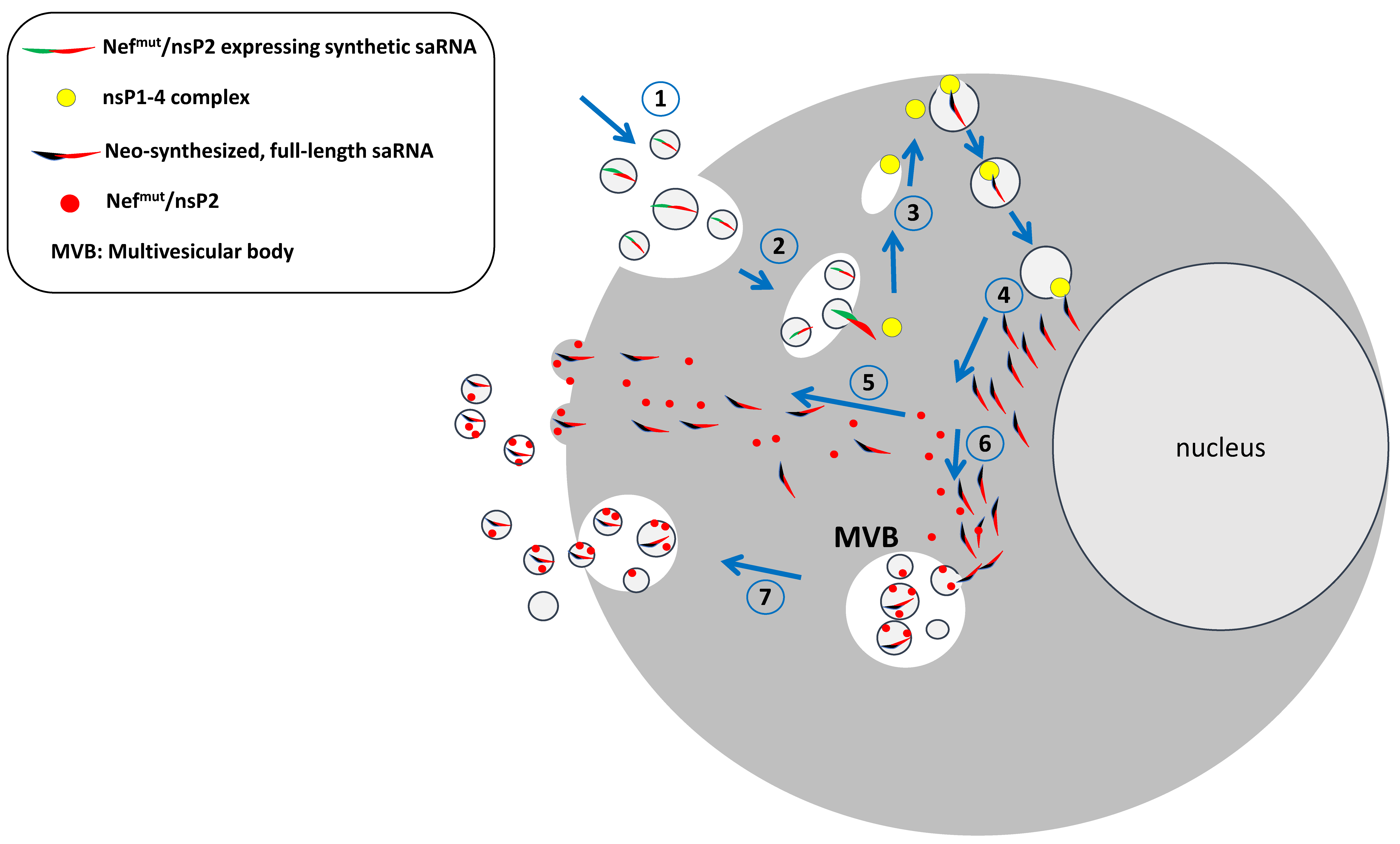
5. A Way to Control the saRNA Spread
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Available online: https://www.ema.europa.eu/en/news/meeting-highlights-committee-medicinal-products-human-use-chmp-9-12-december-2024 (accessed on 27 March 2025).
- Available online: https://www.ema.europa.eu/en/medicines/human/EPAR/kostaive (accessed on 27 March 2025).
- Hồ, N.T.; Hughes, S.G.; Ta, V.T.; Phan, L.T.; Đỗ, Q.; Nguyễn, T.V.; Phạm, A.T.V.; Thị Ngọc Đặng, M.; Nguyễn, L.V.; Trịnh, Q.V.; et al. Safety, Immunogenicity and Efficacy of the Self-Amplifying mRNA ARCT-154 COVID-19 Vaccine: Pooled Phase 1, 2, 3a and 3b Randomized, Controlled Trials. Nat. Commun. 2024, 15, 4081. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Kumagai, Y.; Kanai, M.; Iwama, Y.; Okura, I.; Minamida, T.; Yagi, Y.; Kurosawa, T.; Greener, B.; Zhang, Y.; et al. Immunogenicity and Safety of a Booster Dose of a Self-Amplifying RNA COVID-19 Vaccine (ARCT-154) versus BNT162b2 mRNA COVID-19 Vaccine: A Double-Blind, Multicentre, Randomised, Controlled, Phase 3, Non-Inferiority Trial. Lancet Infect. Dis. 2024, 24, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Pollock, K.M.; Cheeseman, H.M.; Szubert, A.J.; Libri, V.; Boffito, M.; Owen, D.; Bern, H.; McFarlane, L.R.; O’Hara, J.; Lemm, N.-M.; et al. Safety and Immunogenicity of a Self-Amplifying RNA Vaccine against COVID-19: COVAC1, a Phase I, Dose-Ranging Trial. eClinicalMedicine 2022, 44, 101262. [Google Scholar] [CrossRef]
- Szubert, A.J.; Pollock, K.M.; Cheeseman, H.M.; Alagaratnam, J.; Bern, H.; Bird, O.; Boffito, M.; Byrne, R.; Cole, T.; Cosgrove, C.A.; et al. COVAC1 Phase 2a Expanded Safety and Immunogenicity Study of a Self-Amplifying RNA Vaccine against SARS-CoV-2. eClinicalMedicine 2023, 56, 101823. [Google Scholar] [CrossRef]
- Kitonsa, J.; Serwanga, J.; Abaasa, A.; Lunkuse, J.; Cheeseman, H.M.; Ruzagira, E.; Kato, L.; Nambaziira, F.; Oluka, G.K.; Gombe, B.; et al. Safety and Immunogenicity of a Modified Self-Amplifying Ribonucleic Acid (saRNA) Vaccine Encoding SARS-CoV-2 Spike Glycoprotein in SARS-CoV-2 Seronegative and Seropositive Ugandan Individuals; Social Science Research Network: Rochester, NY, USA, 2025. [Google Scholar] [CrossRef]
- Saraf, A.; Gurjar, R.; Kaviraj, S.; Kulkarni, A.; Kumar, D.; Kulkarni, R.; Virkar, R.; Krishnan, J.; Yadav, A.; Baranwal, E.; et al. An Omicron-Specific, Self-Amplifying mRNA Booster Vaccine for COVID-19: A Phase 2/3 Randomized Trial. Nat. Med. 2024, 30, 1363–1372. [Google Scholar] [CrossRef]
- Akahata, W.; Sekida, T.; Nogimori, T.; Ode, H.; Tamura, T.; Kono, K.; Kazami, Y.; Washizaki, A.; Masuta, Y.; Suzuki, R.; et al. Safety and Immunogenicity of SARS-CoV-2 Self-Amplifying RNA Vaccine Expressing an Anchored RBD: A Randomized, Observer-Blind Phase 1 Study. Cell Rep. Med. 2023, 4, 101134. [Google Scholar] [CrossRef]
- Aboshi, M.; Matsuda, K.; Kawakami, D.; Kono, K.; Kazami, Y.; Sekida, T.; Komori, M.; Morey, A.L.; Suga, S.; Smith, J.F.; et al. Safety and Immunogenicity of VLPCOV-02, a SARS-CoV-2 Self-Amplifying RNA Vaccine with a Modified Base, 5-Methylcytosine. iScience 2024, 27, 108964. [Google Scholar] [CrossRef]
- Quintana, V.; Caillava, J.; Byk, L.A.; Mondotte, J.A.; Battini, L.; Tarte, P.; Samsa, M.M.; Filomatori, C.V.; Alvarez, D.E. Improvement of the Potency of a N1-Methylpseudouridine-Modified Self-Amplifying RNA through Mutations in the RNA-Dependent-RNA-Polymerase. bioRxiv 2024. [Google Scholar] [CrossRef]
- Maine, C.J.; Miyake-Stoner, S.J.; Spasova, D.S.; Picarda, G.; Chou, A.C.; Brand, E.D.; Olesiuk, M.D.; Domingo, C.C.; Little, H.J.; Goodman, T.T.; et al. Safety and Immunogenicity of an Optimized Self-Replicating RNA Platform for Low Dose or Single Dose Vaccine Applications: A Randomized, Open Label Phase I Study in Healthy Volunteers. Nat. Commun. 2025, 16, 456. [Google Scholar] [CrossRef]
- WHO Guideline on Nonclinical Evaluation of Vaccines, WHO Technical Report Series, No. 927. 2005. Available online: https://cdn.who.int/media/docs/default-source/biologicals/annex1nonclinical.p31-63.pdf (accessed on 8 May 2025).
- Schmidt, C.; Schnierle, B.S. Self-Amplifying RNA Vaccine Candidates: Alternative Platforms for mRNA Vaccine Development. Pathogens 2023, 12, 138. [Google Scholar] [CrossRef]
- Pietilä, M.K.; Hellström, K.; Ahola, T. Alphavirus Polymerase and RNA Replication. Virus Res. 2017, 234, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Skidmore, A.M.; Bradfute, S.B. The Life Cycle of the Alphaviruses: From an Antiviral Perspective. Antivir. Res. 2023, 209, 105476. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.B.; Lambert, L.; Kinnear, E.; Busse, D.; Erbar, S.; Reuter, K.C.; Wicke, L.; Perkovic, M.; Beissert, T.; Haas, H.; et al. Self-Amplifying RNA Vaccines Give Equivalent Protection against Influenza to mRNA Vaccines but at Much Lower Doses. Mol. Ther. 2018, 26, 446–455. [Google Scholar] [CrossRef]
- Krawczyk, P.S.; Mazur, M.; Orzeł, W.; Gewartowska, O.; Jeleń, S.; Antczak, W.; Kasztelan, K.; Brouze, A.; Matylla-Kulińska, K.; Gumińska, N.; et al. Re-Adenylation by TENT5A Enhances Efficacy of SARS-CoV-2 mRNA Vaccines. Nature 2025, 641, 984–992. [Google Scholar] [CrossRef]
- Ruiz-Guillen, M.; Gabev, E.; Quetglas, J.I.; Casales, E.; Ballesteros-Briones, M.C.; Poutou, J.; Aranda, A.; Martisova, E.; Bezunartea, J.; Ondiviela, M.; et al. Capsid-Deficient Alphaviruses Generate Propagative Infectious Microvesicles at the Plasma Membrane. Cell Mol. Life Sci. 2016, 73, 3897–3916. [Google Scholar] [CrossRef]
- Le, B.C.T.; Burassakarn, A.; Tongchai, P.; Ekalaksananan, T.; Aromseree, S.; Phanthanawiboon, S.; Polsan, Y.; Alexander, N.; Overgaard, H.J.; Pientong, C. Characterization and Involvement of Exosomes Originating from Chikungunya Virus-Infected Epithelial Cells in the Transmission of Infectious Viral Elements. Int. J. Mol. Sci. 2022, 23, 12117. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in Extracellular Vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Das, S.; Extracellular RNA Communication Consortium; Ansel, K.M.; Bitzer, M.; Breakefield, X.O.; Charest, A.; Galas, D.J.; Gerstein, M.B.; Gupta, M.; Milosavljevic, A.; et al. The Extracellular RNA Communication Consortium: Establishing Foundational Knowledge and Technologies for Extracellular RNA Research. Cell 2019, 177, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 Controls the Sorting of miRNAs into Exosomes through Binding to Specific Motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Kumar Patel, S.; Schilling, B.; Kahn, C.R. MicroRNA Sequence Codes for Small Extracellular Vesicle Release and Cellular Retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef]
- Brinkman, K.; Meyer, L.; Bickel, A.; Enderle, D.; Berking, C.; Skog, J.; Noerholm, M. Extracellular Vesicles from Plasma Have Higher Tumour RNA Fraction than Platelets. J. Extracell. Vesicles 2020, 9, 1741176. [Google Scholar] [CrossRef]
- Nawaz, M.; Heydarkhan-Hagvall, S.; Tangruksa, B.; González-King Garibotti, H.; Jing, Y.; Maugeri, M.; Kohl, F.; Hultin, L.; Reyahi, A.; Camponeschi, A.; et al. Lipid Nanoparticles Deliver the Therapeutic VEGFA mRNA In Vitro and In Vivo and Transform Extracellular Vesicles for Their Functional Extensions. Adv. Sci. 2023, 10, e2206187. [Google Scholar] [CrossRef]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between Endosomal Escape of LNP-mRNA and Loading into EVs for Transport to Other Cells. Nat. Commun. 2019, 10, 4333. [Google Scholar] [CrossRef]
- Stokes, A.; Pion, J.; Binazon, O.; Laffont, B.; Bigras, M.; Dubois, G.; Blouin, K.; Young, J.K.; Ringenberg, M.A.; Ben Abdeljelil, N.; et al. Nonclinical Safety Assessment of Repeated Administration and Biodistribution of a Novel Rabies Self-Amplifying mRNA Vaccine in Rats. Regul. Toxicol. Pharmacol. 2020, 113, 104648. [Google Scholar] [CrossRef]
- Cui, X.; Vervaeke, P.; Gao, Y.; Opsomer, L.; Sun, Q.; Snoeck, J.; Devriendt, B.; Zhong, Z.; Sanders, N.N. Immunogenicity and Biodistribution of Lipid Nanoparticle Formulated Self-Amplifying mRNA Vaccines against H5 Avian Influenza. NPJ Vaccines 2024, 9, 138. [Google Scholar] [CrossRef]
- Röltgen, K.; Nielsen, S.C.A.; Silva, O.; Younes, S.F.; Zaslavsky, M.; Costales, C.; Yang, F.; Wirz, O.F.; Solis, D.; Hoh, R.A.; et al. Immune Imprinting, Breadth of Variant Recognition, and Germinal Center Response in Human SARS-CoV-2 Infection and Vaccination. Cell 2022, 185, 1025–1040.e14. [Google Scholar] [CrossRef] [PubMed]
- Krauson, A.J.; Casimero, F.V.C.; Siddiquee, Z.; Stone, J.R. Duration of SARS-CoV-2 mRNA Vaccine Persistence and Factors Associated with Cardiac Involvement in Recently Vaccinated Patients. NPJ Vaccines 2023, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Perincheri, S.; Fleming, T.; Poulson, C.; Tiffany, B.; Bremner, R.M.; Mohanakumar, T. Circulating Exosomes with COVID Spike Protein Are Induced by BNT162b2 (Pfizer-BioNTech) Vaccination Prior to Development of Antibodies: Novel Mechanism for Immune Activation by mRNA Vaccines. J. Immunol. 2021, 207, 2405–2410. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Fleming, T.; Perincheri, S.; Smith, M.; Bremner, R.; Mohanakumar, T. Lung Transplant Recipients with SARS-CoV-2 Infection Induce Circulating Exosomes with SARS-CoV-2 Spike Protein S2 Which Are Immunogenic in Mice. J. Heart Lung Transplant. 2022, 41 (Suppl. S4), S134. [Google Scholar] [CrossRef]
- Parry, P.I.; Lefringhausen, A.; Turni, C.; Neil, C.J.; Cosford, R.; Hudson, N.J.; Gillespie, J. “Spikeopathy”: COVID-19 Spike Protein Is Pathogenic, from Both Virus and Vaccine mRNA. Biomedicines 2023, 11, 2287. [Google Scholar] [CrossRef]
- Federico, M. The Immunologic Downsides Associated with the Powerful Translation of Current COVID-19 Vaccine mRNA Can Be Overcome by Mucosal Vaccines. Vaccines 2024, 12, 1281. [Google Scholar] [CrossRef]
- Arlt, F.A.; Breuer, A.; Trampenau, E.; Boesl, F.; Kirchner, M.; Mertins, P.; Sánchez-Sendín, E.; Nasouti, M.; Mayrhofer, M.; Blüthner, M.; et al. High Serum Prevalence of Autoreactive IgG Antibodies against Peripheral Nerve Structures in Patients with Neurological Post-COVID-19 Vaccination Syndrome. Front. Immunol. 2024, 15, 1404800. [Google Scholar] [CrossRef]
- Bellucci, M.; Bozzano, F.M.; Castellano, C.; Pesce, G.; Beronio, A.; Farshchi, A.H.; Limongelli, A.; Uccelli, A.; Benedetti, L.; De Maria, A. Post-SARS-CoV-2 Infection and Post-Vaccine-Related Neurological Complications Share Clinical Features and the Same Positivity to Anti-ACE2 Antibodies. Front. Immunol. 2024, 15, 1398028. [Google Scholar] [CrossRef]
- Collins, C.P.; Herzog, C.; Vick, L.V.; Nielsen, R.; Harville, Y.I.; Longo, D.L.; Arthur, J.M.; Murphy, W.J. Sequential SARS-CoV-2 mRNA Vaccination Induces Anti-Idiotype (Anti-ACE2) Antibodies in K18 Human ACE2 Transgenic Mice. Vaccines 2025, 13, 224. [Google Scholar] [CrossRef]
- Lattanzi, L.; Federico, M. A Strategy of Antigen Incorporation into Exosomes: Comparing Cross-Presentation Levels of Antigens Delivered by Engineered Exosomes and by Lentiviral Virus-like Particles. Vaccine 2012, 30, 7229–7237. [Google Scholar] [CrossRef]
- Chiozzini, C.; Manfredi, F.; Arenaccio, C.; Ferrantelli, F.; Leone, P.; Federico, M. N-Terminal Fatty Acids of NEFMUT Are Required for the CD8+ T-Cell Immunogenicity of In Vivo Engineered Extracellular Vesicles. Vaccines 2020, 8, 243. [Google Scholar] [CrossRef]
- Karpf, A.R.; Lenches, E.; Strauss, E.G.; Strauss, J.H.; Brown, D.T. Superinfection Exclusion of Alphaviruses in Three Mosquito Cell Lines Persistently Infected with Sindbis Virus. J. Virol. 1997, 71, 7119–7123. [Google Scholar] [CrossRef] [PubMed]
- Nasar, F.; Erasmus, J.H.; Haddow, A.D.; Tesh, R.B.; Weaver, S.C. Eilat Virus Induces Both Homologous and Heterologous Interference. Virology 2015, 484, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Cherkashchenko, L.; Rausalu, K.; Basu, S.; Alphey, L.; Merits, A. Expression of Alphavirus Nonstructural Protein 2 (nsP2) in Mosquito Cells Inhibits Viral RNA Replication in Both a Protease Activity-Dependent and -Independent Manner. Viruses 2022, 14, 1327. [Google Scholar] [CrossRef]
- Martin, P.; Albagli, O.; Poggi, M.C.; Boulukos, K.E.; Pognonec, P. Development of a New Bicistronic Retroviral Vector with Strong IRES Activity. BMC Biotechnol. 2006, 6, 4. [Google Scholar] [CrossRef][Green Version]
- Di Bonito, P.; Chiozzini, C.; Arenaccio, C.; Anticoli, S.; Manfredi, F.; Olivetta, E.; Ferrantelli, F.; Falcone, E.; Ruggieri, A.; Federico, M. Antitumor HPV E7-Specific CTL Activity Elicited by in Vivo Engineered Exosomes Produced through DNA Inoculation. Int. J. Nanomed. 2017, 12, 4579–4591. [Google Scholar] [CrossRef]
- Anticoli, S.; Manfredi, F.; Chiozzini, C.; Arenaccio, C.; Olivetta, E.; Ferrantelli, F.; Capocefalo, A.; Falcone, E.; Ruggieri, A.; Federico, M. An Exosome-Based Vaccine Platform Imparts Cytotoxic T Lymphocyte Immunity Against Viral Antigens. Biotechnol. J. 2018, 13, e1700443. [Google Scholar] [CrossRef]
- Manfredi, F.; Chiozzini, C.; Ferrantelli, F.; Leone, P.; Pugliese, K.; Spada, M.; Di Virgilio, A.; Giovannelli, A.; Valeri, M.; Cara, A.; et al. Antiviral Effect of SARS-CoV-2 N-Specific CD8+ T Cells Induced in Lungs by Engineered Extracellular Vesicles. NPJ Vaccines 2023, 8, 83. [Google Scholar] [CrossRef]
- Pan, K.; Farrukh, H.; Chittepu, V.C.S.R.; Xu, H.; Pan, C.; Zhu, Z. CAR Race to Cancer Immunotherapy: From CAR T, CAR NK to CAR Macrophage Therapy. J. Exp. Clin. Cancer Res. 2022, 41, 119. [Google Scholar] [CrossRef]
- Hick, T.A.H.; Geertsema, C.; Nguyen, W.; Bishop, C.R.; van Oosten, L.; Abbo, S.R.; Dumenil, T.; van Kuppeveld, F.J.M.; Langereis, M.A.; Rawle, D.J.; et al. Safety Concern of Recombination between Self-Amplifying mRNA Vaccines and Viruses Is Mitigated in Vivo. Mol. Ther. 2024, 32, 2519–2534. [Google Scholar] [CrossRef]
- Button, J.M.; Mukhopadhyay, S. Removing the Polyanionic Cargo Requirement for Assembly of Alphavirus Core-Like Particles to Make an Empty Alphavirus Core. Viruses 2020, 12, 846. [Google Scholar] [CrossRef]
- Gould, S.J.; Booth, A.M.; Hildreth, J.E.K. The Trojan Exosome Hypothesis. Proc. Natl. Acad. Sci. USA 2003, 100, 10592–10597. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Han, Q.; Hou, Z.; Zhang, C.; Tian, Z.; Zhang, J. Exosomes Mediate Hepatitis B Virus (HBV) Transmission and NK-Cell Dysfunction. Cell Mol. Immunol. 2017, 14, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnaiah, V.; Thumann, C.; Fofana, I.; Habersetzer, F.; Pan, Q.; de Ruiter, P.E.; Willemsen, R.; Demmers, J.A.A.; Stalin Raj, V.; Jenster, G.; et al. Exosome-Mediated Transmission of Hepatitis C Virus between Human Hepatoma Huh7.5 Cells. Proc. Natl. Acad. Sci. USA 2013, 110, 13109–13113. [Google Scholar] [CrossRef] [PubMed]
- Bello-Morales, R.; Praena, B.; de la Nuez, C.; Rejas, M.T.; Guerra, M.; Galán-Ganga, M.; Izquierdo, M.; Calvo, V.; Krummenacher, C.; López-Guerrero, J.A. Role of Microvesicles in the Spread of Herpes Simplex Virus 1 in Oligodendrocytic Cells. J. Virol. 2018, 92, e00088-18. [Google Scholar] [CrossRef]
- Vora, A.; Zhou, W.; Londono-Renteria, B.; Woodson, M.; Sherman, M.B.; Colpitts, T.M.; Neelakanta, G.; Sultana, H. Arthropod EVs Mediate Dengue Virus Transmission through Interaction with a Tetraspanin Domain Containing Glycoprotein Tsp29Fb. Proc. Natl. Acad. Sci. USA 2018, 115, E6604–E6613. [Google Scholar] [CrossRef]
- Arenaccio, C.; Chiozzini, C.; Ferrantelli, F.; Leone, P.; Olivetta, E.; Federico, M. Exosomes in Therapy: Engineering, Pharmacokinetics and Future Applications. Curr. Drug Targets 2019, 20, 87–95. [Google Scholar] [CrossRef]
- Sinha, A.; Yadav, A.K.; Chakraborty, S.; Kabra, S.K.; Lodha, R.; Kumar, M.; Kulshreshtha, A.; Sethi, T.; Pandey, R.; Malik, G.; et al. Exosome-Enclosed microRNAs in Exhaled Breath Hold Potential for Biomarker Discovery in Patients with Pulmonary Diseases. J. Allergy Clin. Immunol. 2013, 132, 219–222.e7. [Google Scholar] [CrossRef]
- Mitchell, M.I.; Ben-Dov, I.Z.; Ye, K.; Liu, C.; Shi, M.; Sadoughi, A.; Shah, C.; Siddiqui, T.; Okorozo, A.; Gutierrez, M.; et al. Exhaled Breath Condensate Contains Extracellular Vesicles (EVs) That Carry miRNA Cargos of Lung Tissue Origin That Can Be Selectively Purified and Analyzed. J. Extracell. Vesicles 2024, 13, e12440. [Google Scholar] [CrossRef]
- Bano, A.; Yadav, P.; Sharma, M.; Verma, D.; Vats, R.; Chaudhry, D.; Kumar, P.; Bhardwaj, R. Extraction and Characterization of Exosomes from the Exhaled Breath Condensate and Sputum of Lung Cancer Patients and Vulnerable Tobacco Consumers—Potential Noninvasive Diagnostic Biomarker Source. J. Breath Res. 2024, 18, 046003. [Google Scholar] [CrossRef]
- Mueller, S. Existing and Emerging mRNA Vaccines and Their Environmental Impact: A Transdisciplinary Assessment. Environ. Sci. Eur. 2024, 36, 144. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Federico, M. The Potential of Extracellular Vesicle-Mediated Spread of Self-Amplifying RNA and a Way to Mitigate It. Int. J. Mol. Sci. 2025, 26, 5118. https://doi.org/10.3390/ijms26115118
Federico M. The Potential of Extracellular Vesicle-Mediated Spread of Self-Amplifying RNA and a Way to Mitigate It. International Journal of Molecular Sciences. 2025; 26(11):5118. https://doi.org/10.3390/ijms26115118
Chicago/Turabian StyleFederico, Maurizio. 2025. "The Potential of Extracellular Vesicle-Mediated Spread of Self-Amplifying RNA and a Way to Mitigate It" International Journal of Molecular Sciences 26, no. 11: 5118. https://doi.org/10.3390/ijms26115118
APA StyleFederico, M. (2025). The Potential of Extracellular Vesicle-Mediated Spread of Self-Amplifying RNA and a Way to Mitigate It. International Journal of Molecular Sciences, 26(11), 5118. https://doi.org/10.3390/ijms26115118





