Abstract
Roots are the hidden and most important part of plants. They serve as stabilizers and channels for uptaking water and nutrients and play a crucial role in the growth and development of plants. Here, two-dimensional image data were used to identify quantitative trait loci (QTL) controlling root traits in an interspecific mapping population derived from a cross between wild soybean ‘PI366121’ and cultivar ‘Williams 82’. A total of 2830 single-nucleotide polymorphisms were used for genotyping, constructing genetic linkage maps, and analyzing QTLs. Forty-two QTLs were identified on twelve chromosomes, twelve of which were identified as major QTLs, with a phenotypic variation range of 36.12% to 39.11% and a logarithm of odds value range of 12.01 to 17.35. Two significant QTL regions for the average diameter, root volume, and link average diameter root traits were detected on chromosomes 3 and 13, and both wild and cultivated soybeans contributed positive alleles. Six candidate genes, Glyma.03G027500 (transketolase/glycoaldehyde transferase), Glyma.03G014500 (dehydrogenases), Glyma.13G341500 (leucine-rich repeat receptor-like protein kinase), Glyma.13G341400 (AGC kinase family protein), Glyma.13G331900 (60S ribosomal protein), and Glyma.13G333100 (aquaporin transporter) showed higher expression in root tissues based on publicly available transcriptome data. These results will help breeders improve soybean genetic components and enhance soybean root morphological traits using desirable alleles from wild soybeans.
1. Introduction
Worldwide, soybean [Glycine max (L.) Merr.] is a major economic, legume, and oil crop that is essential for food, feed, fodder, and industrial production [1,2,3]. The main soybean-producing countries are the United States of America, Brazil, and Argentina, accounting for 33%, 29%, and 19% of the total global production, respectively (www.soystats.com, accessed on 20 April 2023). It is considered that cultivable soybeans evolved from wild soybeans (Glycine soja Seib.et Zucc) approximately 3000–6000 years ago in China [4,5,6].
Roots are the most important features of a plant for transporting and capturing water, supplying nutrients, and regulating floods [7,8]. Root traits, including root length, diameter, and surface area, act as a harbor for plants, enhancing plant growth by taking up or transporting water and nutrients and regulating nutrient availability through symbiotic association with microbes [9,10]. Root morphology helps regulate plant efficiency under different environmental conditions and plays a vital role in recovering drought avoidance by increasing water absorption from the soil layers [11,12]. Root morphological traits also play a significant role in minimizing the different types of stress. Hence, soybean root growth significantly affects soybean production and adaptation to morphological, physiological, and anatomical changes under stress conditions [13]. It is widely recognized that different root traits play a significant role in withstanding stress conditions in many plant species, such as chickpea [14], common bean [15], soybean [16], rice [17], wheat [18], and maize [19]. For a new green revolution, the root morphological traits of crops have become a target for increasing crop yield and quality [20]. These studies suggest the importance of root morphological traits in crop growth and development.
In conventional breeding, crop improvement is achieved by phenotypic selection, whereby breeders monitor better progeny lines to achieve genetic improvement with target traits [21]. However, in modern times, interspecific crossing is used for genetic improvement, as desired traits are transferred from wild relatives to cultivated species [22]. Naturally, soybean varieties derived from G. soja are a vital wellspring of rich genetic assets, teeming with an abundance of unique alleles [23,24]. Therefore, next-generation sequencing of the wild soybean is crucial for genetic research and breeding [25,26]. In addition, primary features, such as overall root length, volume, and surface area, of wild soybean accessions are relatively smaller than those of commodity soybeans [27,28]. Therefore, it is essential to research root features and the interactions between roots and their environments [29]. In soybean breeding programs, phenotypic traits were taken using morphological features, such as visual root score assistance, for the easiest phenotypic evaluation of soybeans [30].
The regulation of root morphology is a multifaceted process influenced by a combination of genetic and environmental factors [31,32]. To date, several quantitative trait loci (QTL) have been identified and reported for the morphological traits of soybean roots [27,29,33,34,35]. Consequently, they are commonly employed in genetic research endeavors to enhance our understanding of the genetic constituents of the morphological traits of roots. Several QTLs with alleles from wild soybeans have been identified and reported to have various root morphological traits [27,28]. Similarly, a number of genes associated with root development, such as Glyma.07g126400, Glyma.07g127300, Glyma.07g127100, Glyma.08g12320, Glyma.08g121770, Glyma.08g09550, Glyma.08g13900, and Glyma.16g141800, have been found to enhance and control the root morphological traits in an interspecific mapping population of soybean [27,28,35]. Interspecific mapping root studies are relatively underrepresented; therefore, we focused on an interspecific soybean root study. Additionally, the measurement of root morphological traits was very difficult and laborious for many genotypes, and high phenotypic variations were observed in field conditions due to numerous factors, including soil density, distribution of nutrients, and water content [36,37]. Therefore, researchers have developed several software programs for root analyses, such as the SmartRoot (semi-automated image analysis, https://smartroot.github.io/SmartRoot-Installation/, accessed on 11 May 2023) software [38]. In our experiment, we used WinRHIZO Pro version 2019 computer-based image analysis software, which can easily measure root morphological traits, including root length, diameter, and volume.
In this study, we used an interspecific mapping population derived from a crossing between cultivated soybean ‘Williams 82’ and wild soybean accession ‘PI366121’ and used 3K single-nucleotide polymorphism (SNP) markers for linkage map construction. The main objectives of this study were to detect the major QTLs and determine the potential genes underlying the significant genomic regions of soybean seedling root morphological traits in an interspecific mapping population.
2. Results
2.1. Phenotypic Analysis of Root Traits
Table 1 shows the phenotypic measurements of root characteristics in parental genotypes and recombinant inbred lines (RILs). The descriptive phenotypic data are displayed in normal frequency distributions as average diameter (AD), root volume (RV), and link average diameter (LAD) (Figure 1(A1–A3,B1–B3,C1–C3)). In environment 1 planting, the average AD was evenly distributed across the population, with values of 0.33–0.50 mm and a mean value of 0.37 mm. For RV and LAD, we obtained values of 0.28–3.54 cm3 and 0.38–0.57 mm with corresponding mean values of 1.34 cm3 and 0.45 mm, respectively. For environment 2, the values of AD, RV, and LAD were uniformly distributed across the population, with values of 0.31–0.68 mm, 0.30–2.40 cm3, and 0.37–0.87 mm, respectively, and mean values of 0.36 mm, 1.24 cm3, and 0.46 mm, respectively. However, compared with the parents, transgressive segregation was found in AD, RV, and LAD root traits in both environments (Table 1). The coefficient variation (CV) of the root traits was comparable in different environments. Among these traits, CV was the largest for RV (31.10%), followed by AD (13.57%), and LAD (13.78%) in the first environment. A similar CV trend was found in environment 2. Most skewness values ranged from 0.52 to 1.08 in all root traits in environment 1; however, in environment 2, skewness values were 4.64, 0.18, and 4.11 for AD, RV, and LAD, respectively. A negative kurtosis value was observed for the RV (−0.22) root trait in environment 2. Therefore, based on the skewness and kurtosis values, all three root traits were normally distributed (Figure 1).

Table 1.
Descriptive statistics for the three root traits of the soybean interspecific mapping population.
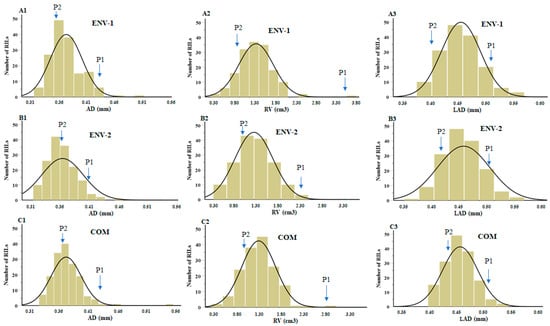
Figure 1.
Frequency distribution with normal curve of soybean root traits in different environments (ENV-1, ENV-2, and COM) of mapping populations. (A1–A3) AD, RV, and LAD for ENV-1. (B1–B3) AD, RV, and LAD for ENV-2. (C1–C3) AD, RV, and LAD for COM. The traits are described in Table 7. Arrows represent the mean values of P1 (‘William 82’) and P2 (‘PI366121’). ENV: environment; COM: combined.
An analysis of variance (ANOVA) for the three root traits was performed to understand the genotype, environment, and their interaction for the traits. Significant variations were observed for genotypes and the interaction between genotype and environment (p < 0.0001) in all root traits (Table 2). Pearson’s correlation coefficient test showed that there was a significant positive correlation between any two root traits in both environments (Table 3). A significant positive correlation was observed between AD and LAD (r = 0.97, p < 0.0001), followed by RV and LAD (r = 0.56, p < 0.0001) in environment 1. Similar results were observed for environment 2.

Table 2.
F-value from analysis of variance for the three root traits of the interspecific mapping population.

Table 3.
Phenotypic Pearson correlation coefficient among the three soybean root traits.
2.2. QTL Detection of Root Traits
A total of 42 significant QTLs were detected on 12 different chromosomes (chr.), including 17 for AD, 9 for RV, and 16 for LAD; some were co-located QTLs in this mapping population (Table 4 and Figure 2). Among these QTLs, 12 were considered major QTLs, as their phenotypic variation was >30% [39].

Table 4.
QTLs for the soybean root traits identified using composite interval mapping in an interspecific mapping population.
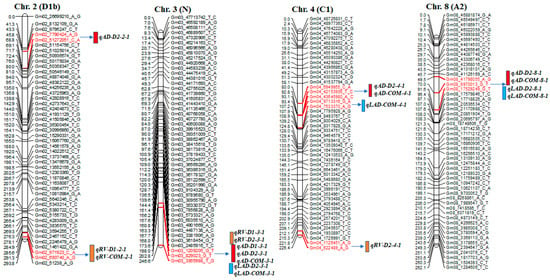
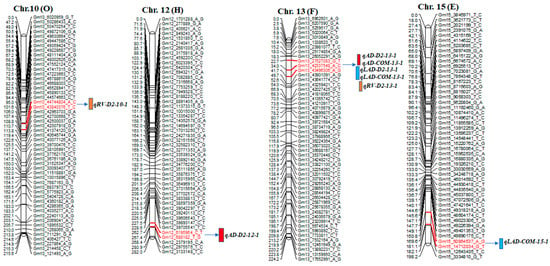
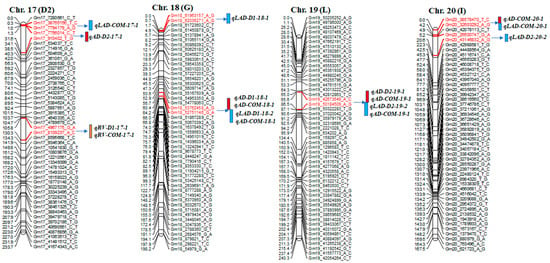
Figure 2.
Diagram showing the location of root QTLs on 12 different chromosomes: Chr. 2, 3, 4, 8, 10, 12, 13, 15, 17, 18, 19, and 20 detected in an interspecific mapping population of soybean. Genetic distance and markers are on the left side of the linkage groups, and marker names are shown on the right side of the linkage groups. Colored bars indicate QTL regions.
For AD, 17 QTLs were identified on chr. 2, 3, 4, 8, 12, 13, 17, 18, 19, and 20. Each of these 17 QTLs contributed to phenotypic variance (R2) ranging from 7.8% to 39.1% and a logarithm of odds (LOD) value of 2.7–18.7. One of the 17 QTLs, qAD_D2-19-1, had an LOD value of 18.7 and a phenotypic variation of 39.0%. The wild soybean parent ‘PI366121’ contributed a positive allele on chr. 2, 4, 8, 12, 13, 19, and 20 for AD; the cultivated soybean, Williams 82, contributed a positive allele on chr. 3, 18, 19, and 20 for the trait (Table 4 and Figure 2).
Nine QTLs for RV were found on chr. 2, 3, 4, 10, 13, and 17, with LOD scores of 2.6–4.4. They had a phenotypic variation of 5.5–16.5%. The qRV_D1-3-1 QTL was on chr. 3, accounting for a phenotypic variation of 16.5% with an LOD value of 4.4 and a positive allele from cultivated soybean ‘William 82’ (Table 4 and Figure 2).
For LAD, 16 QTLs were detected on chr. 3, 4, 8, 13, 15, 17, 18, 19, and 20. The phenotypic variation and LOD score ranged from 6.3% to 36.2% and 2.7 to 16.4, respectively. High LOD values were found for the QTLs qLAD_D2-19-1 (16.4), qLAD_D2-8-1 (15.0), qLAD_D2-3-1 (14.9), qLAD_D2-13-1 (13.8), and qLAD_D2-20-1 (12.0). qLAD_D2-19-1, flanked by Gm19_42673649_A_C and Gm19_50184509_A_G markers, accounted for 36.1% of the phenotypic variation. Favorable alleles of this trait came from both cultivated and wild parents (Table 4 and Figure 2).
SNP markers at 244.8 cM in the interval of Gm3_829023_G_T-~3365988_T_G and SNP markers at 24.7 cM in the interval of Gm13_27527083_G_T~43496306_A_G were associated with AD, RV, and LAD (Figure 2). Moreover, QTL regions for AD and LAD were detected on chr. 8, 18, 19, and 20 at positions 72.0, 64.2, 85.7, and 4.0 cM, respectively (Table 4).
2.3. Putative Candidate Genes and Gene Expression in QTL Regions
Putative candidate genes were identified within the two most hotspot QTL regions on chr. 3 and 13 (Figure S1). A total of 198 putative candidate genes detected for the most effective SNPs associated with root traits and genes are presented in Table S1. Among the 198 genes, 89 genes had allele variations between the resequencing data of ‘William 82’ and ‘PI366121’ (Table S2). Analysis of the sequence of the parental line ‘PI366121’ [40] within the QTL regions on chr. 3 and 13, 32 genes revealed missense and splice variants that cause amino acid changes (Table 5 and Table S3). Moreover, tissue-specific transcriptome data of roots, root stripped, root tips, root hairs, leaves, meristem, green pod, and root nodules were downloaded from the ePlant soybean database (https://bar.-utoronto.ca/eplant soybean/, accessed on 14 June 2023), and used to assess the expression of the candidate genes (Table S4). Based on transcriptome data, six genes having high expression in root tissues: Glyma.03G027500 (transketolase/glycoaldehyde transferase), Glyma.03G014500 (dehydrogenases), Glyma.13G341500 (leucine-rich repeat receptor-like protein kinase), Glyma.13G341400 (AGC kinase family protein), Glyma.13G331900 (60S ribosomal protein), and Glyma.13G333100 (aquaporin transporter) (Table 6, Figure 3). Among the six candidate genes, three genes (Glyma.03G014500, Glyma.13G341400, and Glyma.13G331900) exhibited the highest expression in root tissue compared with other tissues (leaf, seedling, shoot, stem, meristem, flower, pod, nodule, seed, embryo, and endosperm) using RNA-Seq soybean libraries (4085) (http://ipf.sustech.edu.cn/pub/soybean/, accessed on 22 June 2023). Figure S2 shows the differential expression levels of the three candidate genes in the other tissues.

Table 5.
SNPs of annotated genes on chromosomes 3 and 13 based on genomic information of ‘William 82’ and ‘PI366121’.

Table 6.
The six candidate genes have IDs, function annotations, and physical position.
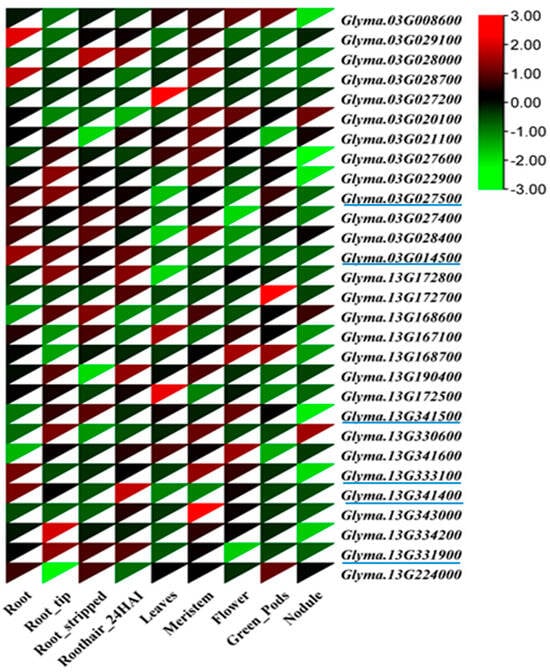
Figure 3.
Heatmap showing the expression level of candidate genes in different plant tissues, including soybean roots. The red color indicates a high expression greater than 0, and the green color indicates a low expression less than 0 among the parameters Blue color underline indicates the final candidate genes.
3. Discussion
The interspecific mapping population used in this study was first genotyped using the GoldenGate® assay, which contained 1536 SNP loci with 169 F4:5 RILs derived from ‘Williams 82’ × ‘PI366121’ [41]. The genetic map was constructed with 414 polymorphic and filtered SNPs to detect the QTL regions for foxglove aphid resistance [42,43], 100-seed weight [44], and seed starch content [40]. In this study, a single plant was randomly selected from 157 F10 RILs to increase homozygosity and genotyped using the BARCSoySNP3K SNP array, which contained a subset of 2830 SNP loci distributed across 20 soybean chromosomes from BARCSoySNP6K [45]. In this study, >1400 SNPs were used to construct the improved genetic map, resulting in a better estimation of QTL positions for the linkage mapping study with this interspecific population.
The root is the hidden part of a plant that plays a crucial role in plant development by serving as a stabilizer, absorbing water and nutrients, and interacting with the soils microorganisms. Our study employed an interspecific crossing between ‘Williams 82’ and ‘PI366121’ and produced an RIL population to map QTLs for root traits. Parents and the RILs showed wide phenotypic variation for root attributes. In contrast to both parents, the cultivated soybean ‘Williams 82’ has a well-developed root system compared with the wild soybean ‘PI366121’, and transgressive segregation was observed in this study (Table 1 and Figure 4A–E). A rice QTL, DEEPER ROOTING1 (DRO1), regulates the root system architecture, including root angle and root tips, and increases rice yield [46]. Several studies have reported that root traits, including root length, surface area, and volume, have been found on different chromosomes, including chr. 3, 7, 8, and 20, which improve seedling growth in soybean [27,28,29]. Fibrous root-related QTLs have been observed on chr. 3, 4, 8, and 20 [34]. Furthermore, average root diameter, lateral root number, and RV-related QTLs were found on chr. 7, 17, and 20, which enhanced soybean root growth and development [47,48].
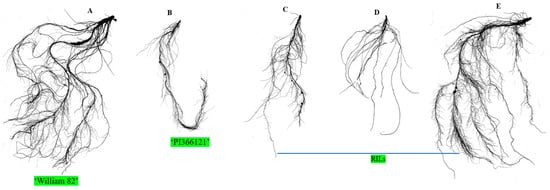
Figure 4.
Variation in 2D root image of the morphology of soybean seedlings; five randomly selected root samples, including parents (A) ‘William 82’, (B) ‘PI366121’, and (C–E) RILs.
In this study, we found nine important genomic regions on 12 chromosomes that were strongly linked with root morphological traits (Table 4). In more than 60% of the total root trait QTLs, a positive allele was contributed by the wild soybean accession ‘PI 366121’, with phenotypic variations of 6.9–39.0%, which significantly affected root attributes. Remarkably, AD QTL was mapped in this population on chr. 3 with positive alleles contributed by the cultivated soybean ‘William 82’ and co-located with LAD and RV QTLs, accounting for phenotypic variations of 16.2–39.1%. A positive allele contributed by inferior parental lines of both wild and cultivated soybeans has been reported in some root trait-related studies [28,49]. In our genomic regions, 12 QTLs had phenotypic variations of >30% and were considered major QTLs [39]. QTL qAD_19-1 was found on chr. 19 at position 85.6 cM, accounting for 39.0% phenotypic variation. Similar root traits of Qrd 14-1 and Qrd 12-1 were reported with 15% and 22% phenotypic variation and 3.36 and 6.15 LOD scores, respectively [50], and a single AD QTL was found with 7% phenotypic variation [49].
Based on our results, two QTL groups were mapped on chr. 3 and 13, along with co-located QTLs for AD, RV, and LAD (Figure S1). qAD-3, qRV-3, qLAD-3, qAD-13, qRV-13, and qLAD-13 were located in the same intervals on chr. 3 and 13, with a wide phenotypic variation (Table 4). The six QTLs may regulate soybean seedling root morphology, such as AD, RV, and LAD, as well as root development. Surprisingly, AD, RV, and LAD-related QTLs on chr. 3 have not been found in any other root studies. However, other root traits such as root length, distribution, and score-related QTLs were found in chr. 3 [28,29,34]. In chr. 13, only root weight QTL was found [29]; however, AD, RV, and LAD-related QTLs have not been reported yet. Additionally, some co-located QTLs were observed for soybean mapping populations, such as seedling root [27], plant height [51,52], and leaf morphology [53]. A significantly positive correlation was detected between AD and LAD (r = 0.97), followed by RV and LAD (r = 0.56) and AD and RV (r = 0.55) in our study, influencing root growth, development, and phenotypic variation of soybean seedlings. In the early growth stage of soybean, the total root length and photosynthetic efficiency correlated positively, influencing the lateral and fibrous rooting capacity under limited water conditions [54]. A weak positive correlation was observed between AD and RV (r = 0.32) and LAD and RV (r = 0.34) in soybean [49].
The SNP markers associated with root features, including AD, RV, and LAD, on chr. 3 and 13, were within the same marker intervals. This genomic region may harbor some root-functioning genes, thereby prompting the root growth and development of soybean seedlings. Soybean root length in chr. 3 with marker interval NCSB_000550–SNP5617_Magellan was mentioned as a potential candidate gene [28]. Several researchers have reported that the maker regions improve soybean root development and stress resistance [55,56,57]. Furthermore, root architectural traits minimize the drought tolerance of rice [17,57], and marker-assisted breeding improves the root system architecture of maize [58].
In this study, we performed analyses on an interspecific mapping population along with the entire genome transcriptome data of soybeans to identify genes associated with potential root traits and examine their differences in tissue/organ-specific expression. Based on our results, 32 genes for SNP variations were identified, including missense and splice variants that altered amino acids (Table 5). A similar method was reported, and variants were observed in soybean root traits [16,27], meristems in Arabidopsis [59], and lateral roots in rice [60]. In our study, the transcriptome data were used for gene expression with different tissue-specific expressions (Figure 3). A similar tactic was reported, and candidate genes for soybean root traits were identified [48]. We identified five candidate genes, including Glyma.03G027500 (transketolase/glycoaldehyde transferase), Glyma.03G014500 (dehydrogenases), Glyma.13G341500 (leucine-rich repeat receptor-like protein kinase), Glyma.13G341400 (AGC kinase family protein), and Glyma.13G331900 (60S ribosomal protein), which showed greater expression in the root. Compared with other plant tissues, these genes may enhance soybean seedling root morphological traits as well as AD, RV, and LAD. The leucine-rich repeat protein (LRP) class was observed within the root QTL of soybean, significantly influencing other root properties, including the number of lateral roots, root diameter, and RV [28]. Furthermore, LRP regulates Medicago truncatula roots [61]. The AGC protein kinase regulates auxin transport polarity [62] and organ (root) development in Arabidopsis [63]. The 60S ribosomal protein L14-2 reportedly enhances the drought and salt tolerance of cotton [64]. Additionally, we found one aquaporin functioning gene, Glyma.13G333100 (Aquaporin Transporter), which probably induces root development and enhances AD and RV. The same gene was reported in soybeans, which regulated seedling root growth under heat stress [65], and the same gene ID AT4G01740 (TIP1;3) controlled drought stress in Arabidopsis [66]. Six potential genes were recently identified in seven root traits, including AD, LAD, and the number of tips in soybean landraces [67]. Therefore, we conclude that the common QTL regions will benefit breeders in improving soybean root morphological traits as well as AD, RV, and LAD.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
In this study, an interspecific mapping population was used. A population of 157 F10:11 RILs was developed from a crossing between cultivated soybean ‘Williams 82’ and wild soybean ‘PI366121’ [42]. These RILs and parental lines were grown in polyvinyl chloride (PVC) pipes (6 cm (diameter) × 40 cm (height)). Furthermore, all (636) PVC pipes were placed on 16 trays (35 cm (wide) × 65 cm (long)) in a greenhouse at the research center of the Kyungpook National University, Daegu, South Korea. Each tray contained 40 PVC pipes; the trays, including the parents ‘Williams 82’ and ‘PI366121’, served as a control. Sandy soil was used in our experiment. Two seeds of each parent and RIL population were planted in the PVC pipes on 25 March 2022 (environment 1) and 27 April 2022 (environment 2), respectively. The study was conducted in a completely randomized block design (RCBD) with three replications. Greenhouse photoperiod (14 h daytime) and temperature (28 °C ± 2 °C) were maintained for seedling growth. After the germination of seeds, they were thinned, and a single seedling was allowed to grow for root analysis. At 24 days after the third trifoliate leaf developed, i.e., at the V3 stage, the seedlings were harvested for root samples in both environments.
4.2. Root Phenotypic Evaluation
Root phenotypic parameters were evaluated at the V3 stage of soybean seedlings in different environments (Figure 5A). During harvesting, all sandy soil was removed from the PVC pipes very carefully, and root samples were separated from the soil. The root samples were then softly washed with clean tap water and kept in medium (20 cm long × 15 cm wide) plastic bags containing a small amount (15–20 mL) of water to maintain moisture in the samples (Figure 5D). Next, a scanner was used to capture clear 2D root images (Epson, Expression 12,000XL, Nagano, Japan). A transparent plastic tray (30 cm long × 20 cm wide), which contained normal clean water, was used for root sample scanning (Figure 5E). The scanned root images were analyzed using WinRHIZO Pro software version 2019 (Regent Instruments Inc., Québec City, QC, Canada) (Figure 5F). In this study, three root trait parameters, AD, RV, and LAD, were measured (Table 7). LAD was considered a minor root trait. According to the WinRHIZO description (https://regent.qc.ca/assets/winrhizo_software.html accessed on 4 June 2023), LAD is a link of the root part between two forks or a fork and a tip. It is a study of the morphology and basic interaction of root segments measured by the AD of links that belong to an order. Therefore, we hypothesized that LAD affected AD and helped in root development by uptaking water and nutrients [10].
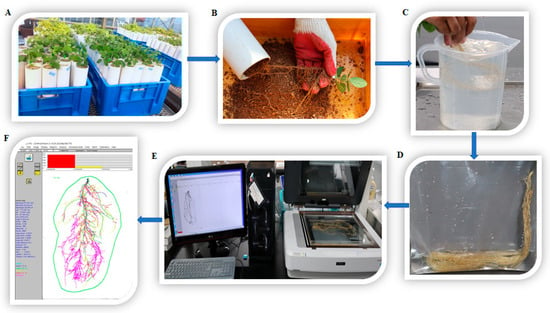
Figure 5.
Procedure for soybean seedling root analysis using WinRHIZO software. (A) Harvesting at the V3 stage of soybean seedlings. (B) Removal of soil and separation of roots from the pipe. (C) Washing of the root sample. (D) Clean roots are kept in plastic bags with a small amount of water. (E) Scanning of the clean root sample. (F) Analysis of the root sample.

Table 7.
List of soybean root morphological traits evaluated in the soybean mapping population.
Table 7.
List of soybean root morphological traits evaluated in the soybean mapping population.
| Trait Abbreviation | Description (Units) |
| AD | Average diameter (mm) |
| RV | Root volume (cm3) |
| LAD | Link average diameter (mm) |
4.3. Genotyping and Linkage Map Construction
The single plant of 157 F10 RILs and their parental lines (‘Williams 82’ and ‘PI366121’) were randomly selected, and genomic DNA from leaves were genotyped using the BARCSoySNP3K SNP array, which contained a subset of 2830 SNP loci distributed across 20 soybean chromosomes from BARCSoySNP6K [45]. This SNP genotype was developed at the soybean genomics and improvement laboratory at the USDA. After filtering, 1408 of the 2830 SNP markers were polymorphic between the parents and were used to construct a genetic linkage map for the 20 linkage groups. The SNP markers were binned based on their segregation patterns among the RIL population, employing the bin function in IciMapping 4.2 (http://www.isbreeding.net, accessed on 4 June 2023) [68]. Using the Kosambi mapping function, bin markers were systematically grouped and categorized using IciMapping 4.2 [69]. The total length of the soybean mapping population map was 4426.7 cM, with an average distance between the adjacent markers of 3.14 cM (Table S5). The average length of individual chromosomes or linkage groups was 221.33 cM, with an average number of markers per linkage group of 70.4 (Table S5). MapChart 2.2 (http://www.biometris.nl/uk/Software/MapChart/, accessed on 10 June 2023) software was used to draw the genetic linkage map [70]. Genomic DNA was isolated from the fresh soybean young leaves using a modified cetyl trimethylammonium bromide method [71].
4.4. QTL Mapping Analysis
The QTLs for soybean root traits were identified using WinQTLCart 2.5 based on the composite interval mapping procedure [72]. The software program uses an enhanced algorithm for composite interval mapping, which has a heightened ability to identify QTLs, lower false detection rates, and provide less skewed QTL effect estimates. The model 6 was used, and the window size was set at 10 cM with background cofactors. To declare significant QTLs with a more rigorous LOD threshold, a permutation (p = 0.05) test was performed with 1000 runs for the mapping software packages for all traits. The forward regression approach was employed to determine the walking speed, which was set at 2 cM. The QTL analysis incorporated the mean values of the three distinct soybean root morphological traits. The QTL map positions on the linkage maps were depicted using the Mapchart program.
4.5. Candidate Gene Identification and Expression Analysis
We used the two most significant QTL regions (Table 4 and Figure S1) on chr. 3 and 13 to identify potential candidate genes based on annotation using Soybase (https://soybase.org/SequenceIntro.php, accessed on 5 July 2023) according to the ‘Wm82.a2.v1’ soybean reference genome. The genome browser was used to identify potential genes within each significant SNP flanking region, and the Phytozome database [73] was used to perform functional annotation of the genes (Table S1). Furthermore, we identified the SNP and INDEL variations with genomes ‘Williams 82’ and ‘PI366121’, using the Soykb database (https://soykb.org/SNPViz2/, accessed on 18 July 2023) [74] (Table S2). We used the ePlant soybean database (https://bar.utoronto.ca/eplant-soybean/, accessed on 18 July 2023) to identify the root trait-related potential gene expression for putative candidate genes, and a heatmap was constructed with root tissue data by the Tbtools software (https://github.com/CJ-Chen/Tbtools, accessed on 29 July 2023) (Table S4). The selected genes were used for distinction expression analysis in diverse tissues using web-based, publicly available RNA-Seq soybean library data (4085) with default settings (http://ipf.sustech.edu.cn/pub/soybean/, accessed on 21 July 2023) [75].
4.6. Statistical Analysis
The experiment used a randomized complete block design (RCBD) with three replications. The root phenotypic traits of the parental lines and RIL population were tested for descriptive statistics with a normal frequency distribution using IBM SPSS statistics 25. To assess statistical significance, we performed an ANOVA with SAS (SAS release 9.4; SAS, Gary, NC, USA). Correlation (Pearson correlation coefficients) analysis was performed using SAS PROC CORR to determine the relationship between root traits. In Soybase, the chr. numbers corresponded to the soybean genetic linkage group (http://www.soybase.org, accessed on 20 April 2023).
5. Conclusions
In this study, we performed QTL mapping for soybean root traits in an interspecific population derived from the ‘Williams 82’ and ‘PI366121’ crosses. We identified 42 QTLs, including 12 major QTLs, distributed across 12 chromosomes. The QTL regions on chr. 3 and 13 were found to control AD, RV, and LAD, with positive alleles derived from both ‘PI366121’ and ‘Williams 82’. These QTL regions have the potential to enhance root development. Additionally, we detected six candidate genes within the most significant QTL regions that influence soybean root morphological traits. In conclusion, these significant regions are valuable for breeders seeking to improve soybean root morphological traits, leveraging alleles inherited from wild soybean accessions.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25094687/s1.
Author Contributions
Y.K. conceptualization. M.S.I. prepared a draft of the manuscript. A.G., L.L. and W.K. performed root phenotype analysis and data collection. M.S.I. and H.J. performed formal analysis and validation. Q.S. genotyped the RIL population, and J.-D.L. provided the planting materials. Y.K. supervised the experimental design and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1I1A3040280).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated in this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Medic, J.; Atkinson, C.; Hurburgh, C.R. Current knowledge in soybean composition. J. Am. Oil Chem. Soc. 2014, 91, 363–384. [Google Scholar] [CrossRef]
- Khojely, D.M.; Ibrahim, S.E.; Sapey, E.; Han, T. History, current status, and prospects of soybean production and research in sub-Saharan Africa. Crop J. 2018, 6, 226–235. [Google Scholar] [CrossRef]
- Rahman, S.U.; McCoy, E.; Raza, G.; Ali, Z.; Mansoor, S.; Amin, I. Improvement of soybean; A way forward transition from genetic engineering to new plant breeding technologies. Mol. Biotechnol. 2023, 65, 162–180. [Google Scholar] [CrossRef] [PubMed]
- Carter, T.E., Jr.; Nelson, R.L.; Clay, S.H.; Cui, Z. Genetic diversity in soybean. In Soybeans: Improvement, Production, and Uses; Shibles, R.M., Harper, J.E., Wilson, R.F., Shoemaker, R.C., Eds.; Wiley Online Library: Hoboken, NJ, USA, 2004; Volume 16, pp. 303–416. [Google Scholar]
- Lee, G.-A.; Crawford, G.W.; Liu, L.; Sasaki, Y.; Chen, X. Archaeological soybean (Glycine max) in East Asia: Does size matter? PLoS ONE 2011, 6, e26720. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.-C.; Moon, J.-K.; Park, S.-K.; Kim, M.-S.; Lee, K.; Lee, S.R.; Jeong, N.; Choi, M.S.; Kim, N.; Kang, S.-T. Genetic diversity patterns and domestication origin of soybean. Theor. Appl. Genet. 2019, 132, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Gregory, P.J. RUSSELL REVIEW Are plant roots only “in” soil or are they “of” it? Roots, soil formation and function. Eur. J. Soil Sci. 2022, 73, e13219. [Google Scholar] [CrossRef]
- Lynch, J.P. Harnessing root architecture to address global challenges. Plant J. 2022, 109, 415–431. [Google Scholar] [CrossRef] [PubMed]
- Hodge, A.; Berta, G.; Doussan, C.; Merchan, F.; Crespi, M. Plant root growth, architecture and function. Plant Soil 2009, 321, 153–187. [Google Scholar] [CrossRef]
- Tripathi, P.; Abdullah, J.S.; Kim, J.; Chung, Y.-S.; Kim, S.-H.; Hamayun, M.; Kim, Y. Investigation of root morphological traits using 2D-imaging among diverse soybeans (Glycine max L.). Plants 2021, 10, 2535. [Google Scholar] [CrossRef] [PubMed]
- Manschadi, A.M.; Christopher, J.; deVoil, P.; Hammer, G.L. The role of root architectural traits in adaptation of wheat to water-limited environments. Funct. Plant Biol. 2006, 33, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, L.P.; Guttikonda, S.K.; Nguyen, V.T.; Shannon, J.G.; Nguyen, H.T. Evaluation of diverse soybean germplasm for root growth and architecture. Plant Soil 2010, 330, 503–514. [Google Scholar] [CrossRef]
- Valliyodan, B.; Van Toai, T.T.; Alves, J.D.; de Fátima P. Goulart, P.; Lee, J.D.; Fritschi, F.B.; Rahman, M.A.; Islam, R.; Shannon, J.G.; Nguyen, H.T. Expression of root-related transcription factors associated with flooding tolerance of soybean (Glycine max). Int. J. Mol. Sci. 2014, 15, 17622–17643. [Google Scholar] [CrossRef]
- Varshney, R.K.; Pazhamala, L.; Kashiwagi, J.; Gaur, P.M.; Krishnamurthy, L.; Hoisington, D. Genomics and physiological approaches for root trait breeding to improve drought tolerance in chickpea (Cicer arietinum L.). In Root Genomics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 233–250. [Google Scholar]
- Sponchiado, B.N.; White, J.W.; Castillo, J.A.; Jones, P.G. Root growth of four common bean cultivars in relation to drought tolerance in environments with contrasting soil types. Exp. Agric. 1989, 25, 249–257. [Google Scholar] [CrossRef]
- Prince, S.J.; Valliyodan, B.; Ye, H.; Yang, M.; Tai, S.; Hu, W.; Murphy, M.; Durnell, L.A.; Song, L.; Joshi, T. Understanding genetic control of root system architecture in soybean: Insights into the genetic basis of lateral root number. Plant Cell Environ. 2019, 42, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Suji, K.K.; Prince, K.S.J.; Mankhar, P.S.; Kanagaraj, P.; Poornima, R.; Amutha, K.; Kavitha, S.; Biji, K.R.; Gomez, S.M.; Babu, R.C. Evaluation of rice (Oryza sativa L.) near iso-genic lines with root QTLs for plant production and root traits in rainfed target populations of environment. Field Crops Res. 2012, 137, 89–96. [Google Scholar] [CrossRef]
- Wasson, A.P.; Richards, R.A.; Chatrath, R.; Misra, S.C.; Prasad, S.V.S.; Rebetzke, G.J.; Kirkegaard, J.A.; Christopher, J.; Watt, M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012, 63, 3485–3498. [Google Scholar] [CrossRef] [PubMed]
- Tuberosa, R.; Salvi, S.; Giuliani, S.; Sanguineti, M.C.; Frascaroli, E.; Conti, S.; Landi, P. Genomics of root architecture and functions in maize. In Root Genomics; Springer: Berlin/Heidelberg, Germany, 2010; pp. 179–204. [Google Scholar]
- Beeckman, T.; De Smet, I.; Den Herder, G.; Van Isterdael, G. The roots of a new green revolution. Trends Plant Sci. 2010, 15, 600–607. [Google Scholar]
- Wang, X.; Xu, Y.; Hu, Z.; Xu, C. Genomic selection methods for crop improvement: Current status and prospects. Crop J. 2018, 6, 330–340. [Google Scholar] [CrossRef]
- Lidder, P.; Sonnino, A. Biotechnologies for the management of genetic resources for food and agriculture. Adv. Genet. 2012, 78, 1–167. [Google Scholar] [PubMed]
- Lee, J.D.; Yu, J.K.; Hwang, Y.H.; Blake, S.; So, Y.S.; Lee, G.J.; Nguyen, H.T.; Shannon, J.G. Genetic diversity of wild soybean (Glycine soja Sieb. and Zucc.) accessions from South Korea and other countries. Crop Sci. 2008, 48, 606–616. [Google Scholar] [CrossRef]
- Mammadov, J.; Buyyarapu, R.; Guttikonda, S.K.; Parliament, K.; Abdurakhmonov, I.Y.; Kumpatla, S.P. Wild relatives of maize, rice, cotton, and soybean: Treasure troves for tolerance to biotic and abiotic stresses. Front. Plant Sci. 2018, 9, 886. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.S.; Diers, B.W.; Hyten, D.L.; Rouf Mian, M.A.; Shannon, J.G.; Nelson, R.L. Identification of positive yield QTL alleles from exotic soybean germplasm in two backcross populations. Theor. Appl. Genet. 2012, 125, 1353–1369. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Valliyodan, B.; Wu, J.-H.; Lee, S.-H.; Xu, D.; Nguyen, H.T. Genomic differences between cultivated soybean, G. max and its wild relative G. soja. BMC Genom. 2013, 14, S5. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, L.P.; Prince, S.J.; Musket, T.A.; Chaky, J.; Deshmukh, R.; Vuong, T.D.; Song, L.; Cregan, P.B.; Nelson, J.C.; Shannon, J.G. Identification of novel QTL governing root architectural traits in an interspecific soybean population. PLoS ONE 2015, 10, e0120490. [Google Scholar] [CrossRef] [PubMed]
- Prince, S.J.; Vuong, T.D.; Wu, X.; Bai, Y.; Lu, F.; Kumpatla, S.P.; Valliyodan, B.; Shannon, J.G.; Nguyen, H.T. Mapping quantitative trait loci for soybean seedling shoot and root architecture traits in an inter-specific genetic population. Front. Plant Sci. 2020, 11, 1284. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Yu, Y.; Yang, H.; Xu, L.; Dong, W.; Du, H.; Cui, W.; Zhang, H. Inheritance and QTL mapping of related root traits in soybean at the seedling stage. Theor. Appl. Genet. 2014, 127, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Pantalone, V.R.; Rebetzke, G.J.; Burton, J.W.; Carter, T.E., Jr. Phenotypic evaluation of root traits in soybean and applicability to plant breeding. Crop Sci. 1996, 36, 456–459. [Google Scholar] [CrossRef]
- Slovak, R.; Ogura, T.; Satbhai, S.B.; Ristova, D.; Busch, W. Genetic control of root growth: From genes to networks. Ann. Bot. 2016, 117, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Waidmann, S.; Sarkel, E.; Kleine-Vehn, J. Same same, but different: Growth responses of primary and lateral roots. J. Exp. Bot. 2020, 71, 2397–2411. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Chen, H.; Wang, X.; Wu, B.; Chen, S.; Zhang, X.; Wu, X.; Yang, Z.; Qiu, D.; Jiang, M. QTL analysis of root traits of soybean at seedling stage. Acta Agron. Sin. 2011, 37, 1151–1158. [Google Scholar] [CrossRef]
- Abdel-Haleem, H.; Lee, G.-J.; Boerma, R.H. Identification of QTL for increased fibrous roots in soybean. Theor. Appl. Genet. 2011, 122, 935–946. [Google Scholar] [CrossRef]
- Chen, H.; Kumawat, G.; Yan, Y.; Fan, B.; Xu, D. Mapping and validation of a major QTL for primary root length of soybean seedlings grown in hydroponic conditions. BMC Genom. 2021, 22, 132. [Google Scholar] [CrossRef] [PubMed]
- Poorter, H.; Nagel, O. The role of biomass allocation in the growth response of plants to different levels of light, CO2, nutrients and water: A quantitative review. Funct. Plant Biol. 2000, 27, 1191. [Google Scholar] [CrossRef]
- Manschadi, A.M.; Hammer, G.L.; Christopher, J.T.; Devoil, P. Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 2008, 303, 115–129. [Google Scholar] [CrossRef]
- Lobet, G.; Pagès, L.; Draye, X. A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 2011, 157, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, H.D., Jr.; Otto, K.G.; Frewen, B.E.; McKay, J.K.; Schemske, D.W. Quantitative trait loci affecting differences in floral morphology between two species of monkeyflower (Mimulus). Genetics 1998, 149, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Dhungana, S.K.; Kulkarni, K.P.; Park, C.W.; Jo, H.; Song, J.T.; Shin, D.H.; Lee, J.D. Mapping quantitative trait loci controlling soybean seed starch content in an interspecific cross of ‘Williams 82’ (Glycine max) and ‘PI 366121’ (Glycine soja). Plant Breed. 2017, 136, 379–385. [Google Scholar] [CrossRef]
- Lenis, J.M. Genetics of Soybean Seed Lipoxygenases and Linolenic Acid Content in Seeds of the Soybean Wild Ancestor; University of Missouri-Columbia: Columbia, MO, USA, 2011. [Google Scholar]
- Lee, J.S.; Yoo, M.-h.; Jung, J.K.; Bilyeu, K.D.; Lee, J.-D.; Kang, S. Detection of novel QTLs for foxglove aphid resistance in soybean. Theor. Appl. Genet. 2015, 128, 1481–1488. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-S.; Kim, J.-M.; Jung, J.; Shin, I.; Park, S.; Lee, J.S.; Jeong, S.-C.; Lee, J.-D.; Jung, J.K.; Ha, B.-K. Fine-mapping and candidate gene analysis for the foxglove aphid resistance gene Raso2 from wild soybean PI 366121. Theor. Appl. Genet. 2021, 134, 2687–2698. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, K.P.; Asekova, S.; Lee, D.-H.; Bilyeu, K.; Song, J.T.; Lee, J.-D. Mapping QTLs for 100-seed weight in an interspecific soybean cross of Williams 82 (Glycine max) and PI 366121 (Glycine soja). Crop Pasture Sci. 2017, 68, 148–155. [Google Scholar] [CrossRef]
- Song, Q.; Yan, L.; Quigley, C.; Fickus, E.; Wei, H.; Chen, L.; Dong, F.; Araya, S.; Liu, J.; Hyten, D. Soybean BARCSoySNP6K: An assay for soybean genetics and breeding research. Plant J. 2020, 104, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Huang, C.; Niu, Y.; Yung, W.-S.; Xiao, Z.; Wong, F.-L.; Huang, M.; Wang, X.; Man, C.-K.; Sze, C.-C. QTL analyses of soybean root system architecture revealed genetic relationships with shoot-related traits. Theor. Appl. Genet. 2022, 135, 4507–4522. [Google Scholar] [CrossRef] [PubMed]
- Seck, W.; Torkamaneh, D.; Belzile, F. Comprehensive genome-wide association analysis reveals the genetic basis of root system architecture in soybean. Front. Plant Sci. 2020, 11, 590740. [Google Scholar] [CrossRef]
- Prince, S.J.; Song, L.; Qiu, D.; Maldonado dos Santos, J.V.; Chai, C.; Joshi, T.; Patil, G.; Valliyodan, B.; Vuong, T.D.; Murphy, M. Genetic variants in root architecture-related genes in a Glycine soja accession, a potential resource to improve cultivated soybean. BMC Genom. 2015, 16, 132. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, L.; Takahashi, R.; Githiri, S.M.; Rodriguez, T.O.; Tsutsumi, N.; Kajihara, S.; Sayama, T.; Ishimoto, M.; Harada, K.; Suematsu, K. Mapping quantitative trait loci for root development under hypoxia conditions in soybean (Glycine max L. Merr.). Theor. Appl. Genet. 2017, 130, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Guzman, P.S.; Diers, B.W.; Neece, D.J.; St. Martin, S.K.; LeRoy, A.R.; Grau, C.R.; Hughes, T.J.; Nelson, R.L. QTL associated with yield in three backcross-derived populations of soybean. Crop Sci. 2007, 47, 111–122. [Google Scholar] [CrossRef]
- Du, W.; Wang, M.; Fu, S.; Yu, D. Mapping QTLs for seed yield and drought susceptibility index in soybean (Glycine max L.) across different environments. J. Genet. Genom. 2009, 36, 721–731. [Google Scholar] [CrossRef]
- Orf, J.H.; Chase, K.; Adler, F.R.; Mansur, L.M.; Lark, K.G. Genetics of soybean agronomic traits: II. Interactions between yield quantitative trait loci in soybean. Crop Sci. 1999, 39, 1652–1657. [Google Scholar] [CrossRef]
- Prince, S.J.; Murphy, M.; Mutava, R.N.; Zhang, Z.; Nguyen, N.; Kim, Y.H.; Pathan, S.M.; Shannon, G.J.; Valliyodan, B.; Nguyen, H.T. Evaluation of high yielding soybean germplasm under water limitation. J. Integr. Plant Biol. 2016, 58, 475–491. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fujita, T.; Yan, Z.-H.; Sakamoto, S.; Xu, D.; Abe, J. QTL mapping of domestication-related traits in soybean (Glycine max). Ann. Bot. 2007, 100, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Ying, L.; Junyi, G.; Huineng, L. Identification of rhizosphere abiotic stress tolerance and related root traits in soybean (Glycine max L. Merr.). Acta Agron. Sin. 2005, 31, 1132–1137. [Google Scholar]
- Shouping, Y.; Jiamin, C.; Xiaohong, H.; Deyue, Y.; Junyi, G. Inheritance of drought tolerance and root traits of seedling in soybeans. Soybean Sci. 2005, 24, 275–280. [Google Scholar]
- Ju, C.; Zhang, W.; Liu, Y.; Gao, Y.; Wang, X.; Yan, J.; Yang, X.; Li, J. Genetic analysis of seedling root traits reveals the association of root trait with other agronomic traits in maize. BMC Plant Biol. 2018, 18, 171. [Google Scholar] [CrossRef] [PubMed]
- Meijón, M.; Satbhai, S.B.; Tsuchimatsu, T.; Busch, W. Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 2014, 46, 77–81. [Google Scholar] [CrossRef]
- Lyu, J.; Zhang, S.; Dong, Y.; He, W.; Zhang, J.; Deng, X.; Zhang, Y.; Li, X.; Li, B.; Huang, W. Analysis of elite variety tag SNPs reveals an important allele in upland rice. Nat. Commun. 2013, 4, 2138. [Google Scholar] [CrossRef] [PubMed]
- De Lorenzo, L.; Merchan, F.; Laporte, P.; Thompson, R.; Clarke, J.; Sousa, C.; Crespi, M. A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 2009, 21, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Offringa, R. Regulation of auxin transport polarity by AGC kinases. Curr. Opin. Plant Biol. 2008, 11, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Benjamins, R.; Quint, A.B.; Weijers, D.; Hooykaas, P.; Offringa, R. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 2001, 128, 4057–4067. [Google Scholar] [CrossRef] [PubMed]
- Shiraku, M.L.; Magwanga, R.O.; Cai, X.; Kirungu, J.N.; Xu, Y.; Mehari, T.G.; Hou, Y.; Wang, Y.; Wang, K.; Peng, R. Knockdown of 60S ribosomal protein L14-2 reveals their potential regulatory roles to enhance drought and salt tolerance in cotton. J. Cotton Res. 2021, 4, 27. [Google Scholar] [CrossRef]
- Feng, Z.-J.; Liu, N.; Zhang, G.-W.; Niu, F.-G.; Xu, S.-C.; Gong, Y.-M. Investigation of the AQP family in soybean and the promoter activity of TIP2; 6 in heat stress and hormone responses. Int. J. Mol. Sci. 2019, 20, 262. [Google Scholar] [CrossRef] [PubMed]
- Alexandersson, E.; Fraysse, L.; Sjövall-Larsen, S.; Gustavsson, S.; Fellert, M.; Karlsson, M.; Johanson, U.; Kjellbom, P. Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 2005, 59, 469–484. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Tayade, R.; Kang, B.-H.; Hahn, B.-S.; Ha, B.-K.; Kim, Y.-H. Genome-Wide Association Studies of Seven Root Traits in Soybean (Glycine max L.) Landraces. Int. J. Mol. Sci. 2023, 24, 873. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Kosambi, D.D. The estimation of map distances from recombination values. In DD Kosambi: Selected Works in Mathematics and Statistics; Ramaswamy, R., Ed.; Springer: New Delhi, India, 2016; pp. 125–130. [Google Scholar]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Lodhi, M.A.; Ye, G.-N.; Weeden, N.F.; Reisch, B.I. A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 1994, 12, 6–13. [Google Scholar] [CrossRef]
- Wang, S. Windows QTL Cartographer 2.5. 2006. Available online: https://brcwebportal.cos.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 22 April 2024).
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Škrabišová, M.; Lyu, Z.; Chan, Y.O.; Dietz, N.; Bilyeu, K.; Joshi, T. Application of SNPViz v2.0 using next-generation sequencing data sets in the discovery of potential causative mutations in candidate genes associated with phenotypes. Int. J. Data Min. Bioinform. 2021, 25, 65–85. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, H.; Long, Y.; Shu, Y.; Zhai, J. Plant public RNA-seq database: A comprehensive online database for expression analysis of ~45000 plant public RNA-seq libraries. Plant Biotechnol. J. 2022, 20, 806. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).