Abstract
The type III secretion system (T3SS) is a key factor for the symbiosis between rhizobia and legumes. In this study, we investigated the effect of calcium on the expression and secretion of T3SS effectors (T3Es) in Sinorhizobium fredii NGR234, a broad host range rhizobial strain. We performed RNA-Seq analysis of NGR234 grown in the presence of apigenin, calcium, and apigenin plus calcium and compared it with NGR234 grown in the absence of calcium and apigenin. Calcium treatment resulted in a differential expression of 65 genes, most of which are involved in the transport or metabolism of amino acids and carbohydrates. Calcium had a pronounced effect on the transcription of a gene (NGR_b22780) that encodes a putative transmembrane protein, exhibiting a 17-fold change when compared to NGR234 cells grown in the absence of calcium. Calcium upregulated the expression of several sugar transporters, permeases, aminotransferases, and oxidoreductases. Interestingly, calcium downregulated the expression of nodABC, genes that are required for the synthesis of nod factors. A gene encoding a putative outer membrane protein (OmpW) implicated in antibiotic resistance and membrane integrity was also repressed by calcium. We also observed that calcium reduced the production of nodulation outer proteins (T3Es), especially NopA, the main subunit of the T3SS pilus. Additionally, calcium mediated the cleavage of NopA into two smaller isoforms, which might affect the secretion of other T3Es and the symbiotic establishment. Our findings suggest that calcium regulates the T3SS at a post-transcriptional level and provides new insights into the role of calcium in rhizobia–legume interactions.
1. Introduction
A group of soil proteobacteria, known as rhizobia, are able to establish a mutualistic interaction with leguminous plants. As a result, the plant forms new root organs called nodules where rhizobia are differentiated into bacteroids, a new physiological and morphological state able to fix atmospheric nitrogen to ammonia [1,2]. The success of this process relies on a complex and coordinated molecular signal interchange between both actors, since only compatible rhizobial strains will be able to induce the formation of effective and, consequently, nitrogen-fixing nodules [3].
The molecular dialogue starts with the release of a set of molecules by the plant roots, including flavonoids, phenolic compounds that interact with the bacterial protein regulator NodD [4,5]. This interaction results in the binding of NodD to specific promoter sequences, called nod boxes (NBs), and the consequent induction of genes located downstream of these NBs, which are mostly involved in the symbiotic process [6]. Among these induced genes are the nod genes, whose function is the production and secretion of the symbiotic signals named nod factors (NF). The perception of the NFs by plant root LysM receptors trigger bacterial infection and nodule organogenesis [7,8,9].
In addition to NF, other bacterial molecular signals are involved in the success of the symbiotic process, such as a set of secreted proteins collectively known as Nops (nodulation outer proteins). These proteins are secreted by a type III secretion system (T3SS) in a NodD-flavonoid dependent manner. NodD activates the expression of TtsI, a transcriptional regulator that recognizes and binds specific promoter sequences, called tts boxes, located upstream of T3SS-related genes. Thus, TtsI induces the expression of genes involved in both the T3SS machinery assembly and the production of Nops. These proteins are delivered through the T3SS into host plant cells where they may alter host pathways or suppress plant defense responses. Therefore, Nops are involved in nodulation efficiency and host-range determination [10,11,12,13,14,15].
Sinorhizobium fredii NGR234 (hereafter NGR234) is a fast-growing rhizobial strain isolated from Lablab purpureus nodules from Papua New Guinea [16]. Unlike other Sinorhizobium fredii strains, NGR234 possesses the broadest nodulation host range known so far, nodulating 112 genera including legumes and the non-legume Parasponia andersonii [17]. The symbiotic effects caused by defects in the Nops secretion can be beneficial, detrimental, or have no effect, depending on the specific rhizobia/legume couple. Even the recognition of one Nop can completely block the nodulation [13,18,19]. In the case of a NGR234 rhcN mutant, unable to secrete any Nops, the symbiotic effects can vary depending on the plant tested, producing an increase, no changes, or a decrease in the nodule number [18]. These changes are not only produced among different plant species but also among different cultivars, for example as occurs in the G. soja cultivars CH2, CH3, and CH4 inoculated with S. fredii HH103 T3SS mutants, which are able to induce nitrogen-fixing nodules in CH2 (while the wild type is unable) or cause a nodulation impairment in CH3 and CH4 [20]. In the same strain, the T3SS abolition also provokes an increase in the nodulation host range, allowing the appearance of nitrogen-fixing nodules in the plant model legume Lotus japonicus Gifu (the wild type only produces ineffective nodules) and switching the infection mode from intercellular infection to a more evolved one by infection thread formation in L. burttii [18].
In a previous report, it was demonstrated that Nops production was drastically reduced in the extracellular media of flavonoid-induced culture of S. fredii USDA257 in the presence of calcium. This effect seems to occur at post-transcriptional level and is calcium specific since other cations such as magnesium or manganese had no effect on Nops production in the same conditions [21]. In this work, we have investigated the effect of calcium on the overall NGR234 gene expression in the absence and presence of the inducer flavonoid apigenin through RNA-seq experiments. Our results show that calcium supplemented at 0.5 mM concentration is able to modulate the expression of a specific set of NGR234 genes, even in the presence of apigenin, since the gene expression profile differs in the presence of both elements. In addition, we have investigated the expression of genes involved in symbiotic signals, such as NF or Nops production, upon calcium, apigenin, and both treatments together. We detected that Nops production is drastically reduced in the presence of high amounts of calcium. Surprisingly, we identified that the NopA protein exhibited a cleavage process in the presence of apigenin and calcium, indicating that calcium may have an important role in Nops secretion.
2. Results
2.1. Calcium Inhibits the Secretion of NGR 234 Nops
Sinorhizobium fredii NGR 234, S. fredii USDA257, and S. fredii HH103 secretes several nodulation outer proteins (Nops) into the extracellular milieu, when grown in the presence of flavonoids in the growth media [19,22,23]. In a previous work, it has been demonstrated that flavonoid-induced S. fredii USDA257 cultures in the presence of increasing amounts of calcium-reduced Nops production secreted into the extracellular media [21]. In this work, we decided to investigate whether this reduction also occurs in NGR234. For this purpose, we isolated NGR234 Nops produced in the presence of 1 µM of apigenin and increasing levels of Ca2+ and analyzed them by SDS-PAGE and subsequent silver staining (Figure 1). When NGR 234 was grown in the presence of 1 µM apigenin without any added calcium, several Nops including NopX, NopL, NopP, NopB, NopC, and NopA accumulated in the extracellular milieu (Figure 1). Interestingly, the addition of 1 mM Ca2+ to the growth medium resulted in a drastic reduction in the Nops accumulation observed when NGR 234 was grown in the presence of 1 mM calcium (Figure 1). A densitometer scan of the stained gel clearly demonstrated the severe reduction of Nops accumulation in culture media containing 1 mM Ca2+ (Supplementary Figure S1).
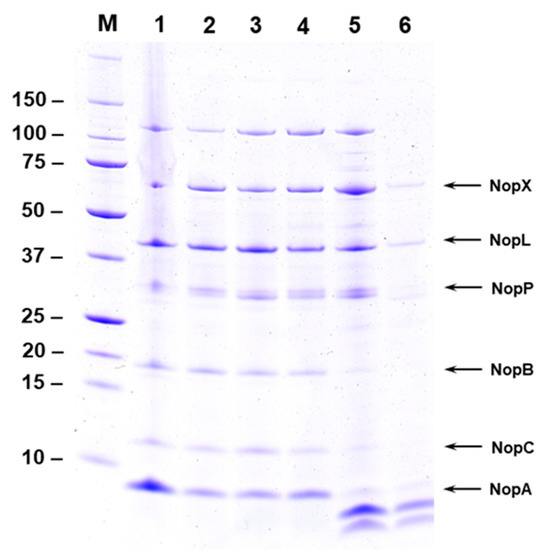
Figure 1.
Effect of calcium on Nops secretion. Nops from S. fredii NGR234 grown for 48 h in presence of 1 µM apigenin and increasing concentration of calcium were isolated and separated by SDS-PAGE. Resolved proteins were visualized by silver stain. NGR234 was cultured in YEM medium containing 1 µM apigenin and 1 µM (lane 2), 10 µM (lane 3), 100 µM (lane 4), 500 µM (lane 5), and 1000 µM (lane 6) of calcium. Nops isolated from NGR234 grown in the presence of 1 µM apigenin without any added calcium is shown in lane 1. The identity of Nops and the sizes of molecular weight markers are indicated on the sides of the figure.
2.2. Calcium Promotes the Cleavage of NopA
Calcium at higher concentration (1 mM) inhibited the secretion of NGR234 Nops into the extracellular media (Figure 1). Interestingly, two new proteins were also detected in the extracellular media when NGR234 was grown in the presence of apigenin and 0.5–1 mM calcium (Figure 1 and Figure 2). The new proteins appeared to have low molecular weight and were not seen in the culture supernatants when calcium concentration was lower than 0.5 mM (Figure 1). We wanted to examine if these low-molecular-weight proteins represent cleaved products of NopA or new calcium-induced proteins. To test our hypothesis, we performed immunoblot analysis using antibodies specific for NopA. The NopA antibodies recognized the 8 kDa protein when NGR234 was grown in the presence of 1 µM apigenin (Figure 2A, labeled ‘A’). In addition to this 8 kDa protein, one additional low-molecular-weight protein (Figure 2A, labeled ‘B’) was also recognized by NopA antibodies when NGR234 was grown in the presence of 1 µM apigenin and 0.5 mM calcium (Figure 2A). However, the NopA antibodies failed to recognize the lower molecular weight protein (Figure 2A, labeled ‘C’) indicating that either this peptide does not contain the epitope recognized by the NopA antibody or it could be a new calcium-induced protein. To verify the identity of these low-molecular-weight proteins, we excised gel slices corresponding to NopA (Band A) and the two other peptides (Bands B and C) and analyzed them by mass spectrometry. This analysis revealed that band A, B, and C showed significant sequence homology to the S. fredii NGR234 NopA (Supplementary Table S3). Several peptides from band A, B, and C gave statistically significant protein scores for the matches with NopA, with MOWSE scores above the 95% confidence level (Supplementary Table S3). Band A was composed of peptides that covered the full length of NopA while the peptides included in band B and C matched 85% and 87% amino acid sequences of NopA. Our analysis confirmed that band B and C both correspond to NopA fragments although band C was not recognized by the NopA antibody (Figure 2).
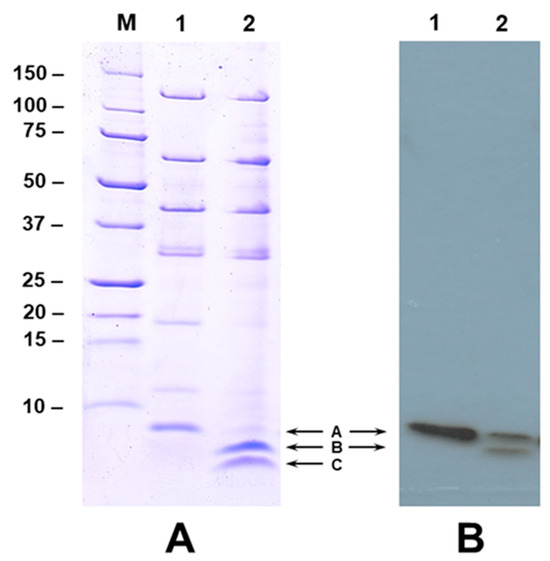
Figure 2.
Calcium promotes the cleavage of NopA. Panel A. SDS-PAGE analysis of NGR234 Nops produced in the presence of 1 µM apigenin (lane 1) and 1 µM apigenin and 500 µM of calcium (lane 2). Nops were separated on a 15% acrylamide gel and visualized by Coomassie Blue stain. Panel B. Immunoblot analysis of NopA cleavage. Proteins shown in Panel A were transferred to nitrocellulose membrane and incubated with NopA specific polyclonal antibodies. The immunoreactive polypeptides were detected by the chemiluminescent detection method. The position of NopA and cleaved NopA products are shown with arrows.
2.3. Global Gene Expression Analysis
In order to analyze the impact of the addition of calcium and apigenin on the global gene expression of NGR234, we performed RNA-seq experiments. We compared the transcriptome profile at the early stationary growth phase of NGR234 cultures that were grown in YEM under the following conditions: (1) in the presence of calcium at 0.5 mM concentration, (2) apigenin at 1 µM, (3) 0.5 mM calcium and 1 µM apigenin, and (4) control condition (no calcium or apigenin added). For each condition, three independent biological sample experiments were performed, and their corresponding RNA samples were obtained. Thus, 12 cDNA libraries were generated and sequenced, obtaining between 39 and 86 million reads in each condition. In general, all the samples showed about 99.6% alignment when they were mapped to the NGR234 genome (Genome assembly GCF_000018545.1).
The differentially expressed genes (DEGs) were considered as statistically significant when the fold change ratio between two conditions was ≥3 and had an adjusted p value of ≤0.05. The DEGs of each comparison are listed in the Supplementary Datasets S1–S4, where the shown data are the mean of the three replicates in each condition. The transcriptome of NGR234 in the presence of apigenin, calcium, and both compounds together showed a total of 150 (131 upregulated and 19 downregulated), 65 (39 upregulated and 26 downregulated), and 187 (162 upregulated and 25 downregulated) DEGs, respectively, compared to the NGR234 grown in control conditions (Table 1).

Table 1.
Summary of S. fredii NGR234 DEGs in each condition and replicon. WT, control conditions; AP, apigenin; Ca, calcium.
The NGR234 genome is composed of 6443 genes; the supplementation of apigenin, calcium and the combination of both compounds affected the expression of about 2.3%, 1.0%, and 2.9% of the genome of this strain, respectively. Depending on the condition, the number of DEGs in each replicon is different. For example, the presence of apigenin showed 46.7% out of the total number of DEGs in the symbiotic plasmid (plasmid a), while 56.9% of the DEGs observed in the presence of calcium are located on the chromosome. In the case of the DEGs obtained by the presence of both compounds together, apigenin and calcium, about 81% out of the total number of DEGs are located on the chromosome and plasmid a (Table 2). Curiously, 87.3% of the DEGs seen in the presence of apigenin were upregulated (independently of the replicon analyzed), while calcium treatment upregulated and repressed 60% and 40% of the DEGs, respectively.

Table 2.
Summary of DEGs percentage of S. fredii NGR234 in each condition and replicon. WT, control conditions; AP, apigenin; Ca, calcium.
2.4. Effect of Apigenin on the NGR234 Transcriptome
Previous transcriptomic studies carried out in the presence of inducer flavonoids demonstrated a profound effect on the global gene expression in several rhizobia and have identified numerous differentially expressed genes [18]. Most of the genes that are upregulated upon flavonoid treatment were located on the symbiotic plasmids while a limited number of genes of this plasmid were also down regulated [24,25]. In accordance with previous reports, we also found that apigenin regulated the expression of 150 genes, out of which 131 were upregulated and 19 were downregulated (Table 1). The upregulated genes included several nod genes involved in NF production (nodABC, nodS, nodU, nodZ), genes involved in the type III secretion system (rhcJ, nolU, rhcQ, rhcR, rhcS, rhcT, rhcV, rhcU), and genes encoding Nops (nopA, nopB, nopL, nopP, nopT, nopC, nopM). These DEGs can be grouped into different classes, namely, genes controlled by a nod box (NB), tts box (TB), and genes that do not contain either NBs or TBs [25]. In the case of Sinorhizobium fredii NGR234, 19 NBs (NB1 to NB19) have been identified; 18 out of these 19 NBs were inducible by flavonoid in a NodD1-dependent manner. The induction of four NB-containing genes was found to be dependent on NodD2 [26,27]. Our transcriptome analysis of NGR234 also clearly showed that several nod genes were highly induced by the addition of apigenin (Supplementary Dataset S1). We also analyzed the genes whose expression is driven by the 19 NBs described in NGR234 [28] and the 7 TBs located on the symbiotic plasmid. Clearly, the addition of apigenin upregulated the expression of all the 19 NB- and 7 TB-containing genes (Table 3 and Table 4).

Table 3.
Induction on NGR234 ORFs regulated by apigenin, calcium, and both treatments located downstream of nod boxes.

Table 4.
Induction on NGR234 ORFs regulated by apigenin, calcium, and both treatments located downstream of tts boxes.
2.5. Calcium Represses the Expression of nodABC Genes
Transcriptome profiling of NGR234 upon calcium treatment revealed 65 differentially expressed genes (Table 1). These DEGs can be assigned into several functional categories based on the KEGG database (http://www.genome.jp/kegg/pathway.html; accessed on 29 May 2023) along with some insertion sequences and hypothetical proteins without known function. Regarding the 39 upregulated genes, two main groups can be extracted based on the processes that they are involved. A set of 12 DEGs was related to amino acid transport and metabolism, and another 12 DEGs were involved in carbohydrate transport and metabolism (Table 5). Calcium had a pronounced effect on the transcription of NGR_b22780, which exhibits a 17-fold change when compared to NGR234 cells grown in the absence of calcium. This DEG encodes a putative transmembrane protein consisting of 88 amino acids. This small protein contains a conserved DUF3311 domain and belongs to a family of short bacterial proteins of unknown function. Calcium also upregulated the expression of several sugar transporters, permeases, aminotransferases, and oxidoreductases (Table 5).

Table 5.
Functional characterization on the NGR234 DEGs upon calcium treatment compared to the control conditions.
The 26 downregulated genes in the presence of calcium carry out diverse putative functions. One of them, the NGR_a02570 gene, codes for OmpW, an outer membrane protein similar to Omp22. Consistent with our observation, it was earlier shown that the expression of Omp22 under high levels of calcium was reduced [29]. Surprisingly, we found that nodABCs, which are involved in NF production in response to plant root flavonoid induction, were included into the repressed genes (−4.53, −4.41, and −3.62, respectively) (Table 4). Additionally, calcium also repressed the expression of several proteins including a cytochrome c-type biogenesis protein, a cytochrome bd ubiquinol oxidase subunit I, and a cytochrome bd ubiquinol oxidase subunit II (Supplementary Dataset S2).
2.6. Effect of Calcium plus Apigenin on the NGR234 Transcriptome
Comparative analysis of the DEGs among the calcium, apigenin, and calcium-plus-apigenin treatments with respect to the control conditions showed 16 shared genes, while 42 were uniquely regulated in the presence of apigenin and 16 specifically expressed in the presence of calcium (Supplementary Dataset S3). Surprisingly, the simultaneous presence of both compounds differentially expressed 45 genes which were not shared when either calcium or apigenin were added to the culture. The higher amount of shared DEGs (92) was found between the apigenin and calcium-plus-apigenin conditions (Figure 3). We carried out another interesting analysis by comparing the RNA-Seq data between samples treated with calcium plus apigenin with respect to apigenin (Supplementary Dataset S4). Our analysis showed that most of the DEGs were related to the sugar metabolism and transport (tripartite ATP-independent periplasmic transporters), as well as genes related to the glycerol metabolism.
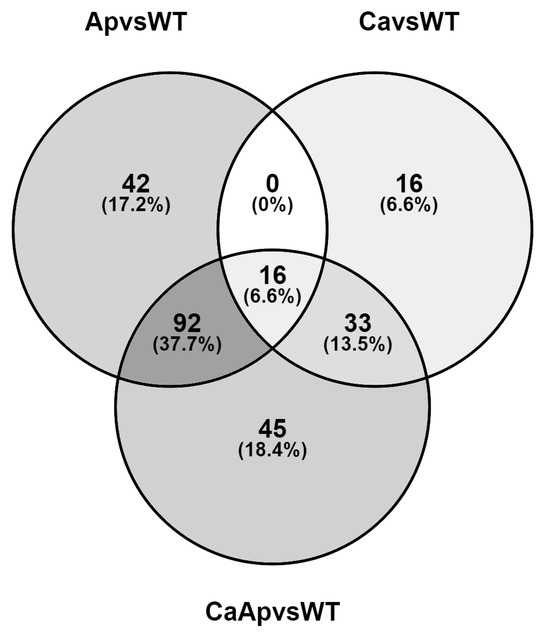
Figure 3.
Venn diagram showing the overlap of differentially expressed genes among the calcium, apigenin, and calcium-plus-apigenin treatments with respect to the control condition. The diagram shows the number of uniquely regulated genes and the number of commonly regulated genes in each treatment compared to the control condition.
Analyzing the shared and specific up- or downregulated genes in more detail, the results showed that the calcium treatment and the combination of calcium plus apigenin shared more percentage of downregulated genes than upregulated genes with respect to the global comparison. On the contrary, the presence of apigenin mainly revealed upregulated shared genes with the calcium-plus-apigenin treatment (Supplementary Figures S2–S4). As mentioned previously, we observed that calcium treatment repressed the expression of the nodABC genes. This observation prompted us to check if other NB- or TB-containing genes were also affected by the calcium addition or the combination of both compounds. We analyzed the genes whose expression is driven by the 19 NBs described in NGR234 [28] and the 7 TBs located on the symbiotic plasmid. Unlike the nodABC repression by the presence of calcium, we did not find changes in the expression of genes included in the nod regulon or the genes under TB control (Table 3). In addition, the combination of the two conditions did not substantially modify the expression of those genes, only the nolO, y4hM, and fixBC genes increased their expression and are therefore included as DEGs (Supplementary Dataset S3).
2.7. Quantitative RT-PCR Analysis Verification of RNAseq Transcriptome Data
To validate the RNA-Seq results, we randomly selected 16 DEGs and examined their transcriptional profile by performing qRT-PCR (Figure 4). Primers that are specific for the amplification of conserved regions of nodA, nodZ, rhcJ, syrM2, nopA, tts1, transcriptional regulators (NGR_b17530; NGR_b03240), sugar ABC transporter, putative outer membrane protein, glycerol-3-phosphate dehydrogenase, aspartate racemase, cytochrome c-type biogenesis protein, dihydrolipoamide dehydrogenase, and succinoglycan biosynthesis protein exoA were synthesized (Supplementary Table S1) and were used for performing qRT-PCR analysis (Figure 4). Our results are in agreement with the RNA-seq data. In most cases we found a linear correlation between the fold change values from the RNA-seq and qPCR experiments. Even though the fold changes were not identical, both methods yielded similar expression trends under different treatments.
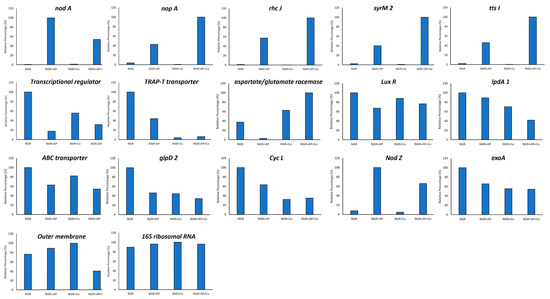
Figure 4.
qRT-PCR validation of select differentially expressed genes identified by RNAseq analysis.
3. Discussion
In the symbiotic process, several molecular signals must be exchanged between the host plant and the bacterial symbiont in a coordinated way. Among these signals, the inducer flavonoids, together with the bacterial regulator NodD, play a critical role since they trigger the expression of genes under the control of NB sequences [6,22]. Among the upregulated genes are those related to NF production as well as several regulators such as ttsI, whose encoded product activates the expression of genes involved in the T3SS, both structural and Nop-coding genes [13,14]. Both NF and Nops act as molecular determinants for the nodulation specificity between the host plant and the rhizobium. Therefore, they are involved in the nodulation host range [5,30,31]. However, little is known about the impact of environmental conditions, physical or chemical, on the production of rhizobial symbiotic signals. One of these elements is calcium availability, which may vary depending on the type of soil. Calcium is a crucial element in the symbiotic process, since it is involved in signal transmission through its concentration oscillations, commonly known as “calcium spiking”, in root cells [8,9,32]. The calcium spiking not only occurs during nodule organogenesis but also in rhizobial cells in response to the detection of plant flavonoids, which allows the expression of nod genes since their transcription is calcium dependent [33].
In this work, we have studied the role of calcium on the global gene expression of NGR234 in the absence and presence of the inducer flavonoid apigenin, and its effect on T3SS function. Our results indicate that calcium only affects the expression of 1% of genes within the entire NGR234 genome with respect to the control condition (Supplementary Dataset S2). The DEGs observed in the presence of 0.5 mM of calcium predominantly belong to specific functional groups, including sugar and amino acid metabolism and transport, although there are others involved in protein and transporter production within rhizobial membranes (Supplementary Dataset S2). Regarding genes related to symbiosis, only the genes responsible for NF core production, nodABC genes, appeared as downregulated in comparison to the control (Supplementary Dataset S3). As expected, the presence of apigenin induces all genes belonging to the nod regulon (Supplementary Dataset S1), even in the presence of both treatments, where only some genes slightly increase their expression (less than three-fold between the two conditions, calcium versus apigenin plus calcium (Supplementary Dataset S3). These results are in agreement with previous works in S. fredii USDA257, where the expression of T3SS-related genes was not affected by calcium treatment in the presence of inducer flavonoids [21]. As reported in S. fredii USDA257, we also detected in NGR234 that calcium downregulated the expression of an outer membrane protein, putative OmpW (NGR_a02570) (Table 3). OmpW might be involved in the membrane integrity in USDA257 [29], but in the case of pathogenic bacteria, such as Acinetobacter baumannii, the increased expression of this protein seems to be involved in the antibiotic resistance mechanism together with OmpA [34]. As mentioned before, NGR234_b22780 is the most upregulated gene in the presence of calcium and codes for a putative small transmembrane protein with an unknown functional domain. In addition, genes flanking the NGR234_22780 (NGR234_b22770 and b22790) are also upregulated upon calcium treatment. These genes encode a predicted monocarboxylic acid permease and a putative protein erfK/srfK precursor. NGR234_b22780 contains two predicted domains, a domain with unknown function (DUF5313) and another domain responsible for sodium/glucose cotransporter. In the case of the NGR234_b2290, it contains a predicted L, D-transpeptidase catalytic domain which can act as an L, D-transpeptidase facilitating an alternative pathway for peptidoglycan cross-linking [35].
SDS-PAGE analysis of the NGR234 extracellular proteins in the presence of apigenin and increasing amounts of calcium revealed a reduction of Nops production at the highest calcium concentration (Figure 1, line 6). Surprisingly, the NopA protein not only reduced its protein content but also appeared in two smaller isoforms. Western blot analysis using an antibody against NopA protein indicated the presence of at least two proteins as NopA, a native isoform and another smaller one (band A and B in Figure 2 panel B). Even though band C was not recognized by the NopA antibody (band C in Figure 2 panel A and B), mass spectrometry analysis of the excised band confirmed that band C is also NopA. These results indicate that calcium does not exert its regulatory function at a transcriptional level but post-transcriptionally. As was commented in previous works, T3SS is regulated by proteases in Yersinia, Salmonella, and Pseudomonas [36,37] under stress conditions. A similar process might be happening in NGR234 in the presence of calcium treatment. However, no protease or peptidase present in the NGR234 genome was found to be upregulated in both calcium and calcium plus apigenin conditions even though they could be activated by the presence of calcium. The novel discovery of calcium-mediated cleavage sites in the NopA protein provides new insights about the reduction of the Nops production in this condition. Since NopA is the main subunit of the T3SS pilus [13], the lack of a native NopA protein could block the secretion of the rest of Nops and therefore it could affect the first steps of the symbiotic establishment. This process might be a mechanism by which the increased concentration of calcium in the symbiosome [38] restricts the T3SS production through the NopA cleavage and disassembles its structure blocking the Nops secretion. On the other hand, the high concentration of calcium would activate the expression of sugar and amino acid transporters that might be necessary for the metabolite exchanges between the host plant and the rhizobium in bacteroid state [3].
The concentration of calcium in different soil types varies significantly. This variation primarily depends on the parent material and the extent to which weathering and leaching have shaped soil development. The essential role of calcium in nodulation and nitrogen fixation was documented more than 90 years ago [39]. Calcium availability directly influences the establishment of legume–rhizobia symbiosis. Legumes rely on calcium for proper root development and signaling during nodule formation. Deficiency of calcium has been known to adversely affect the infection and nodulation of several legumes. Calcium deficiency was shown to impair nitrogen fixation while high calcium levels increased the number of nodules [40,41]. Richardson and his associates [42] have demonstrated that high Ca2+ increased the amount of nod gene-inducing compounds in root exudates. Calcium has also been shown to play a direct role in the formation and growth of the infection thread [43] and in the induction of a normal distribution of nodules on the taproot and lateral roots of soybean [44]. Several studies have demonstrated that the initiation of nodule formation involves a calcium-dependent signal transduction system [8,9,32]. This system triggers metabolic changes that culminate in cell division and nodule development. The effectiveness of nodule formation depends on the optimal concentration of calcium. Too much or too little calcium can disrupt the Nops function and affect symbiosis. Our study demonstrates the important role of calcium in regulating Nops production in NGR234. In summary, Nops are critical players in symbiosis, and the effect of calcium on Nops production may serve as a regulatory mechanism ensuring harmony between rhizobia and leguminous plants in soil.
4. Materials and Methods
4.1. Basic Molecular and Microbiological Techniques
Sinorhizobium fredii NGR234 [16] was grown at 30 °C in yeast extract/mannitol (YEM) medium. Apigenin was dissolved in ethanol at a concentration of 1 mg/mL and used at 1 μg/mL. A sterile stock solution of 100 mM of Calcium chloride (Sigma, St. Louis, MO, USA) was prepared and used at various concentrations ranging from 1 µM to 1 mM. All primer pairs used in this work are listed in Supplementary Table S1.
4.2. Culture Conditions and RNA Extraction
Sinorhizobium fredii NGR234 was grown in YEM media at 30 °C until stationary phase (OD600 ≈ 1.2). One mL of stationary phase NGR234 culture was then transferred to 250 mL Erlenmeyer’s flasks, each containing 100 mL of sterile YEM media. Nops production was triggered by the addition of 1 µM apigenin to the culture media. The effect of calcium on Nops production was studied by the addition of 500 µM calcium to YEM media that was also supplemented with 1 µM apigenin.
Total RNA was isolated using the High Pure RNA Isolation Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Verification of the amount and quality of the resulting total RNA samples was carried out by using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and a Qubit 2.0 Fluorometer (Invitrogen, Waltham, MA, USA). Three independent total RNA extractions were obtained for each condition.
4.3. RNA Isolation and Sequencing
Ribosomal RNA was depleted using a MICROB Express Bacterial mRNA Purification kit (Ambion, Austin, TX, USA), following the manufacturer’s protocol. The integrity and quality of the ribosomal depleted RNA were checked with Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA sequencing was carried out by a company with the Next Generation Sequence (NGS) platform Illumina using the Illumina HiSeq 2000 sequencing instrument (Illumina, San Diego, CA, USA). Ribosomal-depleted samples were used to generate whole transcriptome libraries following the manufacturer’s recommendations for sequencing on this NGS platform. Amplified cDNA quality was analyzed by the Bioanalyzer 2100 DNA 1000 kit (Agilent Technologies) and quantified using the Qubit 2.0 Fluorometer (Invitrogen).
4.3.1. Mapping of the RNA-Seq Data
The initial whole transcriptome paired-end reads obtained from sequencing were mapped against the latest version of the S. fredii NGR234 genome (https://www.ncbi.nlm.nih.gov/assembly/GCF_000018545.1/; accessed on 1 December 2022) using the Life Technologies mapping algorithm version 1.3 (http://www.lifetechnologies.com/).
4.3.2. Assessment of Differentially Expressed Genes
The gene expression levels were calculated using the Bioconductor Packages (DESeq2) version 1.36.0 [45], Rbowtie2 version 2.2.0 [46], Rsamtools version 2.2.3 [47], and Rsubread version 2.10.4 [48], bedtools version 2.30.0 [49], and samtools version 1.15.1 [50] software. Differentially expressed genes were defined as those genes with a fold-change lower or higher than −3 or 3, respectively, with a p value lower than 0.05.
4.3.3. General Features of the Total Sequenced and Mapped Reads
Reads were mapped and paired using the software mentioned above. Three biological and independent experiments were carried out for each condition (Supplementary Table S2).
4.3.4. RNA-Seq Data Accession Number
The RNA-seq data discussed in this publication have been deposited in the Sequence Read Archive of NCBI (BioProject database) under the BioProject ID PRJNA1024659.
4.4. Quantitative Reverse Transcription PCR
Results obtained in the RNA-Seq analysis were validated by quantitative reverse transcription PCR (qRT-PCR) of 16 selected genes, which represented differentially and non-differentially expressed genes in the four conditions (control, Apigenin, calcium, apigenin + calcium). Total RNA was isolated using the High Pure RNA Isolation kit (Roche, Basel, Switzerland) and RNase Free DNA Set (Qiagen, Chuo City, Tokyo) according to the manufacturer’s instructions. This (DNA free) RNA was reverse transcribed to cDNA by using PrimeScript RT reagent kit with gDNA Eraser (Takara, San Jose, CA, USA). Quantitative PCR was performed using a LightCycler 480 (Roche, Switzerland) with the following conditions: 95 °C, 10 min; 95 °C, 30 s; 50 °C, 30 s; 72 °C, 20 s; forty cycles, followed by the melting curve profile from 60 to 95 °C to verify the specificity of the reaction. The S. fredii NGR234 16S rRNA was used as an internal control to normalize gene expression. The selected genes and primers are listed in Supplementary Table S1.
4.5. Purification and Analysis of Nops
Nodulation outer proteins (Nops) were isolated as described earlier [22]. Nops were separated by SDS-PAGE using the discontinuous buffer system of Laemmli [51]. An equal volume of protein was loaded in each lane taking into consideration that protein extractions were carried out from the same volume of cultures at the same growth stage with similar cell number. Electrophoresis was performed on 15%(w/v) SDS polyacrylamide gels. After the electrophoresis, proteins were stained with Coomassie Blue.
4.6. Western Blot Analysis
Nodulation outer proteins (Nops) of NGR234 that were grown in the presence or absence of apigenin and calcium were separated by 1D SDS-PAGE. Resolved proteins were electrophoretically transferred to a nitrocellulose membrane. The membrane was incubated with polyclonal antibodies raised against S. fredii USDA257 NopA protein that was diluted 1:10,000 in Tris-buffered saline (TBS; 10 mM Tris-HCl [pH 7.5], 500 mM NaCl) containing 5% nonfat dry milk. Following overnight incubation, the membrane was washed three times with TBST (TBS containing 0.3% Tween 20). The membrane was then incubated with goat anti-rabbit IgG–horseradish peroxidase conjugate which was diluted 1:10,000 in TBST containing 5% nonfat dry milk. After several rinses in TBST, immunoreactive polypeptides were detected with an enhanced chemiluminescent substrate (Super Signal West Pico kit; Pierce Biotechnology, Rockford, IL, USA) according to the manufacturer’s instructions.
4.7. Mass Spectrometry Analysis
Protein bands corresponding to NopA and the processed peptides were excised from the acrylamide gel, washed in distilled water, and then destained in a 50% solution of acetonitrile (v/v) containing 25 mM of ammonium bicarbonate. After destaining, a 100% acetonitrile wash was performed and digested with 20 μL (10 μg/mL) of modified porcine trypsin in 25 mM ammonium bicarbonate (Promega, Madison, WI, USA). The peptides resulting from tryptic digestion were then analyzed by liquid chromatography mass spectrometry (LCMS). Database searches were conducted using Proteome discoverer.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms25063443/s1.
Author Contributions
H.B.K. conceived and planned the experiments; W.K., S.K. and H.B.K. performed the experiments; S.A.-J. and W.K. analyzed the data; S.A.-J. and H.B.K. wrote the original draft; W.K. and S.K. were involved in writing—review and editing; H.B.K. was responsible for project administration and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for this research was provided by Agricultural Research Service, USDA (8042-21220-234-00D).
Data Availability Statement
The RNA-seq data discussed in this publication have been deposited in the Sequence Read Archive of NCBI (BioProject database) under the ID PRJNA1024659. The original contributions presented in the study are included in the article and Supplementary Material. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to thank Nathan Oehrle for his help in creating figures included in this manuscript. Mention of a trademark, vendor, or proprietary product does not constitute a guarantee or warranty of the product by the USDA and does not imply its approval to the exclusion of other products or vendors that may also be suitable. The US Department of Agriculture, Agricultural Research Service, Midwest Area, is an equal opportunity, affirmative action employer and all agency services are available without discrimination.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Spaink, H.P. Root nodulation and infection factors produced by rhizobial bacteria. Annu. Rev. Microbiol. 2000, 54, 257–288. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nature Reviews. Microbiology 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.E. Early interactions between legumes and rhizobia: Disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 2007, 103, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.W.; Murray, J.D. The role of flavonoids in nodulation host-range specificity: An update. Plants 2016, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Peck, M.C.; Fisher, R.F.; Bliss, R.; Long, S.R. Isolation and characterization of mutant Sinorhizobium meliloti NodD1 proteins with altered responses to luteolin. J. Bacteriol. 2013, 195, 3714–3723. [Google Scholar] [CrossRef] [PubMed]
- Khokhani, D.; Carrera Carriel, C.; Vayla, S.; Irving, T.B.; Stonoha-Arther, C.; Keller, N.P.; Ané, J.M. Deciphering the chitin code in plant symbiosis, defense, and microbial networks. Annu. Rev. Microbiol. 2021, 75, 583–607. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms underlying legume-rhizobium symbioses. J. Integr. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef] [PubMed]
- Krause, A.; Doerfel, A.; Göttfert, M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact 2002, 15, 1228–1235. [Google Scholar] [CrossRef]
- López-Baena, F.J.; Vinardell, J.M.; Pérez-Montaño, F.; Crespo-Rivas, J.C.; Bellogín, R.A.; Espuny, M.d.R.; Ollero, F.J. Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiology 2008, 154, 1825–1836. [Google Scholar] [CrossRef] [PubMed]
- Wassem, R.; Kobayashi, H.; Kambara, K.; Le Quéré, A.; Walker, G.C.; Broughton, W.J.; Deakin, W.J. TtsI regulates symbiotic genes in Rhizobium sp. NGR234 by binding to tts boxes. Mol. Microbiol. 2008, 78, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, C.; Krishnan, H.B. Nodulation outer proteins: Double-edged swords of symbiotic rhizobia. Biochem. J. 2015, 470, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.S.; Sadowsky, M.J. Secretion systems and signal exchange between nitrogen-fixing rhizobia and legumes. Front. Plant Sci. 2015, 6, 491. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. NopC is a Rhizobium-specific Type 3 Secretion System effector secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS ONE 2015, 10, e0142866. [Google Scholar] [CrossRef] [PubMed]
- Trinick, M.J. Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa sp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J. Appl. Bacteriol. 1980, 49, 39–53. [Google Scholar] [CrossRef]
- Pueppke, S.G.; Broughton, W.J. Rhizobium sp. Strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant-Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Medina, C.; Vinardell, J.M.; Ollero, F.J.; López-Baena, F.J. The Rhizobial Type 3 Secretion System: The Dr. Jekyll and Mr. Hyde in the Rhizobium-Legume Symbiosis. Int. J. Mol. Sci. 2022, 23, 11089. [Google Scholar] [CrossRef]
- Viprey, V.; del Greco, A.; Golinowski, W.; Broughton, W.J.; Perret, X. Symbiotic implications of type III protein secretion machinery in Rhizobium. Mol. Microbiol. 1998, 28, 1381–1389. [Google Scholar] [CrossRef]
- Temprano-Vera, F.; Rodríguez-Navarro, D.N.; Acosta-Jurado, S.; Perret, X.; Fossou, R.K.; Navarro-Gómez, P.; Zhen, T.; Yu, D.; An, Q.; Buendía-Clavería, A.M.; et al. Sinorhizobium fredii strains HH103 and NGR234 form nitrogen fixing nodules with diverse wild soybeans (Glycine soja) from central China are ineffective on northern china accessions. Front. Microbiol. Front. Microbiol. 2018, 9, 2843. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Kim, W.S.; Sun-Hyung, J. Calcium regulates the production of nodulation outer proteins (Nops) and precludes pili formation by Sinorhizobium fredii USDA257, a soybean symbiont. FEMS Microbiol. Lett. 2007, 271, 59–64. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Krishnan, H.B.; Lorio, J.; Kim, W.S.; Jiang, G.; Kim, K.Y.; DeBoer, M.; Pueppke, S.G. Extracellular proteins involved in soybean cultivar-specific nodulation are associated with pilus-like surface appendages and exported by a type III protein secretion system in Sinorhizobium fredii USDA257. Mol. Plant-Microbe Interact. 2003, 16, 617–625. [Google Scholar] [CrossRef] [PubMed]
- de Lyra Mdo, C.; Lopez-Baena, F.J.; Madinabeitia, N.; Vinardell, J.M.; Espuny Mdel, R.; Cubo, M.T.; Belloguin, R.A.; Ruiz-Sainz, J.E.; Ollero, F.J. Inactivation of the Sinorhizobium fredii HH103 rhcJ gene abolishes nodulation outer proteins (Nops) secretion and decreases the symbiotic capacity with soybean. Int. Microbiol. 2006, 9, 125–133. [Google Scholar] [PubMed]
- Pérez-Montaño, F.; Del Cerro, P.; Jiménez-Guerrero, I.; López-Baena, F.J.; Cubo, M.T.; Hungría, M.; Megías, M.; Ollero, F.J. RNA-seq analysis of the Rhizobium tropici CIAT 899 transcriptome shows similarities in the activation patterns of symbiotic genes in the presence of apigenin and salt. BMC Genom. 2016, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Acosta-Jurado, S.; Navarro-Gómez, P.; Ollero, F.J.; Ruiz-Sainz, J.E.; López-Baena, F.J.; Vinardell, J.M. A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 2016, 6, 1592. [Google Scholar] [CrossRef] [PubMed]
- Fellay, R.; Hanin, M.; Montorzi, G.; Frey, J.; Freiberg, C.; Golinowski, W.; Staehelin, C.; Broughton, W.J.; Jabbouri, S. nodD2 of Rhizobium sp. NGR234 is involved in the repression of the nodABC operon. Mol. Microbiol. 1998, 27, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Theunis, M.; Kobayashi, H.; Broughton, W.J.; Prinsen, E. Flavonoids, NodD1, NodD2, and nod-box NB15 modulate expression of the y4wEFG locus that is required for indole-3-acetic acid synthesis in Rhizobium sp. strain NGR234. Mol. Plant-Microbe Interact. 2004, 17, 1153–1161. [Google Scholar] [CrossRef]
- Perret, X.; Freiberg, C.; Rosenthal, A.; Broughton, W.J.; Fellay, R. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 1999, 32, 415–425. [Google Scholar] [CrossRef]
- Kim, W.S.; Sun-Hyung, J.; Park, R.D.; Kim, K.Y.; Krishnan, H.B. Sinorhizobium fredii USDA257 releases a 22-kDa outer membrane protein (Omp22) to the extracellular milieu when grown in calcium-limiting conditions. Mol. Plant-Microbe Interact. 2005, 18, 808–818. [Google Scholar] [CrossRef]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef]
- Walker, L.; Lagunas, B.; Gifford, M.L. Determinants of host range specificity in legume-rhizobia symbiosis. Front. Microbiol. 2020, 11, 585749. [Google Scholar] [CrossRef]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Moscatiello, R.; Squartini, A.; Mariani, P.; Navazio, L. Flavonoid-induced calcium signalling in Rhizobium leguminosarum bv. viciae. New Phytol. 2010, 188, 814–823. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, B.L.; Leal, B.F.; Leyser, M.; de Barros, M.P.; Trentin, D.S.; Ferreira, C.A.S.; de Oliveira, S.D. Increased ompW and ompA expression and higher virulence of Acinetobacter baumannii persister cells. BMC Microbiol. 2023, 23, 157. [Google Scholar] [CrossRef] [PubMed]
- Biarrotte-Sorin, S.; Hugonnet, J.-E.; Delfosse, V.; Mainardi, J.-L.; Gutmann, L.; Arthur, M.; Mayer, C. Crystal Structure of a novel β-Lactam-insensitive peptidoglycan transpeptidase. J. Mol. Biol. 2006, 359, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.W.; Siva-Herzog, E.; Plano, G.V. The ATPdependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 2004, 54, 1364–1378. [Google Scholar] [CrossRef]
- Bretz, L.; Losada, L.; Lisboa, K.; Hutcheson, S.W. Lon protease functions as a negative regulator of type III protein secretion in Pseudomonas syringae. Mol. Microbiol. 2002, 45, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.K.; Day, D.A. Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 493–523. [Google Scholar] [CrossRef]
- Albrecht, W.A.; Davis, F.L. Physiological importance of Ca2+ in legume inoculation. Bot. Gaz. 1929, 88, 310–321. [Google Scholar] [CrossRef]
- Lowter, W.L.; Loneragan, J.F. Effects of calcium deficiency on symbiotic nitrogen fixation. Plant Physiol. 1968, 43, 1362–1366. [Google Scholar]
- Munns, D.N. Nodulation of Medicago sativa in solution culture. V. Calcium and pH requirements during infection. Plant Soil 1970, 32, 90–102. [Google Scholar] [CrossRef]
- Richardson, A.E.; Djordjevic, M.A.; Rolfe, B.G.; Simpson, R.J. Effects of pH, Ca and Al on the exudation from clover seedlings of compounds that induce the expression of nodulation genes in Rhizobium trifolii. Plant Soil 1988, 109, 37–47. [Google Scholar] [CrossRef]
- Sethi, R.S.; Reporter, M. Calcium localization pattern in clover root hair cells associated with the infection process: Studies with aureomycin. Protoplasma 1981, 105, 321–325. [Google Scholar] [CrossRef]
- Balatti, P.A.; Krishnan, H.B.; Pueppke, S.G. Calcium regulates growth of rhizobium fredii and its ability to nodulate soybean cv. peking. Can. J. Microbiol. 1991, 37, 542–548. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Zhang, W.; Fang, H.; Li, Y.; Wang, X. esATAC: An easy-to-use systematic pipeline for ATAC-seq data analysis. Bioinformatics 2018, 34, 2664–2665. [Google Scholar] [CrossRef]
- Morgan, M.; Pages, H.; Obenchain, V.; Hayden, N. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R Package Version 2016, 1, 677–689. [Google Scholar]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).