Chrononutrition: Potential, Challenges, and Application in Managing Obesity
Abstract
1. Introduction
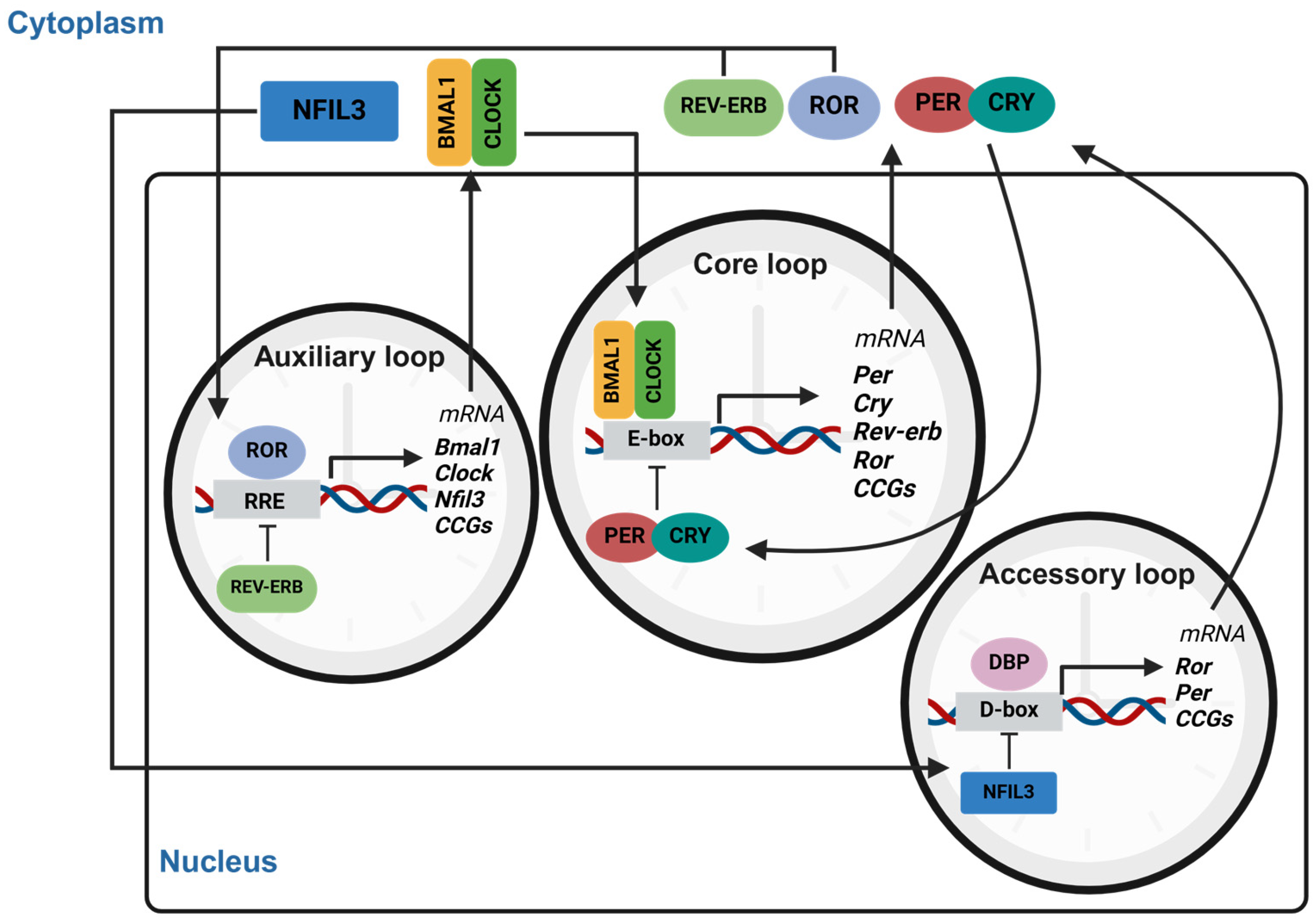
2. The Mammalian Circadian System
2.1. The Circadian Central Clock and Its Regulatory Components
2.2. The Peripheral Clocks
2.3. Adipose Tissue
2.4. Liver
2.5. Skeletal Muscle
2.6. Gut Microbiota
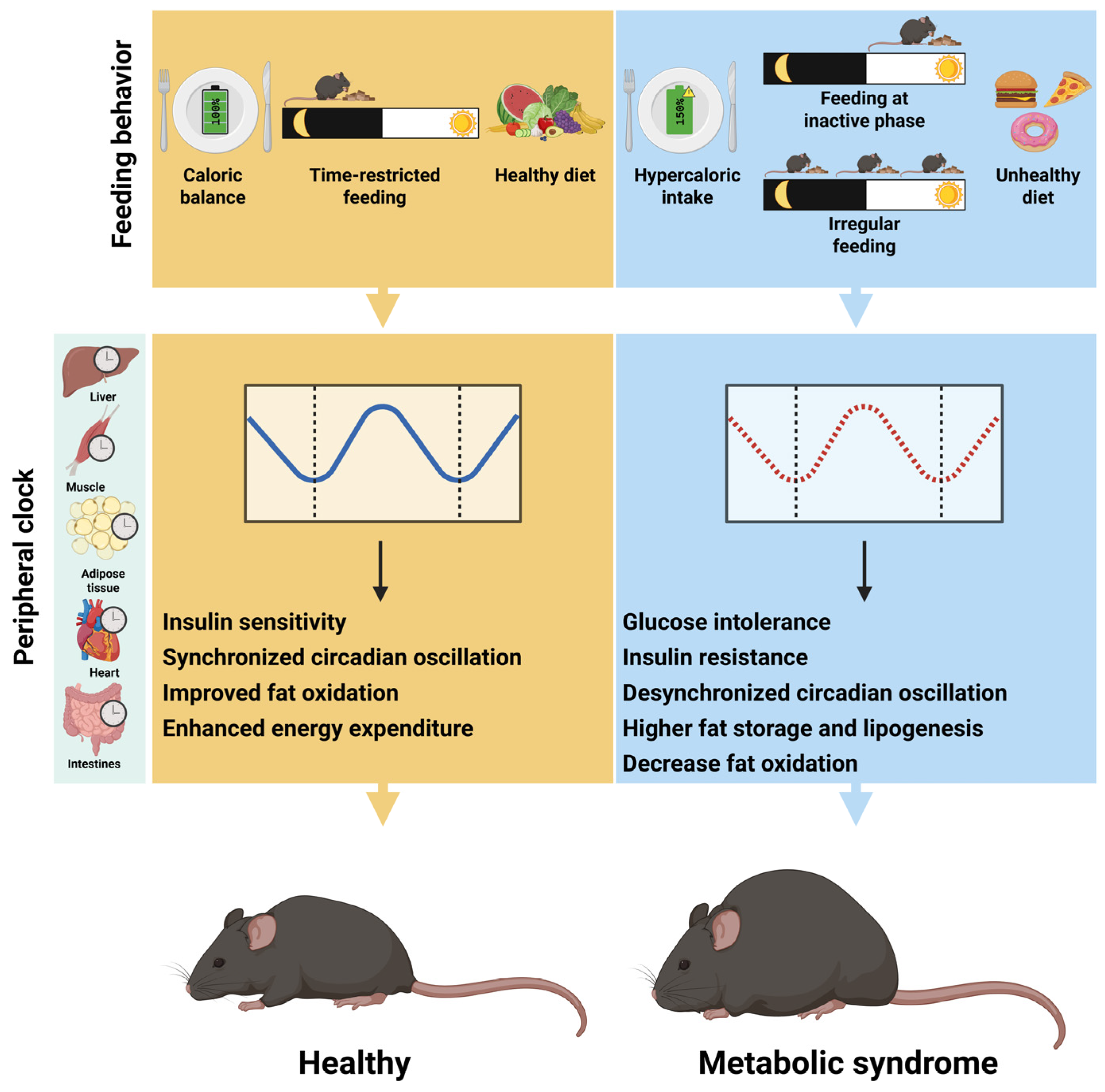
3. Feeding Behavior as a Key Driver of Peripheral Rhythms Metabolic Health
4. Comparison of Chronometabolism Regulation in Mouse and Human Models
4.1. Anatomical Differences in Mice and Humans in Relation to Chronobiology
4.1.1. SCN and Pineal Gland: The Central Clock and Its Role in Metabolic Control
4.1.2. Melatonin: A Bidirectional Signal Linking Light, Darkness, and Metabolism
4.1.3. Liver: Circadian Control of Glucose and Lipid Homeostasis
4.1.4. Adipose Tissue: Species-Specific Regulation of Energy Storage and Thermogenesis
5. Bridging the Translational Gap: Limitations of Murine Models and Emerging Diurnal Alternatives
Translating Pre-Clinical Findings to Human Trials
6. Human-Centric Limitation in Chrononutritional Research
6.1. Inconsistent Methodologies
6.2. Individual Differences
6.3. Long-Term Feasibility
6.4. Socio-Ecological Factor
7. Future Direction of Chrononutrition Research for Obesity Management
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SCN | Suprachiasmatic Nucleus |
| TRF | Time-restricted Feeding |
| TTFL | Transcriptional–translational Feedback Loop |
| ROREs | ROR Response Elements |
| eWAT | Epididymal White Adipose Tissue |
| iWAT | Inguinal White Adipose Tissue |
| BAT | Brown Adipose Tissue |
| LDL | Low-density Lipoprotein |
| VLDL | Very Low-density Lipoprotein |
| PSCK9 | Proprotein Convertase Subtilisin/kexin Type 9 |
| CREB | cAMP-responsive Element Binding Protein |
| SGLT1 | Sodium–glucose Transporter |
| PEPT1 | Peptide Transporter |
| NAFLD | Non-alcoholic Fatty Liver Disease |
| RHT | Retinohypothalamic Tract |
| SES | Socioeconomic Status |
| PACAP | Pituitary adenylate cyclase-activating polypeptide |
References
- Marcheva, B.; Ramsey, K.M.; Peek, C.B.; Affinati, A.; Maury, E.; Bass, J. Circadian Clocks and Metabolism; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 127–155. [Google Scholar] [CrossRef]
- Fagiani, F.; Di Marino, D.; Romagnoli, A.; Travelli, C.; Voltan, D.; Di Cesare Mannelli, L.; Racchi, M.; Govoni, S.; Lanni, C. Molecular regulations of circadian rhythm and implications for physiology and diseases. Signal Transduct. Target. Ther. 2022, 7, 41. [Google Scholar] [CrossRef] [PubMed]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Damiola, F.; Le Minh, N.; Preitner, N.; Kornmann, B.; Fleury-Olela, F.; Schibler, U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes. Dev. 2000, 14, 2950–2961. [Google Scholar] [CrossRef] [PubMed]
- Regmi, P.; Heilbronn, L.K. Time-Restricted Eating: Benefits, Mechanisms, and Challenges in Translation. iScience 2020, 23, 101161. [Google Scholar] [CrossRef]
- Duez, H.; Staels, B. Circadian Disruption and the Risk of Developing Obesity. Curr. Obes. Rep. 2025, 14, 20. [Google Scholar] [CrossRef]
- Schrader, L.A.; Ronnekleiv-Kelly, S.M.; Hogenesch, J.B.; Bradfield, C.A.; Malecki, K.M. Circadian disruption, clock genes, and metabolic health. J. Clin. Investig. 2024, 134, e170998. [Google Scholar] [CrossRef]
- Brum, M.C.; Filho, F.F.; Schnorr, C.C.; Bottega, G.B.; Rodrigues, T.C. Shift work and its association with metabolic disorders. Diabetol. Metab. Syndr. 2015, 7, 45. [Google Scholar] [CrossRef]
- Boege, H.L.; Bhatti, M.Z.; St-Onge, M.P. Circadian rhythms and meal timing: Impact on energy balance and body weight. Curr. Opin. Biotechnol. 2021, 70, 1–6. [Google Scholar] [CrossRef]
- Peters, B.; Vahlhaus, J.; Pivovarova-Ramich, O. Meal timing and its role in obesity and associated diseases. Front. Endocrinol. 2024, 15, 1359772. [Google Scholar] [CrossRef]
- Gill, S.; Panda, S. A Smartphone App Reveals Erratic Diurnal Eating Patterns in Humans that Can Be Modulated for Health Benefits. Cell Metab. 2015, 22, 789–798. [Google Scholar] [CrossRef]
- Kwak, J.; Jang, K.A.; Kim, H.R.; Kang, M.S.; Lee, K.W.; Shin, D. Identifying the Associations of Nightly Fasting Duration and Meal Timing with Type 2 Diabetes Mellitus Using Data from the 2016-2020 Korea National Health and Nutrition Survey. Nutrients 2023, 15, 1385. [Google Scholar] [CrossRef] [PubMed]
- Jamshed, H.; Beyl, R.A.; Della Manna, D.L.; Yang, E.S.; Ravussin, E.; Peterson, C.M. Early Time-Restricted Feeding Improves 24-Hour Glucose Levels and Affects Markers of the Circadian Clock, Aging, and Autophagy in Humans. Nutrients 2019, 11, 1234. [Google Scholar] [CrossRef] [PubMed]
- Sutton, E.F.; Beyl, R.; Early, K.S.; Cefalu, W.T.; Ravussin, E.; Peterson, C.M. Early Time-Restricted Feeding Improves Insulin Sensitivity, Blood Pressure, and Oxidative Stress Even without Weight Loss in Men with Prediabetes. Cell Metab. 2018, 27, 1212–1221.e3. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Zadourian, A.; Lo, H.C.; Gutierrez, N.R.; Shoghi, A.; Rosander, A.; Pazargadi, A.; Ormiston, C.K.; Wang, X.; Sui, J.; et al. Feasibility of time-restricted eating and impacts on cardiometabolic health in 24-h shift workers: The Healthy Heroes randomized control trial. Cell Metab. 2022, 34, 1442–1456.e7. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Zadourian, A.; Lo, H.C.; Gutierrez, N.R.; Shoghi, A.; Rosander, A.; Pazargadi, A.; Wang, X.; Fleischer, J.G.; Golshan, S.; et al. Protocol for a randomised controlled trial on the feasibility and effects of 10-hour time-restricted eating on cardiometabolic disease risk among career firefighters doing 24-hour shift work: The Healthy Heroes Study. BMJ Open 2021, 11, e045537. [Google Scholar] [CrossRef]
- Aragona, F.; Fazio, F.; Piccione, G.; Giannetto, C. Chronophysiology of domestic animals. Chronobiol. Int. 2024, 41, 888–903. [Google Scholar] [CrossRef]
- Reghunandanan, V.; Reghunandanan, R. Neurotransmitters of the suprachiasmatic nuclei. J. Circadian Rhythm. 2006, 4, 2. [Google Scholar] [CrossRef]
- Brenna, A.; Ripperger, J.A.; Saro, G.; Glauser, D.A.; Yang, Z.; Albrecht, U. PER2 mediates CREB-dependent light induction of the clock gene Per1. Sci. Rep. 2021, 11, 21766. [Google Scholar] [CrossRef]
- Uriu, K.; Tei, H. Complementary phase responses via functional differentiation of dual negative feedback loops. PLoS Comput. Biol. 2021, 17, e1008774. [Google Scholar] [CrossRef]
- Blume, C.; Garbazza, C.; Spitschan, M. Effects of light on human circadian rhythms, sleep and mood. Somnologie 2019, 23, 147–156. [Google Scholar] [CrossRef]
- Duffy, J.F.; Czeisler, C.A. Effect of Light on Human Circadian Physiology. Sleep Med. Clin. 2009, 4, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Canton, J.L.; Smith, M.R.; Choi, H.S.; Eastman, C.I. Phase delaying the human circadian clock with a single light pulse and moderate delay of the sleep/dark episode: No influence of iris color. J. Circadian Rhythm. 2009, 7, 8. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Touitou, Y.; Reinberg, A.; Touitou, D. Association between light at night, melatonin secretion, sleep deprivation, and the internal clock: Health impacts and mechanisms of circadian disruption. Life Sci. 2017, 173, 94–106. [Google Scholar] [CrossRef]
- DelRosso, L.M.; Hoque, R.; James, S.; Gonzalez-Toledo, E.; Chesson, A.L. Sleep-wake pattern following gunshot suprachiasmatic damage. J. Clin. Sleep Med. 2014, 10, 443–445. [Google Scholar] [CrossRef]
- Healy, K.L.; Morris, A.R.; Liu, A.C. Circadian Synchrony: Sleep, Nutrition, and Physical Activity. Front. Netw. Physiol. 2021, 1, 732243. [Google Scholar] [CrossRef]
- Mathew, D.; Luo, Q.; Bhatwadekar, A.D. Circadian rhythm disruption results in visual dysfunction. FASEB BioAdvances 2022, 4, 364–378. [Google Scholar] [CrossRef]
- Ramkisoensing, A.; Meijer, J.H. Synchronization of Biological Clock Neurons by Light and Peripheral Feedback Systems Promotes Circadian Rhythms and Health. Front. Neurol. 2015, 6, 128. [Google Scholar] [CrossRef]
- Finger, A.M.; Dibner, C.; Kramer, A. Coupled network of the circadian clocks: A driving force of rhythmic physiology. FEBS Lett. 2020, 594, 2734–2769. [Google Scholar] [CrossRef]
- Manoogian, E.N.C.; Panda, S. Circadian rhythms, time-restricted feeding, and healthy aging. Ageing Res. Rev. 2017, 39, 59–67. [Google Scholar] [CrossRef]
- Potter, G.D.; Skene, D.J.; Arendt, J.; Cade, J.E.; Grant, P.J.; Hardie, L.J. Circadian Rhythm and Sleep Disruption: Causes, Metabolic Consequences, and Countermeasures. Endocr. Rev. 2016, 37, 584–608. [Google Scholar] [CrossRef]
- Yanagihara, H.; Ando, H.; Hayashi, Y.; Obi, Y.; Fujimura, A. High-fat feeding exerts minimal effects on rhythmic mRNA expression of clock genes in mouse peripheral tissues. Chronobiol. Int. 2006, 23, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Adamczak, M.; Wiecek, A. The adipose tissue as an endocrine organ. Semin. Nephrol. 2013, 33, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.; Oliveira, T.; Fernandes, R. Biochemistry of adipose tissue: An endocrine organ. Arch. Med. Sci. 2013, 9, 191–200. [Google Scholar] [CrossRef]
- Scheja, L.; Heeren, J. The endocrine function of adipose tissues in health and cardiometabolic disease. Nat. Rev. Endocrinol. 2019, 15, 507–524. [Google Scholar] [CrossRef]
- Civelek, E.; Ozturk Civelek, D.; Akyel, Y.K.; Kaleli Durman, D.; Okyar, A. Circadian Dysfunction in Adipose Tissue: Chronotherapy in Metabolic Diseases. Biology 2023, 12, 1077. [Google Scholar] [CrossRef]
- Shimizu, I.; Yoshida, Y.; Minamino, T. A role for circadian clock in metabolic disease. Hypertens. Res. 2016, 39, 483–491. [Google Scholar] [CrossRef]
- Wang, S.; Lin, Y.; Gao, L.; Yang, Z.; Lin, J.; Ren, S.; Li, F.; Chen, J.; Wang, Z.; Dong, Z.; et al. PPAR-gamma integrates obesity and adipocyte clock through epigenetic regulation of Bmal1. Theranostics 2022, 12, 1589–1606. [Google Scholar] [CrossRef]
- Paschos, G.K.; Ibrahim, S.; Song, W.L.; Kunieda, T.; Grant, G.; Reyes, T.M.; Bradfield, C.A.; Vaughan, C.H.; Eiden, M.; Masoodi, M.; et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 2012, 18, 1768–1777. [Google Scholar] [CrossRef]
- Kettner, N.M.; Mayo, S.A.; Hua, J.; Lee, C.; Moore, D.D.; Fu, L. Circadian Dysfunction Induces Leptin Resistance in Mice. Cell Metab. 2015, 22, 448–459. [Google Scholar] [CrossRef]
- Martin, S.S.; Qasim, A.; Reilly, M.P. Leptin resistance: A possible interface of inflammation and metabolism in obesity-related cardiovascular disease. J. Am. Coll. Cardiol. 2008, 52, 1201–1210. [Google Scholar] [CrossRef]
- Mechanick, J.I.; Zhao, S.; Garvey, W.T. Leptin, An Adipokine With Central Importance in the Global Obesity Problem. Glob. Heart 2018, 13, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Nam, D.; Guo, B.; Chatterjee, S.; Chen, M.H.; Nelson, D.; Yechoor, V.K.; Ma, K. The adipocyte clock controls brown adipogenesis through the TGF-beta and BMP signaling pathways. J. Cell Sci. 2015, 128, 1835–1847. [Google Scholar] [CrossRef] [PubMed]
- Ribas-Latre, A.; Santos, R.B.; Fekry, B.; Tamim, Y.M.; Shivshankar, S.; Mohamed, A.M.T.; Baumgartner, C.; Kwok, C.; Gebhardt, C.; Rivera, A.; et al. Cellular and physiological circadian mechanisms drive diurnal cell proliferation and expansion of white adipose tissue. Nat. Commun. 2021, 12, 3482. [Google Scholar] [CrossRef]
- Sato, S.; Sakurai, T.; Ogasawara, J.; Takahashi, M.; Izawa, T.; Imaizumi, K.; Taniguchi, N.; Ohno, H.; Kizaki, T. A circadian clock gene, Rev-erbalpha, modulates the inflammatory function of macrophages through the negative regulation of Ccl2 expression. J. Immunol. 2014, 192, 407–417. [Google Scholar] [CrossRef]
- Rull, A.; Camps, J.; Alonso-Villaverde, C.; Joven, J. Insulin resistance, inflammation, and obesity: Role of monocyte chemoattractant protein-1 (or CCL2) in the regulation of metabolism. Mediat. Inflamm. 2010, 2010, 326580. [Google Scholar] [CrossRef]
- He, W.; Wang, H.; Yang, G.; Zhu, L.; Liu, X. The Role of Chemokines in Obesity and Exercise-Induced Weight Loss. Biomolecules 2024, 14, 1121. [Google Scholar] [CrossRef]
- Xu, H.; Li, H.; Woo, S.L.; Kim, S.M.; Shende, V.R.; Neuendorff, N.; Guo, X.; Guo, T.; Qi, T.; Pei, Y.; et al. Myeloid cell-specific disruption of Period1 and Period2 exacerbates diet-induced inflammation and insulin resistance. J. Biol. Chem. 2014, 289, 16374–16388. [Google Scholar] [CrossRef]
- Soon, G.S.T.; Torbenson, M. The Liver and Glycogen: In Sickness and in Health. Int. J. Mol. Sci. 2023, 24, 6133. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef]
- Koronowski, K.B.; Kinouchi, K.; Welz, P.S.; Smith, J.G.; Zinna, V.M.; Shi, J.; Samad, M.; Chen, S.; Magnan, C.N.; Kinchen, J.M.; et al. Defining the Independence of the Liver Circadian Clock. Cell 2019, 177, 1448–1462.e14. [Google Scholar] [CrossRef]
- Ferrell, J.M. Circadian rhythms and inflammatory diseases of the liver and gut. Liver Res. 2023, 7, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Xiong, Y.; Borck, P.C.; Jang, C.; Doulias, P.T.; Papazyan, R.; Fang, B.; Jiang, C.; Zhang, Y.; Briggs, E.R.; et al. Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 2018, 174, 831–842.e12. [Google Scholar] [CrossRef] [PubMed]
- Adamovich, Y.; Aviram, R.; Asher, G. The emerging roles of lipids in circadian control. Biochim. Biophys. Acta 2015, 1851, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Tognini, P.; Murakami, M.; Liu, Y.; Eckel-Mahan, K.L.; Newman, J.C.; Verdin, E.; Baldi, P.; Sassone-Corsi, P. Distinct Circadian Signatures in Liver and Gut Clocks Revealed by Ketogenic Diet. Cell Metab. 2017, 26, 523–538.e5. [Google Scholar] [CrossRef]
- Gaucher, J.; Kinouchi, K.; Ceglia, N.; Montellier, E.; Peleg, S.; Greco, C.M.; Schmidt, A.; Forne, I.; Masri, S.; Baldi, P.; et al. Distinct metabolic adaptation of liver circadian pathways to acute and chronic patterns of alcohol intake. Proc. Natl. Acad. Sci. USA 2019, 116, 25250–25259. [Google Scholar] [CrossRef]
- Sozio, M.; Crabb, D.W. Alcohol and lipid metabolism. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E10–E16. [Google Scholar] [CrossRef]
- Jeon, S.; Carr, R. Alcohol effects on hepatic lipid metabolism. J. Lipid Res. 2020, 61, 470–479. [Google Scholar] [CrossRef]
- Lu, Z.; Li, X.; Wang, M.; Zhang, X.; Zhuang, R.; Wu, F.; Li, W.; Zhu, W.; Zhang, B. Liver-Specific Bmal1 Depletion Reverses the Beneficial Effects of Nobiletin on Liver Cholesterol Homeostasis in Mice Fed with High-Fat Diet. Nutrients 2023, 15, 2547. [Google Scholar] [CrossRef]
- Guan, D.; Lazar, M.A. Interconnections between circadian clocks and metabolism. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Ma, D.; Liu, T.; Chang, L.; Rui, C.; Xiao, Y.; Li, S.; Hogenesch, J.B.; Chen, Y.E.; Lin, J.D. The Liver Clock Controls Cholesterol Homeostasis through Trib1 Protein-mediated Regulation of PCSK9/Low Density Lipoprotein Receptor (LDLR) Axis. J. Biol. Chem. 2015, 290, 31003–31012. [Google Scholar] [CrossRef]
- Ma, X.; Zhou, Z.; Chen, Y.; Wu, Y.; Liu, Y. RBP4 functions as a hepatokine in the regulation of glucose metabolism by the circadian clock in mice. Diabetologia 2016, 59, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.E.; Yang, Q.; Bluher, M.; Hammarstedt, A.; Ciaraldi, T.P.; Henry, R.R.; Wason, C.J.; Oberbach, A.; Jansson, P.A.; Smith, U.; et al. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N. Engl. J. Med. 2006, 354, 2552–2563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.E.; Liu, Y.; Dentin, R.; Pongsawakul, P.Y.; Liu, A.C.; Hirota, T.; Nusinow, D.A.; Sun, X.; Landais, S.; Kodama, Y.; et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 2010, 16, 1152–1156. [Google Scholar] [CrossRef]
- Narasimamurthy, R.; Hatori, M.; Nayak, S.K.; Liu, F.; Panda, S.; Verma, I.M. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 12662–12667. [Google Scholar] [CrossRef]
- Yang, Z.; Tsuchiya, H.; Zhang, Y.; Lee, S.; Liu, C.; Huang, Y.; Vargas, G.M.; Wang, L. REV-ERBalpha Activates C/EBP Homologous Protein to Control Small Heterodimer Partner-Mediated Oscillation of Alcoholic Fatty Liver. Am. J. Pathol. 2016, 186, 2909–2920. [Google Scholar] [CrossRef]
- Vaca-Dempere, M.; Kumar, A.; Sica, V.; Munoz-Canoves, P. Running skeletal muscle clocks on time- the determining factors. Exp. Cell Res. 2022, 413, 112989. [Google Scholar] [CrossRef]
- McCarthy, J.J.; Andrews, J.L.; McDearmon, E.L.; Campbell, K.S.; Barber, B.K.; Miller, B.H.; Walker, J.R.; Hogenesch, J.B.; Takahashi, J.S.; Esser, K.A. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genom. 2007, 31, 86–95. [Google Scholar] [CrossRef]
- Wolff, G.; Esser, K.A. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med. Sci. Sports Exerc. 2012, 44, 1663–1670. [Google Scholar] [CrossRef]
- Nakao, R.; Shimba, S.; Oishi, K. Ketogenic diet induces expression of the muscle circadian gene Slc25a25 via neural pathway that might be involved in muscle thermogenesis. Sci. Rep. 2017, 7, 2885. [Google Scholar] [CrossRef]
- Kondratov, R.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Vykhovanets, O.V.; Antoch, M.P. Early aging and age-related pathologies in mice deficient in BMAL1, the core componentof the circadian clock. Genes. Dev. 2006, 20, 1868–1873. [Google Scholar] [CrossRef]
- Dyar, K.A.; Ciciliot, S.; Wright, L.E.; Bienso, R.S.; Tagliazucchi, G.M.; Patel, V.R.; Forcato, M.; Paz, M.I.; Gudiksen, A.; Solagna, F.; et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 2014, 3, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Schroder, E.A.; Harfmann, B.D.; Zhang, X.; Srikuea, R.; England, J.H.; Hodge, B.A.; Wen, Y.; Riley, L.A.; Yu, Q.; Christie, A.; et al. Intrinsic muscle clock is necessary for musculoskeletal health. J. Physiol. 2015, 593, 5387–5404. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Arthur, S.; Haynes, J.; Butts, M.R.; Nepal, N.; Sundaram, U. The Role of Gut Microbiota and Metabolites in Obesity-Associated Chronic Gastrointestinal Disorders. Nutrients 2022, 14, 624. [Google Scholar] [CrossRef]
- Boulange, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.E. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef]
- Zhang, M.; Zhou, C.; Li, X.; Li, H.; Han, Q.; Chen, Z.; Tang, W.; Yin, J. Interactions between Gut Microbiota, Host Circadian Rhythms, and Metabolic Diseases. Adv. Nutr. 2025, 16, 100416. [Google Scholar] [CrossRef]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Zilberman-Schapira, G.; Suez, J.; Tengeler, A.C.; Abramson, L.; Katz, M.N.; Korem, T.; Zmora, N.; et al. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell 2014, 159, 514–529. [Google Scholar] [CrossRef]
- Frazier, K.; Chang, E.B. Intersection of the Gut Microbiome and Circadian Rhythms in Metabolism. Trends Endocrinol. Metab. 2020, 31, 25–36. [Google Scholar] [CrossRef]
- Hills, R.D., Jr.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrients 2019, 11, 1613. [Google Scholar] [CrossRef]
- Gutierrez Lopez, D.E.; Lashinger, L.M.; Weinstock, G.M.; Bray, M.S. Circadian rhythms and the gut microbiome synchronize the host’s metabolic response to diet. Cell Metab. 2021, 33, 873–887. [Google Scholar] [CrossRef]
- Liang, X.; Bushman, F.D.; FitzGerald, G.A. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. USA 2015, 112, 10479–10484. [Google Scholar] [CrossRef]
- Soliz-Rueda, J.R.; Cuesta-Marti, C.; O’Mahony, S.M.; Clarke, G.; Schellekens, H.; Muguerza, B. Gut microbiota and eating behaviour in circadian syndrome. Trends Endocrinol. Metab. 2025, 36, 15–28. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Lazar, M.A. Circadian Regulation of Gene Expression and Metabolism in the Liver. Semin. Liver Dis. 2022, 42, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Pickel, L.; Sung, H.K. Feeding Rhythms and the Circadian Regulation of Metabolism. Front. Nutr. 2020, 7, 39. [Google Scholar] [CrossRef]
- Mukherji, A.; Kobiita, A.; Damara, M.; Misra, N.; Meziane, H.; Champy, M.F.; Chambon, P. Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. USA 2015, 112, E6691–E6698. [Google Scholar] [CrossRef]
- Novakova, M.; Polidarova, L.; Sladek, M.; Sumova, A. Restricted feeding regime affects clock gene expression profiles in the suprachiasmatic nucleus of rats exposed to constant light. Neuroscience 2011, 197, 65–71. [Google Scholar] [CrossRef]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef]
- Froy, O.; Garaulet, M. The Circadian Clock in White and Brown Adipose Tissue: Mechanistic, Endocrine, and Clinical Aspects. Endocr. Rev. 2018, 39, 261–273. [Google Scholar] [CrossRef]
- Adamovich, Y.; Rousso-Noori, L.; Zwighaft, Z.; Neufeld-Cohen, A.; Golik, M.; Kraut-Cohen, J.; Wang, M.; Han, X.; Asher, G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. Cell Metab. 2014, 19, 319–330. [Google Scholar] [CrossRef]
- Maury, E.; Ramsey, K.M.; Bass, J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ. Res. 2010, 106, 447–462. [Google Scholar] [CrossRef]
- Acosta-Rodriguez, V.A.; Rijo-Ferreira, F.; van Rosmalen, L.; Izumo, M.; Park, N.; Joseph, C.; Hepler, C.; Thorne, A.K.; Stubblefield, J.; Bass, J.; et al. Misaligned feeding uncouples daily rhythms within brown adipose tissue and between peripheral clocks. Cell Rep. 2024, 43, 114523. [Google Scholar] [CrossRef]
- Duez, H.; Staels, B. Rev-erb alpha gives a time cue to metabolism. FEBS Lett. 2008, 582, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Jeong, J.H.; Hong, S.C. The impact of sleep and circadian disturbance on hormones and metabolism. Int. J. Endocrinol. 2015, 2015, 591729. [Google Scholar] [CrossRef] [PubMed]
- Bideyan, L.; Nagari, R.; Tontonoz, P. Hepatic transcriptional responses to fasting and feeding. Genes Dev. 2021, 35, 635–657. [Google Scholar] [CrossRef]
- Andreani, T.S.; Itoh, T.Q.; Yildirim, E.; Hwangbo, D.S.; Allada, R. Genetics of Circadian Rhythms. Sleep. Med. Clin. 2015, 10, 413–421. [Google Scholar] [CrossRef]
- Zarrinpar, A.; Chaix, A.; Yooseph, S.; Panda, S. Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 2014, 20, 1006–1017. [Google Scholar] [CrossRef]
- Nobs, S.P.; Tuganbaev, T.; Elinav, E. Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Rep. 2019, 20, e47129. [Google Scholar] [CrossRef]
- Van Drunen, R.; Eckel-Mahan, K. Circadian Rhythms of the Hypothalamus: From Function to Physiology. Clocks Sleep. 2021, 3, 189–226. [Google Scholar] [CrossRef]
- Waterson, M.J.; Horvath, T.L. Neuronal Regulation of Energy Homeostasis: Beyond the Hypothalamus and Feeding. Cell Metab. 2015, 22, 962–970. [Google Scholar] [CrossRef]
- Gillette, M.U. Biological Rhythms and Sleep. In Encyclopedia of Neuroscience; Binder, M.D., Hirokawa, N., Windhorst, U., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 408–414. [Google Scholar] [CrossRef]
- Magni, P.; Dozio, E.; Ruscica, M.; Celotti, F.; Masini, M.A.; Prato, P.; Broccoli, M.; Mambro, A.; More, M.; Strollo, F. Feeding behavior in mammals including humans. Ann. N. Y. Acad. Sci. 2009, 1163, 221–232. [Google Scholar] [CrossRef]
- Scheving, L.A. Biological clocks and the digestive system. Gastroenterology 2000, 119, 536–549. [Google Scholar] [CrossRef]
- Astiz, M.; Heyde, I.; Oster, H. Mechanisms of Communication in the Mammalian Circadian Timing System. Int. J. Mol. Sci. 2019, 20, 343. [Google Scholar] [CrossRef] [PubMed]
- Kumar Jha, P.; Challet, E.; Kalsbeek, A. Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Mol. Cell. Endocrinol. 2015, 418 Pt 1, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Martchenko, A.; Martchenko, S.E.; Biancolin, A.D.; Brubaker, P.L. Circadian Rhythms and the Gastrointestinal Tract: Relationship to Metabolism and Gut Hormones. Endocrinology 2020, 161, bqaa167. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.; Vieira-Silva, S.; Liston, A.; Raes, J. How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef]
- Teichman, E.M.; O’Riordan, K.J.; Gahan, C.G.M.; Dinan, T.G.; Cryan, J.F. When Rhythms Meet the Blues: Circadian Interactions with the Microbiota-Gut-Brain Axis. Cell Metab. 2020, 31, 448–471. [Google Scholar] [CrossRef]
- Van Cauter, E.; Polonsky, K.S.; Scheen, A.J. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr. Rev. 1997, 18, 716–738. [Google Scholar] [CrossRef]
- Balakrishnan, A.; Tavakkolizadeh, A.; Rhoads, D.B. Circadian clock genes and implications for intestinal nutrient uptake. J. Nutr. Biochem. 2012, 23, 417–422. [Google Scholar] [CrossRef][Green Version]
- Hussain, M.M.; Pan, X. Circadian Regulation of Macronutrient Absorption. J. Biol. Rhythm. 2015, 30, 459–469. [Google Scholar] [CrossRef]
- Pan, X.; Terada, T.; Okuda, M.; Inui, K. The diurnal rhythm of the intestinal transporters SGLT1 and PEPT1 is regulated by the feeding conditions in rats. J. Nutr. 2004, 134, 2211–2215. [Google Scholar] [CrossRef]
- Ogasawara, J.; Matsumoto, N.; Takeuchi, Y.; Yamashiro, K.; Yasui, M.; Ikegaya, Y. Lengthened circadian rhythms in mice with self-controlled ambient light intensity. Sci. Rep. 2024, 14, 7778. [Google Scholar] [CrossRef]
- Siepka, S.M.; Takahashi, J.S. Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol. 2005, 393, 230–239. [Google Scholar] [CrossRef] [PubMed]
- de Assis, L.V.M.; Oster, H. The circadian clock and metabolic homeostasis: Entangled networks. Cell Mol. Life Sci. 2021, 78, 4563–4587. [Google Scholar] [CrossRef] [PubMed]
- Kondratov, R.V.; Chernov, M.V.; Kondratova, A.A.; Gorbacheva, V.Y.; Gudkov, A.V.; Antoch, M.P. BMAL1-dependent circadian oscillation of nuclear CLOCK: Posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003, 17, 1921–1932. [Google Scholar] [CrossRef]
- Lee, D.Y.; Jung, I.; Park, S.Y.; Yu, J.H.; Seo, J.A.; Kim, K.J.; Kim, N.H.; Yoo, H.J.; Kim, S.G.; Choi, K.M.; et al. Attention to Innate Circadian Rhythm and the Impact of Its Disruption on Diabetes. Diabetes Metab. J. 2024, 48, 37–52. [Google Scholar] [CrossRef]
- Ono, D.; Honma, S.; Nakajima, Y.; Kuroda, S.; Enoki, R.; Honma, K.I. Dissociation of Per1 and Bmal1 circadian rhythms in the suprachiasmatic nucleus in parallel with behavioral outputs. Proc. Natl. Acad. Sci. USA 2017, 114, E3699–E3708. [Google Scholar] [CrossRef]
- Peschke, E.; Muhlbauer, E. New evidence for a role of melatonin in glucose regulation. Best Pract. Res. Clin. Endocrinol. Metab. 2010, 24, 829–841. [Google Scholar] [CrossRef]
- Richter, H.G.; Torres-Farfan, C.; Rojas-Garcia, P.P.; Campino, C.; Torrealba, F.; Seron-Ferre, M. The circadian timing system: Making sense of day/night gene expression. Biol. Res. 2004, 37, 11–28. [Google Scholar] [CrossRef]
- Takahashi, J.S.; Hong, H.K.; Ko, C.H.; McDearmon, E.L. The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 2008, 9, 764–775. [Google Scholar] [CrossRef]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef]
- Berthier, A.; Johanns, M.; Zummo, F.P.; Lefebvre, P.; Staels, B. PPARs in liver physiology. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166097. [Google Scholar] [CrossRef]
- Dibner, C.; Schibler, U. Circadian timing of metabolism in animal models and humans. J. Intern. Med. 2015, 277, 513–527. [Google Scholar] [CrossRef] [PubMed]
- Saran, A.R.; Dave, S.; Zarrinpar, A. Circadian Rhythms in the Pathogenesis and Treatment of Fatty Liver Disease. Gastroenterology 2020, 158, 1948–1966.e1. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Kim, H.; Ali, A.; Zheng, Z.; Zhang, K. Interaction between stress responses and circadian metabolism in metabolic disease. Liver Res. 2017, 1, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Kazak, L.; Spiegelman, B.M. New Advances in Adaptive Thermogenesis: UCP1 and Beyond. Cell Metab. 2019, 29, 27–37. [Google Scholar] [CrossRef]
- Peng, X.; Chen, Y. The emerging role of circadian rhythms in the development and function of thermogenic fat. Front. Endocrinol. 2023, 14, 1175845. [Google Scholar] [CrossRef]
- Razzoli, M.; Emmett, M.J.; Lazar, M.A.; Bartolomucci, A. beta-Adrenergic receptors control brown adipose UCP-1 tone and cold response without affecting its circadian rhythmicity. FASEB J. 2018, 32, 5640–5646. [Google Scholar] [CrossRef]
- Arendt, J.; Aulinas, A. Physiology of the Pineal Gland and Melatonin. In Endotext; Feingold, K.R., Ahmed, S.F., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Foulkes, N.S.; Borjigin, J.; Snyder, S.H.; Sassone-Corsi, P. Rhythmic transcription: The molecular basis of circadian melatonin synthesis. Trends Neurosci. 1997, 20, 487–492. [Google Scholar] [CrossRef]
- Cardinali, D.P.; Pevet, P. Basic aspects of melatonin action. Sleep Med. Rev. 1998, 2, 175–190. [Google Scholar] [CrossRef]
- Patton, A.P.; Hastings, M.H. The suprachiasmatic nucleus. Curr. Biol. 2018, 28, R816–R822. [Google Scholar] [CrossRef]
- Doghramji, K. Melatonin and its receptors: A new class of sleep-promoting agents. J. Clin. Sleep Med. 2007, 3, S17–S23. [Google Scholar] [CrossRef]
- Begemann, K.; Rawashdeh, O.; Olejniczak, I.; Pilorz, V.; de Assis, L.V.M.; Osorio-Mendoza, J.; Oster, H. Endocrine regulation of circadian rhythms. npj Biol. Timing Sleep 2025, 2, 10. [Google Scholar] [CrossRef]
- Oster, H.; Challet, E.; Ott, V.; Arvat, E.; de Kloet, E.R.; Dijk, D.J.; Lightman, S.; Vgontzas, A.; Van Cauter, E. The Functional and Clinical Significance of the 24-Hour Rhythm of Circulating Glucocorticoids. Endocr. Rev. 2017, 38, 3–45. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, T.; Abe, K.; Mekada, K.; Yoshiki, A.; Kato, T. Genetic variation of melatonin productivity in laboratory mice under domestication. Proc. Natl. Acad. Sci. USA 2010, 107, 6412–6417. [Google Scholar] [CrossRef]
- Zhang, Z.; Silveyra, E.; Jin, N.; Ribelayga, C.P. A congenic line of the C57BL/6J mouse strain that is proficient in melatonin synthesis. J. Pineal Res. 2018, 65, e12509. [Google Scholar] [CrossRef]
- Benloucif, S.; Guico, M.J.; Reid, K.J.; Wolfe, L.F.; L’Hermite-Baleriaux, M.; Zee, P.C. Stability of melatonin and temperature as circadian phase markers and their relation to sleep times in humans. J. Biol. Rhythm. 2005, 20, 178–188. [Google Scholar] [CrossRef]
- Korf, H.W.; von Gall, C. Mouse Models in Circadian Rhythm and Melatonin Research. J. Pineal Res. 2024, 76, e12986. [Google Scholar] [CrossRef]
- von Gall, C.; Lewy, A.; Schomerus, C.; Vivien-Roels, B.; Pevet, P.; Korf, H.W.; Stehle, J.H. Transcription factor dynamics and neuroendocrine signalling in the mouse pineal gland: A comparative analysis of melatonin-deficient C57BL mice and melatonin-proficient C3H mice. Eur. J. Neurosci. 2000, 12, 964–972. [Google Scholar] [CrossRef]
- Hong, S.H.; Lee, D.B.; Yoon, D.W.; Kim, J. Melatonin Improves Glucose Homeostasis and Insulin Sensitivity by Mitigating Inflammation and Activating AMPK Signaling in a Mouse Model of Sleep Fragmentation. Cells 2024, 13, 470. [Google Scholar] [CrossRef]
- Bayon, V.; Berger, M.; Solelhac, G.; Haba-Rubio, J.; Marques-Vidal, P.; Strippoli, M.P.; Preisig, M.; Leger, D.; Heinzer, R. Impact of night and shift work on metabolic syndrome and its components: A cross-sectional study in an active middle-to-older-aged population-based sample. BMJ Open 2022, 12, e053591. [Google Scholar] [CrossRef]
- Chaput, J.P.; McHill, A.W.; Cox, R.C.; Broussard, J.L.; Dutil, C.; da Costa, B.G.G.; Sampasa-Kanyinga, H.; Wright, K.P., Jr. The role of insufficient sleep and circadian misalignment in obesity. Nat. Rev. Endocrinol. 2023, 19, 82–97. [Google Scholar] [CrossRef]
- Guan, Q.; Wang, Z.; Cao, J.; Dong, Y.; Chen, Y. Mechanisms of Melatonin in Obesity: A Review. Int. J. Mol. Sci. 2021, 23, 218. [Google Scholar] [CrossRef] [PubMed]
- James, S.M.; Honn, K.A.; Gaddameedhi, S.; Van Dongen, H.P.A. Shift Work: Disrupted Circadian Rhythms and Sleep-Implications for Health and Well-Being. Curr. Sleep. Med. Rep. 2017, 3, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Fernandez, O.H.; Liu, J.A.; Nelson, R.J. Circadian Rhythms Disrupted by Light at Night and Mistimed Food Intake Alter Hormonal Rhythms and Metabolism. Int. J. Mol. Sci. 2023, 24, 3392. [Google Scholar] [CrossRef] [PubMed]
- Bonmati-Carrion, M.A.; Arguelles-Prieto, R.; Martinez-Madrid, M.J.; Reiter, R.; Hardeland, R.; Rol, M.A.; Madrid, J.A. Protecting the melatonin rhythm through circadian healthy light exposure. Int. J. Mol. Sci. 2014, 15, 23448–23500. [Google Scholar] [CrossRef]
- Korf, H.W.; von Gall, C. Mice, melatonin and the circadian system. Mol. Cell Endocrinol. 2006, 252, 57–68. [Google Scholar] [CrossRef]
- Wehrens, S.M.T.; Christou, S.; Isherwood, C.; Middleton, B.; Gibbs, M.A.; Archer, S.N.; Skene, D.J.; Johnston, J.D. Meal Timing Regulates the Human Circadian System. Curr. Biol. 2017, 27, 1768–1775.e3. [Google Scholar] [CrossRef]
- BaHammam, A.S.; Pirzada, A. Timing Matters: The Interplay between Early Mealtime, Circadian Rhythms, Gene Expression, Circadian Hormones, and Metabolism-A Narrative Review. Clocks Sleep 2023, 5, 507–535. [Google Scholar] [CrossRef]
- McGinnis, G.R.; Young, M.E. Circadian regulation of metabolic homeostasis: Causes and consequences. Nat. Sci. Sleep 2016, 8, 163–180. [Google Scholar] [CrossRef]
- Paoli, A.; Tinsley, G.; Bianco, A.; Moro, T. The Influence of Meal Frequency and Timing on Health in Humans: The Role of Fasting. Nutrients 2019, 11, 719. [Google Scholar] [CrossRef]
- Zani, F.; Breasson, L.; Becattini, B.; Vukolic, A.; Montani, J.P.; Albrecht, U.; Provenzani, A.; Ripperger, J.A.; Solinas, G. PER2 promotes glucose storage to liver glycogen during feeding and acute fasting by inducing Gys2 PTG and G L expression. Mol. Metab. 2013, 2, 292–305. [Google Scholar] [CrossRef]
- Gamble, K.L.; Berry, R.; Frank, S.J.; Young, M.E. Circadian clock control of endocrine factors. Nat. Rev. Endocrinol. 2014, 10, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Gnocchi, D.; Bruscalupi, G. Circadian Rhythms and Hormonal Homeostasis: Pathophysiological Implications. Biology 2017, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Rogers, N.H. Brown adipose tissue during puberty and with aging. Ann. Med. 2015, 47, 142–149. [Google Scholar] [CrossRef]
- Saito, M.; Okamatsu-Ogura, Y.; Matsushita, M.; Watanabe, K.; Yoneshiro, T.; Nio-Kobayashi, J.; Iwanaga, T.; Miyagawa, M.; Kameya, T.; Nakada, K.; et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: Effects of cold exposure and adiposity. Diabetes 2009, 58, 1526–1531. [Google Scholar] [CrossRef]
- van Marken Lichtenbelt, W.D.; Schrauwen, P. Implications of nonshivering thermogenesis for energy balance regulation in humans. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 301, R285–R296. [Google Scholar] [CrossRef]
- Yoneshiro, T.; Aita, S.; Matsushita, M.; Okamatsu-Ogura, Y.; Kameya, T.; Kawai, Y.; Miyagawa, M.; Tsujisaki, M.; Saito, M. Age-related decrease in cold-activated brown adipose tissue and accumulation of body fat in healthy humans. Obesity 2011, 19, 1755–1760. [Google Scholar] [CrossRef]
- Markussen, L.K.; Rondini, E.A.; Johansen, O.S.; Madsen, J.G.S.; Sustarsic, E.G.; Marcher, A.B.; Hansen, J.B.; Gerhart-Hines, Z.; Granneman, J.G.; Mandrup, S. Lipolysis regulates major transcriptional programs in brown adipocytes. Nat. Commun. 2022, 13, 3956. [Google Scholar] [CrossRef]
- Moller, N.; Jorgensen, J.O. Effects of growth hormone on glucose, lipid, and protein metabolism in human subjects. Endocr. Rev. 2009, 30, 152–177. [Google Scholar] [CrossRef]
- Hagstrom-Toft, E.; Bolinder, J.; Ungerstedt, U.; Arner, P. A circadian rhythm in lipid mobilization which is altered in IDDM. Diabetologia 1997, 40, 1070–1078. [Google Scholar] [CrossRef]
- Bus, J.D.; Boumans, I.; Engel, J.; Te Beest, D.E.; Webb, L.E.; Bokkers, E.A.M. Circadian rhythms and diurnal patterns in the feed intake behaviour of growing-finishing pigs. Sci. Rep. 2023, 13, 16021. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, J.; Tai, J.; Yan, L. Circadian Regulation in Diurnal Mammals: Neural Mechanisms and Implications in Translational Research. Biology 2024, 13, 958. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.M. Octodon degus: A diurnal, social, and long-lived rodent. ILAR J. 2004, 45, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Labyak, S.E.; Lee, T.M.; Goel, N. Rhythm chronotypes in a diurnal rodent, Octodon degus. Am. J. Physiol. 1997, 273, R1058–R1066. [Google Scholar] [CrossRef]
- Phillips, A.J.; Fulcher, B.D.; Robinson, P.A.; Klerman, E.B. Mammalian rest/activity patterns explained by physiologically based modeling. PLoS Comput. Biol. 2013, 9, e1003213. [Google Scholar] [CrossRef]
- Gonzalez, L.M.; Moeser, A.J.; Blikslager, A.T. Porcine models of digestive disease: The future of large animal translational research. Transl. Res. 2015, 166, 12–27. [Google Scholar] [CrossRef]
- Guilloteau, P.; Zabielski, R.; Hammon, H.M.; Metges, C.C. Nutritional programming of gastrointestinal tract development. Is the pig a good model for man? Nutr. Res. Rev. 2010, 23, 4–22. [Google Scholar] [CrossRef]
- Roura, E.; Koopmans, S.J.; Lalles, J.P.; Le Huerou-Luron, I.; de Jager, N.; Schuurman, T.; Val-Laillet, D. Critical review evaluating the pig as a model for human nutritional physiology. Nutr. Res. Rev. 2016, 29, 60–90. [Google Scholar] [CrossRef]
- Paternoster, S.; Falasca, M. Dissecting the Physiology and Pathophysiology of Glucagon-Like Peptide-1. Front. Endocrinol. 2018, 9, 584. [Google Scholar] [CrossRef]
- Yan, L.; Smale, L.; Nunez, A.A. Circadian and photic modulation of daily rhythms in diurnal mammals. Eur. J. Neurosci. 2020, 51, 551–566. [Google Scholar] [CrossRef]
- Mendoza, J. Brain circadian clocks timing the 24h rhythms of behavior. npj Biol. Timing Sleep 2025, 2, 13. [Google Scholar] [CrossRef]
- Vosko, A.M.; Hagenauer, M.H.; Hummer, D.L.; Lee, T.M. Period gene expression in the diurnal degu (Octodon degus) differs from the nocturnal laboratory rat (Rattus norvegicus). Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R353–R361. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bauer, C.M.; Correa, L.A.; Ebensperger, L.A.; Romero, L.M. Stress, sleep, and sex: A review of endocrinological research in Octodon degus. Gen. Comp. Endocrinol. 2019, 273, 11–19. [Google Scholar] [CrossRef] [PubMed]
- la Fleur, S.E.; Blancas-Velazquez, A.S.; Stenvers, D.J.; Kalsbeek, A. Circadian influences on feeding behavior. Neuropharmacology 2024, 256, 110007. [Google Scholar] [CrossRef]
- Bano-Otalora, B.; Rol, M.A.; Madrid, J.A. Behavioral and Thermoregulatory Responses to Changes in Ambient Temperature and Wheel Running Availability in Octodon degus. Front. Integr. Neurosci. 2021, 15, 684988. [Google Scholar] [CrossRef]
- Garcia-Allegue, R.; Lax, P.; Madariaga, A.M.; Madrid, J.A. Locomotor and feeding activity rhythms in a light-entrained diurnal rodent, Octodon degus. Am. J. Physiol. 1999, 277, R523–R531. [Google Scholar] [CrossRef]
- Colonnello, V.; Iacobucci, P.; Fuchs, T.; Newberry, R.C.; Panksepp, J. Octodon degus. A useful animal model for social-affective neuroscience research: Basic description of separation distress, social attachments and play. Neurosci. Biobehav. Rev. 2011, 35, 1854–1863. [Google Scholar] [CrossRef]
- Garduno, B.M.; Holmes, T.C.; Deacon, R.M.J.; Xu, X.; Cogram, P. Octodon degus laboratory colony management principles and methods for behavioral analysis for Alzheimer’s disease neuroscience research. Front. Aging Neurosci. 2024, 16, 1517416. [Google Scholar] [CrossRef]
- Lunney, J.K.; Van Goor, A.; Walker, K.E.; Hailstock, T.; Franklin, J.; Dai, C. Importance of the pig as a human biomedical model. Sci. Transl. Med. 2021, 13, eabd5758. [Google Scholar] [CrossRef]
- Ryu, J.; Prather, R.S.; Lee, K. Use of gene-editing technology to introduce targeted modifications in pigs. J. Anim. Sci. Biotechnol. 2018, 9, 5. [Google Scholar] [CrossRef]
- Zhao, J.; Lai, L.; Ji, W.; Zhou, Q. Genome editing in large animals: Current status and future prospects. Natl. Sci. Rev. 2019, 6, 402–420. [Google Scholar] [CrossRef]
- Perleberg, C.; Kind, A.; Schnieke, A. Genetically engineered pigs as models for human disease. Dis. Model. Mech. 2018, 11, dmm030783. [Google Scholar] [CrossRef] [PubMed]
- Homan, R.; Hanselman, J.C.; Bak-Mueller, S.; Washburn, M.; Lester, P.; Jensen, H.E.; Pinkosky, S.L.; Castle, C.; Taylor, B. Atherosclerosis in Octodon degus (degu) as a model for human disease. Atherosclerosis 2010, 212, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Ingram, D.L.; Dauncey, M.J.; Legge, K.F. Synchronization of motor activity in young pigs to a non-circadian rhythm without affecting food intake and growth. Comp. Biochem. Physiol. A Comp. Physiol. 1985, 80, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Boumans, I.; de Boer, I.J.M.; Hofstede, G.J.; la Fleur, S.E.; Bokkers, E.A.M. The importance of hormonal circadian rhythms in daily feeding patterns: An illustration with simulated pigs. Horm. Behav. 2017, 93, 82–93. [Google Scholar] [CrossRef]
- Ingram, D.L.; Dauncey, M.J. Circadian rhythms in the pig. Comp. Biochem. Physiol. A Comp. Physiol. 1985, 82, 1–5. [Google Scholar] [CrossRef]
- Li, H.; Li, K.; Zhang, K.; Li, Y.; Gu, H.; Liu, H.; Yang, Z.; Cai, D. The Circadian Physiology: Implications in Livestock Health. Int. J. Mol. Sci. 2021, 22, 2111. [Google Scholar] [CrossRef]
- Wang, R.; Liao, Y.; Deng, Y.; Shuang, R. Unraveling the Health Benefits and Mechanisms of Time-Restricted Feeding: Beyond Caloric Restriction. Nutr. Rev. 2025, 83, e1209–e1224. [Google Scholar] [CrossRef]
- Boyd, P.; O’Connor, S.G.; Heckman-Stoddard, B.M.; Sauter, E.R. Time-Restricted Feeding Studies and Possible Human Benefit. JNCI Cancer Spectr. 2022, 6, pkac032. [Google Scholar] [CrossRef]
- Zhang, L.M.; Liu, Z.; Wang, J.Q.; Li, R.Q.; Ren, J.Y.; Gao, X.; Lv, S.S.; Liang, L.Y.; Zhang, F.; Yin, B.W.; et al. Randomized controlled trial for time-restricted eating in overweight and obese young adults. iScience 2022, 25, 104870. [Google Scholar] [CrossRef]
- Chaix, A.; Lin, T.; Le, H.D.; Chang, M.W.; Panda, S. Time-Restricted Feeding Prevents Obesity and Metabolic Syndrome in Mice Lacking a Circadian Clock. Cell Metab. 2019, 29, 303–319.e4. [Google Scholar] [CrossRef]
- Hatori, M.; Vollmers, C.; Zarrinpar, A.; DiTacchio, L.; Bushong, E.A.; Gill, S.; Leblanc, M.; Chaix, A.; Joens, M.; Fitzpatrick, J.A.; et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 2012, 15, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Sun, Y.; Ye, Y.; Hu, D.; Zhang, H.; He, Z.; Zhao, H.; Yang, H.; Mao, Y. Randomized controlled trial for time-restricted eating in healthy volunteers without obesity. Nat. Commun. 2022, 13, 1003. [Google Scholar] [CrossRef] [PubMed]
- Manoogian, E.N.C.; Chow, L.S.; Taub, P.R.; Laferrere, B.; Panda, S. Time-restricted Eating for the Prevention and Management of Metabolic Diseases. Endocr. Rev. 2022, 43, 405–436. [Google Scholar] [CrossRef]
- Jamshed, H.; Steger, F.L.; Bryan, D.R.; Richman, J.S.; Warriner, A.H.; Hanick, C.J.; Martin, C.K.; Salvy, S.J.; Peterson, C.M. Effectiveness of Early Time-Restricted Eating for Weight Loss, Fat Loss, and Cardiometabolic Health in Adults With Obesity: A Randomized Clinical Trial. JAMA Intern. Med. 2022, 182, 953–962. [Google Scholar] [CrossRef]
- Liu, D.; Huang, Y.; Huang, C.; Yang, S.; Wei, X.; Zhang, P.; Guo, D.; Lin, J.; Xu, B.; Li, C.; et al. Calorie Restriction with or without Time-Restricted Eating in Weight Loss. N. Engl. J. Med. 2022, 386, 1495–1504. [Google Scholar] [CrossRef]
- Gabinet, N.M. Effects mediated by melatonin and cortisol of artificial light and noise, alone and in combination, on sleep and health. Explor. Neurosci. 2024, 3, 382–417. [Google Scholar] [CrossRef]
- Phoi, Y.Y.; Rogers, M.; Bonham, M.P.; Dorrian, J.; Coates, A.M. A scoping review of chronotype and temporal patterns of eating of adults: Tools used, findings, and future directions. Nutr. Res. Rev. 2022, 35, 112–135. [Google Scholar] [CrossRef]
- Chakradeo, P.; Rasmussen, H.E.; Swanson, G.R.; Swanson, B.; Fogg, L.F.; Bishehsari, F.; Burgess, H.J.; Keshavarzian, A. Psychometric Testing of a Food Timing Questionnaire and Food Timing Screener. Curr. Dev. Nutr. 2022, 6, nzab148. [Google Scholar] [CrossRef]
- Amoutzopoulos, B.; Page, P.; Roberts, C.; Roe, M.; Cade, J.; Steer, T.; Baker, R.; Hawes, T.; Galloway, C.; Yu, D.; et al. Portion size estimation in dietary assessment: A systematic review of existing tools, their strengths and limitations. Nutr. Rev. 2020, 78, 885–900. [Google Scholar] [CrossRef]
- Malin, S.K.; Syeda, U.S.A.; Remchak, M.E.; Heiston, E.M. Early chronotype favors appetite and reduced later day caloric intake among adults with obesity. Chronobiol. Int. 2024, 41, 427–438. [Google Scholar] [CrossRef]
- Speksnijder, E.M.; Bisschop, P.H.; Siegelaar, S.E.; Stenvers, D.J.; Kalsbeek, A. Circadian desynchrony and glucose metabolism. J. Pineal Res. 2024, 76, e12956. [Google Scholar] [CrossRef] [PubMed]
- Jastran, M.M.; Bisogni, C.A.; Sobal, J.; Blake, C.; Devine, C.M. Eating routines. Embedded, value based, modifiable, and reflective. Appetite 2009, 52, 127–136. [Google Scholar] [CrossRef]
- Hoddy, K.K.; Marlatt, K.L.; Cetinkaya, H.; Ravussin, E. Intermittent Fasting and Metabolic Health: From Religious Fast to Time-Restricted Feeding. Obesity 2020, 28 (Suppl. S1), S29–S37. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, L.J.; Veronese, N.; Ragusa, F.S.; Petralia, V.; Ciriminna, S.; Di Bella, G.; Schiro, P.; Sabico, S.; Al-Daghri, N.M.; Barbagallo, M. Mediterranean diet and spirituality/religion: Eating with meaning. Aging Clin. Exp. Res. 2024, 36, 223. [Google Scholar] [CrossRef]
- Siddiqui, F.; Salam, R.A.; Lassi, Z.S.; Das, J.K. The Intertwined Relationship Between Malnutrition and Poverty. Front. Public Health 2020, 8, 453. [Google Scholar] [CrossRef]
- Lee, A.; Cardel, M.; Donahoo, W.T. Social and Environmental Factors Influencing Obesity. In Endotext; Feingold, K.R., Ahmed, S.F., Anawalt, B., Blackman, M.R., Boyce, A., Chrousos, G., Corpas, E., de Herder, W.W., Dhatariya, K., Dungan, K., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar]
- Darmon, N.; Drewnowski, A. Contribution of food prices and diet cost to socioeconomic disparities in diet quality and health: A systematic review and analysis. Nutr. Rev. 2015, 73, 643–660. [Google Scholar] [CrossRef]
- Darmon, N.; Drewnowski, A. Does social class predict diet quality? Am. J. Clin. Nutr. 2008, 87, 1107–1117. [Google Scholar] [CrossRef]
- Urbano, T.; Vinceti, M.; Filippini, T. Artificial light at night and night-shift work: Emerging threats for human health. Public Health Toxicol. 2023, 3, 12. [Google Scholar] [CrossRef]
- Almoosawi, S.; Vingeliene, S.; Gachon, F.; Voortman, T.; Palla, L.; Johnston, J.D.; Van Dam, R.M.; Darimont, C.; Karagounis, L.G. Chronotype: Implications for Epidemiologic Studies on Chrono-Nutrition and Cardiometabolic Health. Adv. Nutr. 2019, 10, 30–42. [Google Scholar] [CrossRef]
- Dinu, M.; Lotti, S.; Pagliai, G.; Napoletano, A.; Asensi, M.T.; Giangrandi, I.; Marcucci, R.; Amedei, A.; Colombini, B.; Sofi, F. Effects of a chronotype-adapted diet on weight loss, cardiometabolic health, and gut microbiota: Study protocol for a randomized controlled trial. Trials 2024, 25, 152. [Google Scholar] [CrossRef]
- Longo-Silva, G.; Serenini, R.; Pedrosa, A.; Lima, M.; Soares, L.; Melo, J.; Menezes, R. Chrononutrition patterns and their association with body weight: Differences across multiple chronotypes. Endocrinol. Diabetes Nutr. 2025, 72, 4–13. [Google Scholar] [CrossRef]
- Roden, L.; Rudner, T.; Rae, D. Impact of chronotype on athletic performance: Current perspectives. ChronoPhysiology Ther. 2017, 7, 1–6. [Google Scholar] [CrossRef]
- Adafer, R.; Messaadi, W.; Meddahi, M.; Patey, A.; Haderbache, A.; Bayen, S.; Messaadi, N. Food Timing, Circadian Rhythm and Chrononutrition: A Systematic Review of Time-Restricted Eating’s Effects on Human Health. Nutrients 2020, 12, 3770. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, M.K. Chrononutrition-When We Eat Is of the Essence in Tackling Obesity. Nutrients 2022, 14, 5080. [Google Scholar] [CrossRef]
- Franzago, M.; Alessandrelli, E.; Notarangelo, S.; Stuppia, L.; Vitacolonna, E. Chrono-Nutrition: Circadian Rhythm and Personalized Nutrition. Int. J. Mol. Sci. 2023, 24, 2571. [Google Scholar] [CrossRef]
- Amiri, M.; Li, J.; Hasan, W. Personalized Flexible Meal Planning for Individuals With Diet-Related Health Concerns: System Design and Feasibility Validation Study. JMIR Form. Res. 2023, 7, e46434. [Google Scholar] [CrossRef]
- Escoto, K.H.; Laska, M.N.; Larson, N.; Neumark-Sztainer, D.; Hannan, P.J. Work hours and perceived time barriers to healthful eating among young adults. Am. J. Health Behav. 2012, 36, 786–796. [Google Scholar] [CrossRef]
- Huseynov, S.; Palma, M.A. Food decision-making under time pressure. Food Qual. Prefer. 2021, 88, 104072. [Google Scholar] [CrossRef]
- Pelletier, J.E.; Laska, M.N. Balancing healthy meals and busy lives: Associations between work, school, and family responsibilities and perceived time constraints among young adults. J. Nutr. Educ. Behav. 2012, 44, 481–489. [Google Scholar] [CrossRef]
- Costello, R.B.; Lentino, C.V.; Boyd, C.C.; O’Connell, M.L.; Crawford, C.C.; Sprengel, M.L.; Deuster, P.A. The effectiveness of melatonin for promoting healthy sleep: A rapid evidence assessment of the literature. Nutr. J. 2014, 13, 106. [Google Scholar] [CrossRef]
- Emens, J.S.; Burgess, H.J. Effect of Light and Melatonin and Other Melatonin Receptor Agonists on Human Circadian Physiology. Sleep Med. Clin. 2015, 10, 435–453. [Google Scholar] [CrossRef] [PubMed]
- Pechey, R.; Jebb, S.A.; Kelly, M.P.; Almiron-Roig, E.; Conde, S.; Nakamura, R.; Shemilt, I.; Suhrcke, M.; Marteau, T.M. Socioeconomic differences in purchases of more vs. less healthy foods and beverages: Analysis of over 25,000 British households in 2010. Soc. Sci. Med. 2013, 92, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Giskes, K.; Avendano, M.; Brug, J.; Kunst, A.E. A systematic review of studies on socioeconomic inequalities in dietary intakes associated with weight gain and overweight/obesity conducted among European adults. Obes. Rev. 2010, 11, 413–429. [Google Scholar] [CrossRef] [PubMed]
- Pechey, R.; Monsivais, P. Socioeconomic inequalities in the healthiness of food choices: Exploring the contributions of food expenditures. Prev. Med. 2016, 88, 203–209. [Google Scholar] [CrossRef]
- Murakami, K.; Shinozaki, N.; McCaffrey, T.A.; Livingstone, M.B.E.; Masayasu, S.; Sasaki, S. Relative validity of the Chrono-Nutrition Behavior Questionnaire (CNBQ) against 11-day event-based ecological momentary assessment diaries of eating. Int. J. Behav. Nutr. Phys. Act. 2025, 22, 46. [Google Scholar] [CrossRef]
- Luz, C.; Fonseca, A.; Santos, J.S.; Araujo, J.F.; Duarte, L.L.; Moreno, C.R.C. Association of Meal Timing with Sleep Quality and Anxiety According to Chronotype: A Study of University Students. Clocks Sleep. 2024, 6, 156–169. [Google Scholar] [CrossRef]
- Holmback, U.; Forslund, A.; Forslund, J.; Hambraeus, L.; Lennernas, M.; Lowden, A.; Stridsberg, M.; Akerstedt, T. Metabolic responses to nocturnal eating in men are affected by sources of dietary energy. J. Nutr. 2002, 132, 1892–1899. [Google Scholar] [CrossRef]
- Yoshitake, R.; Park, I.; Ogata, H.; Omi, N. Meal Timing and Sleeping Energy Metabolism. Nutrients 2023, 15, 763. [Google Scholar] [CrossRef]
- van der Merwe, C.; Munch, M.; Kruger, R. Chronotype Differences in Body Composition, Dietary Intake and Eating Behavior Outcomes: A Scoping Systematic Review. Adv. Nutr. 2022, 13, 2357–2405. [Google Scholar] [CrossRef]
- Dashti, H.S.; Scheer, F.; Saxena, R.; Garaulet, M. Timing of Food Intake: Identifying Contributing Factors to Design Effective Interventions. Adv. Nutr. 2019, 10, 606–620. [Google Scholar] [CrossRef]
- Stinson, E.J.; Votruba, S.B.; Venti, C.; Perez, M.; Krakoff, J.; Gluck, M.E. Food Insecurity is Associated with Maladaptive Eating Behaviors and Objectively Measured Overeating. Obesity 2018, 26, 1841–1848. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.; Telke, S.; Larson, N.; Mason, S.M.; Neumark-Sztainer, D. Household food insecurity: Associations with disordered eating behaviours and overweight in a population-based sample of adolescents. Public Health Nutr. 2020, 23, 3126–3135. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.P.; Majewski, M.; Schaefer, D.; Tierney, A. Chronic experience with unpredictable food availability promotes food reward, overeating, and weight gain in a novel animal model of food insecurity. Appetite 2022, 176, 106120. [Google Scholar] [CrossRef]
- Peters, U.; Turner, B.; Alvarez, D.; Murray, M.; Sharma, A.; Mohan, S.; Patel, S. Considerations for Embedding Inclusive Research Principles in the Design and Execution of Clinical Trials. Ther. Innov. Regul. Sci. 2023, 57, 186–195. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Pacanowski, M.A.; Bull, J.; Zhang, L. Racial/ethnic differences in drug disposition and response: Review of recently approved drugs. Clin. Pharmacol. Ther. 2015, 97, 263–273. [Google Scholar] [CrossRef]
- Crawley, F.P.; Kurz, R.; Nakamura, H. Testing medications in children. N. Engl. J. Med. 2003, 348, 763–764. [Google Scholar] [CrossRef]
- Rohatgi, K.W.; Humble, S.; McQueen, A.; Hunleth, J.M.; Chang, S.H.; Herrick, C.J.; James, A.S. Medication Adherence and Characteristics of Patients Who Spend Less on Basic Needs to Afford Medications. J. Am. Board Fam. Med. 2021, 34, 561–570. [Google Scholar] [CrossRef]
- Ribas-Latre, A.; Fernandez-Veledo, S.; Vendrell, J. Time-restricted eating, the clock ticking behind the scenes. Front. Pharmacol. 2024, 15, 1428601. [Google Scholar] [CrossRef]
- Parr, E.B.; Devlin, B.L.; Radford, B.E.; Hawley, J.A. A Delayed Morning and Earlier Evening Time-Restricted Feeding Protocol for Improving Glycemic Control and Dietary Adherence in Men with Overweight/Obesity: A Randomized Controlled Trial. Nutrients 2020, 12, 505. [Google Scholar] [CrossRef]
- Samaddar, A.; Cuevas, R.P.; Custodio, M.C.; Ynion, J.; Ray Chakravarti, A.; Mohanty, S.K.; Demont, M. Capturing diversity and cultural drivers of food choice in eastern India. Int. J. Gastron. Food Sci. 2020, 22, 100249. [Google Scholar] [CrossRef]
- Choe, S. Food, Contamination, and Race-Thinking: Culinary Encounters in Late Medieval Missionary Accounts of Asia. Speculum 2024, 99, 458–479. [Google Scholar] [CrossRef]
- Godos, J.; Scazzina, F.; Paterno Castello, C.; Giampieri, F.; Quiles, J.L.; Briones Urbano, M.; Battino, M.; Galvano, F.; Iacoviello, L.; de Gaetano, G.; et al. Underrated aspects of a true Mediterranean diet: Understanding traditional features for worldwide application of a “Planeterranean” diet. J. Transl. Med. 2024, 22, 294. [Google Scholar] [CrossRef] [PubMed]
- Sidossis, L.S.; Lawson, R.; Aprilakis, E.; Barata, B.C.; Baska, A.; Beneka, A.; Bird, R.; Birrell, F.; Chatzinikola, C.; Chondronikola, M.; et al. Defining the Traditional Mediterranean Lifestyle: Joint International Consensus Statement. Lifestyle Med. 2024, 5, e115. [Google Scholar] [CrossRef]
- Link, B.G.; Phelan, J. Social conditions as fundamental causes of disease. J. Health Soc. Behav. 1995, 36, 80–94. [Google Scholar] [CrossRef]
- Billari, F.C.; Hiekel, N.; Liefbroer, A.C. The Social Stratification of Choice in the Transition to Adulthood. Eur. Sociol. Rev. 2019, 35, 599–615. [Google Scholar] [CrossRef]
- Adler, N.E.; Boyce, T.; Chesney, M.A.; Cohen, S.; Folkman, S.; Kahn, R.L.; Syme, S.L. Socioeconomic status and health. The challenge of the gradient. Am. Psychol. 1994, 49, 15–24. [Google Scholar] [CrossRef]
- Bermingham, K.M.; Linenberg, I.; Polidori, L.; Asnicar, F.; Arre, A.; Wolf, J.; Badri, F.; Bernard, H.; Capdevila, J.; Bulsiewicz, W.J.; et al. Effects of a personalized nutrition program on cardiometabolic health: A randomized controlled trial. Nat. Med. 2024, 30, 1888–1897. [Google Scholar] [CrossRef]
- de Hoogh, I.M.; Winters, B.L.; Nieman, K.M.; Bijlsma, S.; Krone, T.; van den Broek, T.J.; Anderson, B.D.; Caspers, M.P.M.; Anthony, J.C.; Wopereis, S. A Novel Personalized Systems Nutrition Program Improves Dietary Patterns, Lifestyle Behaviors and Health-Related Outcomes: Results from the Habit Study. Nutrients 2021, 13, 1763. [Google Scholar] [CrossRef]
- Singar, S.; Nagpal, R.; Arjmandi, B.H.; Akhavan, N.S. Personalized Nutrition: Tailoring Dietary Recommendations through Genetic Insights. Nutrients 2024, 16, 2673. [Google Scholar] [CrossRef]
- Ulusoy-Gezer, H.G.; Rakicioglu, N. The Future of Obesity Management through Precision Nutrition: Putting the Individual at the Center. Curr. Nutr. Rep. 2024, 13, 455–477. [Google Scholar] [CrossRef]
- Lee, B.Y.; Ordovas, J.M.; Parks, E.J.; Anderson, C.A.M.; Barabasi, A.L.; Clinton, S.K.; de la Haye, K.; Duffy, V.B.; Franks, P.W.; Ginexi, E.M.; et al. Research gaps and opportunities in precision nutrition: An NIH workshop report. Am. J. Clin. Nutr. 2022, 116, 1877–1900. [Google Scholar] [CrossRef] [PubMed]
- Amico, K.R.; Mugavero, M.; Krousel-Wood, M.A.; Bosworth, H.B.; Merlin, J.S. Advantages to Using Social-Behavioral Models of Medication Adherence in Research and Practice. J. Gen. Intern. Med. 2018, 33, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, M.; Dinich, J.; Merrow, M.; Roenneberg, T. Social jetlag: Misalignment of biological and social time. Chronobiol. Int. 2006, 23, 497–509. [Google Scholar] [CrossRef]
- Kelly, K.P.; Ellacott, K.L.J.; Chen, H.; McGuinness, O.P.; Johnson, C.H. Time-optimized feeding is beneficial without enforced fasting. Open Biol. 2021, 11, 210183. [Google Scholar] [CrossRef]
- Xie, Z.; He, Z.; Ye, Y.; Mao, Y. Effects of time-restricted feeding with different feeding windows on metabolic health: A systematic review of human studies. Nutrition 2022, 102, 111764. [Google Scholar] [CrossRef]
- Dimitratos, S.M.; German, J.B.; Schaefer, S.E. Wearable Technology to Quantify the Nutritional Intake of Adults: Validation Study. JMIR Mhealth Uhealth 2020, 8, e16405. [Google Scholar] [CrossRef]
- Gioia, S.; Vlasac, I.M.; Babazadeh, D.; Fryou, N.L.; Do, E.; Love, J.; Robbins, R.; Dashti, H.S.; Lane, J.M. Mobile Apps for Dietary and Food Timing Assessment: Evaluation for Use in Clinical Research. JMIR Form. Res. 2023, 7, e35858. [Google Scholar] [CrossRef]
- Malaeb, S.; Harindhanavudhi, T.; Dietsche, K.; Esch, N.; Manoogian, E.N.C.; Panda, S.; Mashek, D.G.; Wang, Q.; Chow, L.S. Time-Restricted Eating Alters Food Intake Patterns, as Prospectively Documented by a Smartphone Application. Nutrients 2020, 12, 3396. [Google Scholar] [CrossRef]
- Prasad, M.; Fine, K.; Gee, A.; Nair, N.; Popp, C.J.; Cheng, B.; Manoogian, E.N.C.; Panda, S.; Laferrere, B. A Smartphone Intervention to Promote Time Restricted Eating Reduces Body Weight and Blood Pressure in Adults with Overweight and Obesity: A Pilot Study. Nutrients 2021, 13, 2148. [Google Scholar] [CrossRef]
- Tofani, G.S.S.; Leigh, S.J.; Gheorghe, C.E.; Bastiaanssen, T.F.S.; Wilmes, L.; Sen, P.; Clarke, G.; Cryan, J.F. Gut microbiota regulates stress responsivity via the circadian system. Cell Metab. 2025, 37, 138–153.e5. [Google Scholar] [CrossRef]
- Dantas Machado, A.C.; Brown, S.D.; Lingaraju, A.; Sivaganesh, V.; Martino, C.; Chaix, A.; Zhao, P.; Pinto, A.F.M.; Chang, M.W.; Richter, R.A.; et al. Diet and feeding pattern modulate diurnal dynamics of the ileal microbiome and transcriptome. Cell Rep. 2022, 40, 111008. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Wang, S.; Chen, Q.; Luo, H.; Lian, X.; Shi, D. Time-restricted feeding affects the fecal microbiome metabolome and its diurnal oscillations in lung cancer mice. Neoplasia 2023, 45, 100943. [Google Scholar] [CrossRef] [PubMed]
- Zeb, F.; Osaili, T.; Obaid, R.S.; Naja, F.; Radwan, H.; Cheikh Ismail, L.; Hasan, H.; Hashim, M.; Alam, I.; Sehar, B.; et al. Gut Microbiota and Time-Restricted Feeding/Eating: A Targeted Biomarker and Approach in Precision Nutrition. Nutrients 2023, 15, 259. [Google Scholar] [CrossRef]
- Hood, S.; Amir, S. The aging clock: Circadian rhythms and later life. J. Clin. Investig. 2017, 127, 437–446. [Google Scholar] [CrossRef]
- Katagiri, R.; Asakura, K.; Kobayashi, S.; Suga, H.; Sasaki, S. Low intake of vegetables, high intake of confectionary, and unhealthy eating habits are associated with poor sleep quality among middle-aged female Japanese workers. J. Occup. Health 2014, 56, 359–368. [Google Scholar] [CrossRef]
- Makarem, N.; Zuraikat, F.M.; Caceres, B.; Sears, D.D.; St-Onge, M.P.; Lai, Y.; Aggarwal, B. Variable Eating Patterns: A Potential Novel Risk Factor for Systemic Inflammation in Women. Ann. Behav. Med. 2023, 57, 93–97. [Google Scholar] [CrossRef]
- Dallmann, R.; Brown, S.A.; Gachon, F. Chronopharmacology: New insights and therapeutic implications. Annu. Rev. Pharmacol. Toxicol. 2014, 54, 339–361. [Google Scholar] [CrossRef]
- Dobrek, L. Chronopharmacology in Therapeutic Drug Monitoring-Dependencies between the Rhythmics of Pharmacokinetic Processes and Drug Concentration in Blood. Pharmaceutics 2021, 13, 1915. [Google Scholar] [CrossRef]
- Kaskal, M.; Sevim, M.; Ulker, G.; Keles, C.; Bebitoglu, B.T. The clinical impact of chronopharmacology on current medicine. Naunyn Schmiedebergs Arch. Pharmacol. 2025. [Google Scholar] [CrossRef]
- Levi, F.A.; Okyar, A.; Hadadi, E.; Innominato, P.F.; Ballesta, A. Circadian Regulation of Drug Responses: Toward Sex-Specific and Personalized Chronotherapy. Annu. Rev. Pharmacol. Toxicol. 2024, 64, 89–114. [Google Scholar] [CrossRef]
- Beaulieu, M.; Schaefer, H.M. The proper time for antioxidant consumption. Physiol. Behav. 2014, 128, 54–59. [Google Scholar] [CrossRef]
- Borja-Magno, A.; Guevara-Cruz, M.; Flores-Lopez, A.; Carrillo-Dominguez, S.; Granados, J.; Arias, C.; Perry, M.; Sears, B.; Bourges, H.; Gomez, F.E. Differential effects of high dose omega-3 fatty acids on metabolism and inflammation in patients with obesity: Eicosapentaenoic and docosahexaenoic acid supplementation. Front. Nutr. 2023, 10, 1156995. [Google Scholar] [CrossRef]



| Chronometabolic Regulation | Mice (Nocturnal) | Humans (Diurnal) |
|---|---|---|
| SCN & Pineal gland regulation [1,21,84] | SCN entrains peripheral clocks primarily via feeding–fasting cycles. | SCN entrains peripheral clocks primarily through light exposure and hormonal signals. |
| Melatonin influence on metabolism [114,115,116,117,118,119,120,121] | Minimal melatonin influence: feeding cues exert stronger control over clock genes | Melatonin peaks at night, suppressing insulin and promoting lipid oxidation. |
| Circadian regulation of liver metabolism [122,123,124,125] | Strong circadian oscillations in hepatic gene expression, driven by meal timing. | Regulation is predominantly hormonal (cortisol, insulin, glucagon) rather than meal timing. |
| Regulation of thermogenesis & lipolysis [126,127,128] | Thermogenesis process is strongly regulated by BMAL1 & CLOCK expression. The lipolysis follows a strict circadian cycle (peaking at night) | Thermogenesis & lipogenesis is less circadian driven; influenced more by hormonal fluctuations |
| Circadian regulation of adipokines [40,90,93] | Leptin and adiponectin secretion follow a strong circadian rhythm, peaking during fasting. | Leptin secretion follows a gradual diurnal rhythm, peaking in the evening, influenced by energy balance. |
| Traits | Pig (Sus scrofa) | Degu (Octodon degus) | Mouse (Mus musculus) |
|---|---|---|---|
| Chronotype [163,164,165,166,167] | Diurnal | Diurnal | Nocturnal |
| Metabolic similarity to humans [168,169,170] | High—similar gastrointestinal, cardiovascular, and metabolic traits | Moderate—robust circadian behavior | Low—opposite feeding/activity cycle |
| Peripheral clock gene expression [170,171,172,173,174] | Rhythmic expression similar to human in liver/adipose tissue | Functional Per1 expression in SCN, peripheral oscillations | Highly studied; strong rhythmicity but reversed phase |
| Postprandial hormonal dynamics [170,171,175,176,177,178] | Responsive insulin, GLP-1, glucagon dynamics | Circadian-aligned feeding and hormonal patterns | Meal timing dominant; less human-like hormonal profiles |
| Ease of handling [165,179,180] | Difficult—large size, high cost | Easier—small, size, short lifespan | Very easy, widely used, low cost |
| Genetic manipulation tools [179,180,181,182,183,184,185] | Limited availability of gene editing tools | Lacks standardized genetic tools | Well-developed gene editing (e.g., CRISPR, Knockouts) |
| Practical limitation [165,170,180,181] | High housing cost, limited scalability | Limited protocol, less well characterized | Circadian mismatch with human |
| Characteristic | Mice (Pre-Clinical Studies) | Humans (Clinical Trials) |
|---|---|---|
| Feeding pattern [190,191] | TRF aligned with active (dark) phase | TRF aligned with active (light) phase |
| Metabolic effect [192,193,194,195] | Strong; ↓ obesity, ↓ insulin resistance, ↓ liver fat | Mild to moderate: ↑ insulin sensitivity, ↓ BP |
| Mechanism assessed [6,196] | Yes—gene expression (Bmal1, Clock, Rev-erbα, Pparα) | Rarely assessed directly |
| Environmental control [13,192,197] | Highly controlled (light, diet, genetics) | Variable (light exposure, chronotype, stress) |
| Clock gene oscillation [13,192,197] | Frequently reported | Rarely reported |
| Limiting factors [13,190,192,195,196,197] | Nocturnal model; limited translation | Poor adherence, high variability |
| Barrier Category | Description | Impact on TRF Adherence |
|---|---|---|
| Methodological Limitation [200,201,202] | Inconsistent dietary assessment tools and lack of standardized protocols reduce comparability and reliability. | High—unreliable intake data and measurement bias undermine study validity. |
| Chronotype Variability [150,203,204] | Differences in individual circadian preferences affect metabolic responses but are rarely accounted for. | High—one-size-fits-all protocols fail to address individual metabolic timing. |
| Cultural & Behavioral Practices [205,206,207] | Traditional meal customs, religious fasting, and family routines may conflict with rigid TRF schedules. | Moderate to high—sociocultural norms often override protocol adherence. |
| Socioeconomic Constraints [208,209,210,211] | Low SES limits food access, meal regularity, and flexibility to follow structured interventions. | Very high—financial, environmental, and access barriers prevent participation. |
| Work Schedules & Urbanization [15,16,31,146,196,212] | Shift work, artificial light, and 24 h lifestyles disrupt circadian alignment and eating patterns. | High—inconsistent daily structure interferes with stable meal timing. |
| Western-Centric Bias [205,206,207] | Early-meal TRF protocols are based on Western routines, limiting global applicability and equity. | High—reduces inclusivity and generalizability of research findings. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuad, S.A.; Ginting, R.P.; Lee, M.-W. Chrononutrition: Potential, Challenges, and Application in Managing Obesity. Int. J. Mol. Sci. 2025, 26, 5116. https://doi.org/10.3390/ijms26115116
Fuad SA, Ginting RP, Lee M-W. Chrononutrition: Potential, Challenges, and Application in Managing Obesity. International Journal of Molecular Sciences. 2025; 26(11):5116. https://doi.org/10.3390/ijms26115116
Chicago/Turabian StyleFuad, Siti Aisyah, Rehna Paula Ginting, and Min-Woo Lee. 2025. "Chrononutrition: Potential, Challenges, and Application in Managing Obesity" International Journal of Molecular Sciences 26, no. 11: 5116. https://doi.org/10.3390/ijms26115116
APA StyleFuad, S. A., Ginting, R. P., & Lee, M.-W. (2025). Chrononutrition: Potential, Challenges, and Application in Managing Obesity. International Journal of Molecular Sciences, 26(11), 5116. https://doi.org/10.3390/ijms26115116







