Abstract
Infections caused by yeasts of the genus Candida are likely to occur not only in immunocompromised patients but also in healthy individuals, leading to infections of the gastrointestinal tract, urinary tract, and respiratory tract. Due to the rapid increase in the frequency of reported Candidiasis cases in recent years, diagnostic research has become the subject of many studies, and therefore, we developed a polyclonal aptamer library-based fluorometric assay with high specificity and affinity towards Candida spec. to quantify the pathogens in clinical samples with high sensitivity. We recently obtained the specific aptamer library R10, which explicitly recognized Candida and evolved it by mimicking an early skin infection model caused by Candida using the FluCell-SELEX system. In the follow-up study presented here, we demonstrate that the aptamer library R10-based bioassay specifically recognizes invasive clinical Candida isolates, including not only C. albicans but also strains like C. tropcialis, C. krusei, or C. glabrata. The next-generation fluorometric bioassay presented here can reliably and easily detect an early Candida infection and could be used for further clinical research or could even be developed into a full in vitro diagnostic tool.
1. Introduction
More than 20 species of the genus Candida are known to cause infections, with Candida albicans being the most prominent [1,2,3]. In the last three decades, candidiasis has emerged as the fourth most common case of blood infections in hospitals, due to the widespread use of antibiotics that destroy the competing bacterial flora and the prolonged systemic immunosuppressive treatments after organ transplantation and chemotherapy [4,5,6,7,8]. Other factors that predispose to candidiasis are pregnancy or the use of hormonal contraceptives that reduce the acidity of the vaginal environment and can lead to Vulvovaginitis candidomycetica, which leads to an annual loss of productivity of up to USD 14–39 billion in high-income countries [9,10]. Furthermore, invasive candidiasis is the cause of more than 250,000 infections and 50,000 deaths worldwide [11,12,13,14,15,16]. This makes Candida’s timely and accurate detection crucial for diagnosis and the subsequent treatment. The conventional diagnostic approach is to accurately identify Candida spp. in clinical samples based on morphological and physiological characteristics, which is complex and can be time-consuming; however, more rapid commercial systems may also inevitably face serious sensitivity problems [17,18]. Newer detection methods, such as matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF), may also be impractical for many laboratories in developing countries as such complex methods mostly rely on pure cultures, and they are not only highly expensive but also very time-consuming [19,20,21,22,23]. Other standard detection methods based on quantitative PCR (qPCR) have their drawbacks due to their restricted sensitivity and limited specificity for only a rather small and pre-defined set of Candida spec. [24,25,26]. Therefore, novel techniques that can identify Candida easily, quickly, and successfully with respect to the economic aspects appear to be of particular importance.
In the last three decades, aptamers have emerged as new ligands that not only have similar properties to antibodies but are more stable and easier to synthesize and modify and have higher affinity towards their dedicated targets [27,28,29,30,31,32,33,34]. Therefore, aptamers can be used as ligands instead of antibodies to detect pathogens like bacteria or fungi [35,36]. Fluorometric aptamer-mediated whole-cell detection methods are recognized as a promising tool for the development of fast, specific, and clinically applicable bioassays [37,38]. In fact, functional aptamer-based assays have been introduced against a variety of health-relevant target organisms, including major pathogens [39,40,41,42,43,44,45,46,47], as well as for potential probiotic human gut bacteria [48]. However, what most of these approaches have in common is that they require additional technical components like different types of (nano)particles or they rely on sandwich-type assay principles [49]. In contrast, more simple assays have been suggested; these were based on the direct labeling of the intended target with enriched (also known as polyclonal) aptamer libraries or individual aptamers without the need for secondary binding molecules or enzyme-mediated signal amplification [47,48,49,50,51,52,53].
With C. albicans, C. auris, and C. parapsilosis as target cells for a whole-cell SELEX process (Systematic Evolution of Ligands by EXponential enrichment) [32,33], we developed an enriched aptamer library against this class of important human pathogens in a previous study [50]. This library was already sufficient to label these Candida species with fluorescence and allowed fungal cells to be distinguished from human dermal fibroblast (HDF) cells via fluorescence microscopy in a skin early infection model [50]. However, the intensity of the unspecific background signals obtained with the HDF cells in a fluorometric suspension assay suggested a considerable potential to improve the sensitivity of such an enriched SELEX library while simultaneously enhancing the specificity for Candida spec. and improving their differentiation from human cells. Thus, additional rounds of SELEX were performed, again with a mixture of the three Candida spec. and counter selection using human fibroblasts. The intended improvements were verified using not only HDF cells but also a set of three additional lines of somatic cells, including colorectal adenocarcinoma cells (HT29), pancreatic cancer cells (MIA-PaCa-2P), and breast cancer cells (MCF7). Enriched aptamer libraries of this type, in their fluorescently labeled versions, offer an efficient option with which to label target cells with high specificity, allowing sensitive measurements simply by using fluorometric suspension assays. Without the need for additional assay components or signal amplification measures, it was possible to distinguish a set of 87 clinical isolates from invasive infections with different Candida spec. from the human cells. This final library was also characterized by high affinities against the Candida spec., with dissociation constants in a low nanomolar range and a low detection limit. The sensitivity, but also the intriguing ease with which the cells of Candida species were efficiently distinguished from human cells in this suspension assay measured with the standard fluorometer equipment may open new avenues for clinical diagnostics of pathogens based on enriched libraries or selected individual aptamers labeled with fluorescent dyes. We believe that this Candida spec.-specific assay may represent a prototype assay, and we suggest the name FluCandA-Assay (“Fluorescence Candida Aptamer”) and hope to inspire other researchers to develop similar concepts, which could rapidly enlarge the portfolio of fast and reliable detection technologies for health-threatening organisms.
2. Results
Based on the published enriched anti-Candida SELEX library R8 [50], two additional rounds of selection were performed to evaluate the resulting affinities against the Candida target strains and their specificities with HDF cells and three further cell lines as controls. As intended, both the affinity and the specificity increased significantly, with the affinity improving up to twofold in the resulting novel final library R10. In addition, R10 showed no binding and thus no fluorescent labeling of the human cell lines (Figure 1a). This allowed a lower detection limit (defined as 50% of the maximal observable fluorescence at the given amount of aptamers) of 2000 cells per milliliter for C. auris, but as low as 20 cells for both C. albicans and C. parapsilosis. Accordingly, the dissociation constants (Kd values) were found to be reasonably low and were in the nanomolar range from 10 to 13 nM, respectively. The curves could be fitted by a typical single site-specific binding model [51] and showed reasonably low dissociation constants (Kd values) of 12.96 nM, 9.715 nM, and 9.611 nM, respectively.
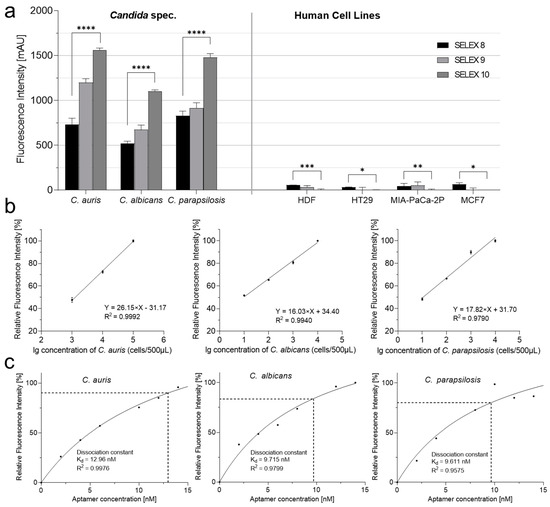
Figure 1.
Increased aptamer affinities towards Candida spec. cells allows their differentiation from four human cell lines. (a) Based on the aptamer library R8 [50], two additional rounds of SELEX delivered libraries R9 and R10. Human cell lines to be differentiated from C. auris, C. albicans, and C. parapsilosis were HDF, HT29, MIA-PaCa-2P, and MCF7. Binding assays were performed using Cy5-labeled R8, R9, and R10 aptamer libraries. The fluorescence intensity can be correlated with the affinity of the aptamers against the given target. p values < 0.05 were considered significant. * p denotes < 0.05, ** denotes p < 0.01, *** denotes p < 0.001 and **** denotes p < 0.0001. (b) Determination of lower detection limits (black lines) of aptamer library R10. Relative fluorescence against logarithmically scaled cell numbers of C. auris, C. albicans, and C. parapsilosis with 10 pmol of Cy5-labeled aptamer. Linearly fitted plot represents lower detection limit X = lg(cell number), Y = (relative fluorescence intensity). (c) Determination of dissociation constants (Kd values) (black dotted lines) of the aptamer library R10 for C. auris, C. albicans, C. parapsilosis by determination of the percentage of bound aptamers (y-axis) against an increasing of aptamer concentration up to 15 pmol. By performing an exponential fit Kd values of 12.96 nM for C. aurius, 9.715 nM for C. albicans, and 9.611 for C. parapsilosis were computed. All experiments were conducted as triplicates (n = 3).
The sensitivity and specificity of the enriched R10 library suggested that our aim to develop an easy but reliable assay to distinguish Candida spec. from human cells might be achievable and realistic. The suggested assay follows the workflow demonstrated in Figure 2a and consists of three principal steps: (1) binding, (2) washing, and (3) elution, prior to the final fluorescence measurement involving only two subsequent centrifugation steps in the experiment (Figure 2a).
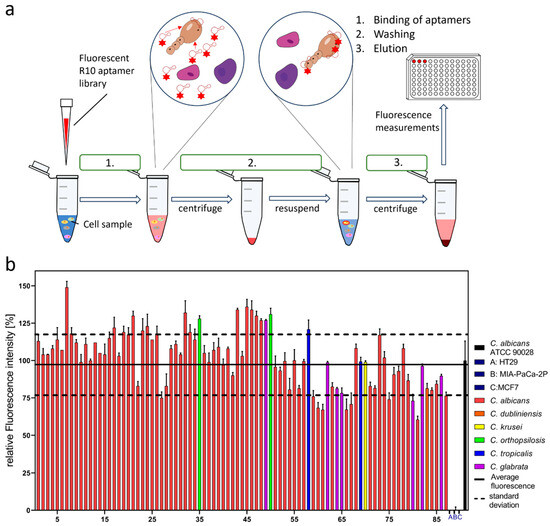
Figure 2.
(a) Schematic of binding assay with Cy5 fluorescent-labeled R10 aptamer library. The fluorescent aptamers are first incubated with the target cells, centrifuged, and resuspended in PBS buffer. The resulting aptamer target complexes are then separated by denaturation at 95 °C for 5 min. The solution is centrifuged, and the fluorescence of the supernatant is measured. (b) Binding assay of aptamer library R10 against a multitude of clinical isolates of Candida (n = 87) of 6 different species, C. albicans, C. dubliniensis, C. krusei, C. orthopsilosis, C. tropicalis, C. glabrata (sorted by sampling number), were conducted, with A, B, C representing the human cell lines (HT29, MIA-PaCa-2P, MCF7). The laboratory strain C. albicans ATCC90028 was used as the control and represents a relative fluorescence of 100%. A detection window was found by determining the average relative fluorescence of the clinical isolates (solid black line) and calculating the 1 sigma interval (dotted black lines) 0.98 ± 0.21.
As a proof of concept, to show the principal functionality of this assay’s workflow, we intended to demonstrate that a considerably large ensemble of real clinical isolates of the genus Candida (Candida spec.) could be differentiated from the human control cells as desired. During a sampling campaign, 87 individual isolates were collected in the university hospital Ulm and were provided anonymously for this study (numbered according to the order of their date of sampling in the campaign, as given in Figure 2b). This ensemble harbored C. albicans as the predominant species, represented by 74 individual strains as well as 5 additional Candida species. Among these, with seven representatives, C. glabrata was the second most frequent species, followed by C. orthopsilosis and C. tropicalis with two isolates each, and C. dublinensis and C. krusei were each represented by only a single isolate. Interestingly, as expected for a larger German hospital, C. auris, which is probably the most relevant and health-threatening Candida pathogen emerging in various other regions of the world, was not present in the cohort of patients who delivered the 87 clinical samples. The assay was performed as described for the 87 clinical isolates and the laboratory strain C. albicans ATCC 90028 as a reference. Moreover, the target negative controls represented by the set of human cell lines were again also included, and it was found, as expected, that they delivered a zero-fluorescence signal. The fluorescence signal of C. albicans ATCC 90028 was judged to be the reference and was set to 100% labeling efficiency, and all the other measurements were normalized accordingly. The arithmetical mean was calculated for all the samples (except the human cell lines), and the positive and negative standard deviations were used to define a detection window (0.98 ± 0.21). The vast majority of isolates delivered fluorescence signals within or above the borders of the detection window, with only four individual C. albicans isolates falling slightly short of the lower border. Interestingly, the non-albicans Candida strains could also be confidently measured and thus distinguished from human cells.
3. Discussion
With the finding that polyclonal aptamer libraries originating directly from SELEX processes may outperform the individual aptamers [52] selected from these libraries, we introduced such enriched or “polyclonal” aptamer libraries as valuable tools for the fluorescent labeling of target structures and thus the specific quantification of a series of microbial symbionts and pathobionts [52,53,54,55,56,57]. Moreover, the functionality of this concept was further demonstrated with different tissues of plant roots [58] and by using specific enriched libraries for the construction of electronic gFET-based biosensors to measure pre-diabetes-related biomarkers [59]. The considerable general potential of oligonucleotide aptamers for the detection of pathogens and, in turn, for the development of assays for monitoring, and therefore disease control involving a variety of aptamer-based bioassays, including lateral flow assay concepts and colorimetric assays, as well as fluorescence-based concepts, is widely accepted and has recently been nicely reviewed by Wan and coworkers [60]. One key challenge in diagnostic assays for pathogen detection is to discriminate the dedicated harmful microbes in the sample preparations for analysis from “contaminating” human cells. We have recently shown that an enriched library against different Candida strains could already be used to distinguish the yeast cells from human cells in this case and was exemplified solely by human dermal fibroblasts (HDF), which served as so-called counterselection targets during the respective SELEX process [50]. It turned out that background detection of HDF cells and other human cell lines was low but significant, particularly in fluorometric binding assays, suggesting a remnant affinity for (experimentally yet undefined) epitopes on human cells. According to the dogma of directed evolution “you get what you screen for”; we decided to use the power of SELEX and to improve the specificity of the library by eliminating the vexatious background affinity with a few additional harsh rounds of selection and counterselection. Interestingly, in addition to this, the affinity of the final library for its dedicated target cells was also significantly enhanced, as measured by the fluorescence-labeling intensity of the different Candida strains. The affinity towards the reference Candida strains C. albicans, C. auris, and C. parapsilosis was in a reasonably low nanomolar range that was comparable to established diagnostic antibodies and aptamers [61,62,63,64,65,66]. This resulting high affinity-enriched aptamer library was used to test the FluCandA concept in a fast and easy assay for the detection of Candida spec. that could distinguish the Candida spec. from the human cells in the background. The strains from the set of clinical isolates were preponderantly encompassed positively by the assay, leaving only 4 of the 87 tested samples as false-negatives, but in these cases, the high fluorescence values lay just outside the detection window at the lower limit. We believe that this proof of the FluCandA concept may serve as the experimental basis with which to approach a feasible diagnostic assay for the further in-depth evaluation of its general potential to detect different Candida species. This evaluation should include larger cohorts of clinical isolates as well as larger ensembles of Candida reference strains. The same enriched aptamer library may also serve as a valuable pool of diverse sequences to isolate individual aptamers against specific Candida species or even strains. We hope that the portfolio of possible applications may also inspire and enable the development of assay systems of higher complexity, like the construction of electronic biosensors.
4. Materials and Methods
4.1. Cell Lines and Cell Culture
Experimental yeast strains, including C. auris (DSMZ-No. 21092), C. albicans (ATCC90028), C. parapsilosis (ATCC22019), and the clinic isolates, were inoculated in 5 mL of RPMI (Roswell Park Memorial Institute) medium (Thermo Fisher Scientific, Waltham, MA, USA) and cultured at 37 °C. All the clinical isolates of the Candida species were identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Maldi-ToF MS, Bruker Corporation, Billerica, MA, USA) and provided by patient samples sent to the microbiology department for diagnostic purposes. The strains were collected anonymously; thus, it was not possible to assign the strains to patients. The accreditation number of the microbiology department is DIN EN ISO15189:2014 (DAkks). The human cells, including HDF, HT29, MCF7, and MIA-PaCa-2P, were incubated in DMEM medium, 1% (v/v) Minimal Essential Medium Non-Essential Amino Acids, MEM NEAA(Life Technologies, Carlsbad, CA, USA), 1% (v/v) Penicillin/Streptavidin (Life Technologies, Carlsbad, CA, USA), 15% (v/v) Fetal Calf Serum (FCS) (Life Technologies, Carlsbad, CA, USA), and 83% (v/v) Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies, Carlsbad, CA, USA) and cultured in a 37 °C cell culture incubator containing 5% CO2.
4.2. Cell Pretreatment
After treatment with Accutase® (Life Technologies, Carlsbad, CA, USA), 20,000 cells were removed, added to a 96-well plate, and incubated at 37 °C for 24 h in a cell incubator.
4.3. Cell SELEX
4.3.1. Pretreatment before Counter SELEX
After being treated with Accutase®, the desired HDF cells were removed and added to 96-well plates (20,000 cells) or 24-well plates (100,000 cells), followed by incubation at 37 °C for 24 h in a cell culture incubator to re-adhere the cells. The cells were carefully washed once with 1× PBS after removing the medium before the screening.
4.3.2. Pretreatment before Target SELEX
C. auris, C. albicans, and C. parapsilosis were centrifuged at 9000× g for 2 min and washed with 1× PBS.
4.3.3. Aptamer Activation
Add the aptamer library to 500 µL of 1× PBS, incubate at 95 °C for 5 min, place in an ice bath for 5 min, and then leave for 20 min at room temperature.
4.3.4. Screening
The screening process for rounds 1–8 can be found in the previous article [50]; this was followed by two additional rounds of screening, as shown in Table 1.

Table 1.
SELEX procedure for rounds 9–10.
The activated library was incubated with adherent HDF cells at 37 °C for 1 h. The supernatant was then carefully aspirated; BSA (100 mg/mL) and tRNA (10 mg/mL) were added to increase stringency and incubated with Candida at 37 °C for 30 min, followed by centrifugation at 9000× g for 2 min, the removal of the supernatant, and the final washing with 1× PBS to remove unbound aptamer from the precipitate (see Table 1).
4.3.5. Elution
The cells from the previous step were resuspended in 100 µL of 1× PBS and incubated at 95 °C for 5 min, followed by centrifugation at 11,000× g for 1 min to collect the Candida-bound aptamer.
4.3.6. Library Amplification
PCR further amplified the aptamers collected in the previous step. The amplification conditions were as follows: 3 min at 95 °C, followed by 25 cycles of 30 s at 94 °C; 30 s at 56 °C; 10 s at 72 °C; and finally, 2 min at 72 °C. Next, the PCR products were purified (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany). The resulting double-stranded DNA was broken down into single-stranded DNA by λ-nucleic acid exonuclease catalysis (New England Biolabs, Ipswich, MA, USA) and finally purified by an optimized PCR purification kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany). The binding buffer required for this purification process was supplemented with 1.5 volumes of isopropanol and 10 µL of natrium acetate solution (pH 5) to increase the yield of single-stranded DNA.
4.3.7. Binding Assay
Candida Binding Assay
After washing the yeast according to Section 4.3.2, 20,000 cells were used for analyses and incubated with 5 pmol of activated aptamer library in 500 µL of PBS for 30 min at 37 °C. Next, the culture was centrifuged at 9000× g for 2 min to remove the supernatant and washed once. The precipitate was resuspended in 100 µL of 1× PBS buffer to obtain the eluted cell junctional aptamers, and the fluorescence intensity was determined by measuring at an excitation wavelength of 637 nm and an emission wavelength of 670 nm using an Infinite M200 spectrophotometer (TECAN, Männedorf, Switzerland).
Cell Binding Assay
After re-culturing 20,000 individual cells in 24-well plates, the cells were incubated with 5 pmol of activated aptamer library in 500 µL of PBS at 37 °C for 30 min. The supernatant was removed and treated with 200 µL of accutase. After centrifugation for 3 min at 2000× g, the cells were washed once with 500 µL PBS and measured according to Section 4.4.
4.4. Affinity Analysis
The binding affinity of the selected aptamer libraries was determined by incubating 20,000 C. auris, C. albicans, and C. parapsilosis in 500 µL of PBS with different concentrations of the aptamers. Finally, the dissociation constants (Kd) of the aptamer libraries were determined by fitting the dependence of the fluorescence intensity on the aptamer concentration to the equation Y = Bmax × X/(Kd + X) using GraphPad PRISM 8. (GraphPad Software, San Diego, CA, USA), with Y = the measured fluorescence, Bmax = the maximal fluorescence, and X = concentration of the aptamers).
4.5. Sensitivity Test
The sensitivity of the aptamers was determined by analyzing the linear relationship between fluorescence intensity and the log value of the cell number for each yeast.
4.6. Detection of Clinic Isolates
One milliliter of clinic isolates with an OD of 0.01 was incubated with 5 pmol of aptamer library, and the fluorescence intensity of the C. albicans strains under the same conditions as above was compared to determine the ability of the aptamer library to detect Candida in practice.
Author Contributions
F.R., supervision, methodology, writing—review and editing; Y.Z., H.X., A.-K.K., B.S., S.S. and F.R.; writing; investigation, H.X., Y.Z., M.K., G.B. and A.-K.K.; formal analysis, H.X., Y.Z., M.K. and A.-K.K.; conceptualization and resources, T.W. and F.R.; investigation, H.X., Y.Z., M.K., A.B. and A.-K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the China Scholarship Council (No.: 202108080084 and 202208080009) and the German Research Society (DFG) project 465229237.
Institutional Review Board Statement
Fungal strains were collected from patient samples sent to the microbiology department for diagnostic purposes. The strains were collected anonymously, and it was not possible to track the source of the yeasts. The accreditation number of the microbiology department is DIN EN ISO15189:2014 (DAkks).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data can be requested from the authors for valid reasons.
Acknowledgments
The authors thank the China Scholarship Council for their financial support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Nobile, C.J.; Johnson, A.D. Candida Albicans Biofilms and Human Disease. Annu. Rev. Microbiol. 2015, 69, 71–92. [Google Scholar] [CrossRef]
- Papon, N.; Courdavault, V.; Clastre, M.; Bennett, R.J. Emerging and Emerged Pathogenic Candida Species: Beyond the Candida Albicans Paradigm. PLoS Pathog. 2013, 9, e1003550. [Google Scholar] [CrossRef]
- Fakhim, H.; Vaezi, A.; Dannaoui, E.; Chowdhary, A.; Nasiry, D.; Faeli, L.; Meis, J.F.; Badali, H. Comparative Virulence of Candida Auris with Candida Haemulonii, Candida Glabrata and Candida Albicans in a Murine Model. Mycoses 2018, 61, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, S.R. Current Epidemiology of Candida Infection. Clin. Microbiol. Newsl. 2014, 36, 131–136. [Google Scholar] [CrossRef]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Perlroth, J.; Choi, B.; Spellberg, B. Nosocomial Fungal Infections: Epidemiology, Diagnosis, and Treatment. Med. Mycol. 2007, 45, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Enoch, D.A.; Yang, H.; Aliyu, S.H.; Micallef, C. The Changing Epidemiology of Invasive Fungal Infections. Methods Mol. Biol. 2017, 1508, 17–65. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Gaitán, A.C.; Moret, A.; López Hontangas, J.L.; Molina, J.M.; Aleixandre López, A.I.; Cabezas, A.H.; Mollar Maseres, J.; Arcas, R.C.; Gómez Ruiz, M.D.; Chiveli, M.Á.; et al. Nosocomial Fungemia by Candida Auris: First Four Reported Cases in Continental Europe. Rev. Iberoam. Micol. 2017, 34, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Sobel, J.D.; Faro, S.; Force, R.W.; Foxman, B.; Ledger, W.J.; Nyirjesy, P.R.; Reed, B.D.; Summers, P.R. Vulvovaginal Candidiasis: Epidemiologic, Diagnostic, and Therapeutic Considerations. Am. J. Obstet. Gynecol. 1998, 178, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.W.; Kneale, M.; Sobel, J.D.; Rautemaa-Richardson, R. Global Burden of Recurrent Vulvovaginal Candidiasis: A Systematic Review. Lancet Infect. Dis. 2018, 18, e339–e347. [Google Scholar] [CrossRef]
- Kullberg, B.J.; Arendrup, M.C. Invasive Candidiasis. N. Engl. J. Med. 2015, 373, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Pappas, P.G.; Lionakis, M.S.; Arendrup, M.C.; Ostrosky-Zeichner, L.; Kullberg, B.J. Invasive Candidiasis. Nat. Rev. Dis. Primers 2018, 4, 18026. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Diekema, D.J. Epidemiology of Invasive Candidiasis: A Persistent Public Health Problem. Clin. Microbiol. Rev. 2007, 20, 133–163. [Google Scholar] [CrossRef] [PubMed]
- Harrington, R.; Kindermann, S.L.; Hou, Q.; Taylor, R.J.; Azie, N.; Horn, D.L. Candidemia and Invasive Candidiasis among Hospitalized Neonates and Pediatric Patients. Curr. Med. Res. Opin. 2017, 33, 1803–1812. [Google Scholar] [CrossRef] [PubMed]
- Quindós, G.; Marcos-Arias, C.; San-Millán, R.; Mateo, E.; Eraso, E. The Continuous Changes in the Aetiology and Epidemiology of Invasive Candidiasis: From Familiar Candida Albicans to Multiresistant Candida Auris. Int. Microbiol. 2018, 21, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Xiao, M.; Liao, K.; Kudinha, T.; Wang, H.; Zhang, L.; Hou, X.; Kong, F.; Xu, Y.-C. Notable Increasing Trend in Azole Non-Susceptible Candida Tropicalis Causing Invasive Candidiasis in China (August 2009 to July 2014): Molecular Epidemiology and Clinical Azole Consumption. Front. Microbiol. 2017, 8, 464. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Preston, T.; Bale, M.; Koontz, F.P.; Body, B.A. Comparison of the Quantum II, API Yeast Ident, and AutoMicrobic Systems for Identification of Clinical Yeast Isolates. J. Clin. Microbiol. 1988, 26, 2054–2058. [Google Scholar] [CrossRef]
- Shankland, G.S.; Hopwood, V.; Forster, R.A.; Evans, E.G.; Richardson, M.D.; Warnock, D.W. Multicenter Evaluation of Microring YT, a New Method of Yeast Identification. J. Clin. Microbiol. 1990, 28, 2808–2810. [Google Scholar] [CrossRef]
- Dhiman, N.; Hall, L.; Wohlfiel, S.L.; Buckwalter, S.P.; Wengenack, N.L. Performance and Cost Analysis of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for Routine Identification of Yeast. J. Clin. Microbiol. 2011, 49, 1614–1616. [Google Scholar] [CrossRef]
- Pinto, A.; Halliday, C.; Zahra, M.; van Hal, S.; Olma, T.; Maszewska, K.; Iredell, J.R.; Meyer, W.; Chen, S.C.-A. Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry Identification of Yeasts Is Contingent on Robust Reference Spectra. PLoS ONE 2011, 6, e25712. [Google Scholar] [CrossRef]
- Jamal, W.; Saleem, R.; Rotimi, V.O. Rapid Identification of Pathogens Directly from Blood Culture Bottles by Bruker Matrix-Assisted Laser Desorption Laser Ionization-Time of Flight Mass Spectrometry versus Routine Methods. Diagn. Microbiol. Infect. Dis. 2013, 76, 404–408. [Google Scholar] [CrossRef]
- Jamal, W.Y.; Ahmad, S.; Khan, Z.U.; Rotimi, V.O. Comparative Evaluation of Two Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) Systems for the Identification of Clinically Significant Yeasts. Int. J. Infect. Dis. 2014, 26, 167–170. [Google Scholar] [CrossRef]
- Stevenson, L.G.; Drake, S.K.; Shea, Y.R.; Zelazny, A.M.; Murray, P.R. Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Clinically Important Yeast Species. J. Clin. Microbiol. 2010, 48, 3482–3486. [Google Scholar] [CrossRef]
- Felix, G.N.; de Freitas, V.L.T.; da Silva Junior, A.R.; Magri, M.M.C.; Rossi, F.; Sejas, O.N.E.; Abdala, E.; Malbouisson, L.M.S.; Guimarães, T.; Benard, G.; et al. Performance of a Real-Time PCR Assay for the Detection of Five Candida Species in Blood Samples from ICU Patients at Risk of Candidemia. J. Fungi 2023, 9, 635. [Google Scholar] [CrossRef] [PubMed]
- Busser, F.D.; Coelho, V.C.; de Abreu Fonseca, C.; Del Negro, G.M.B.; Shikanai-Yasuda, M.A.; Lopes, M.H.; Magri, M.M.C.; de Freitas, V.L.T. A Real Time PCR Strategy for the Detection and Quantification of Candida Albicans in Human Blood. Rev. Inst. Med. Trop. Sao Paulo 2020, 62, e9. [Google Scholar] [CrossRef] [PubMed]
- Camp, I.; Spettel, K.; Willinger, B. Molecular Methods for the Diagnosis of Invasive Candidiasis. J. Fungi 2020, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Ilgu, M.; Nilsen-Hamilton, M. Aptamers in Analytics. Analyst 2016, 141, 1551–1568. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as Therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Chopra, A.; Shukla, R.; Sharma, T.K. Aptamers as an Emerging Player in Biology. Aptamers Synth. Antibodies 2014, 1, 1–11. [Google Scholar]
- Ali, M.H.; Elsherbiny, M.E.; Emara, M. Updates on Aptamer Research. Int. J. Mol. Sci. 2019, 20, 2511. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Yeung, W.; Hili, R. In Vitro Selection of Diversely Functionalized Aptamers. J. Am. Chem. Soc. 2017, 139, 13977–13980. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Huang, R.; Deng, Y.; He, N. Progress in Selection and Biomedical Applications of Aptamers. J. Biomed. Nanotechnol. 2014, 10, 3043–3062. [Google Scholar] [CrossRef] [PubMed]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA Aptamers Using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 440. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Shaban, S.M.; Kim, D.-H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef]
- Ku, T.-H.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.-W.; Han, Y.; Lo, Y.-H. Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef]
- Kusumawati, A.; Mustopa, A.Z.; Wibawan, I.W.T.; Setiyono, A.; Sudarwanto, M.B. A Sequential Toggle Cell-SELEX DNA Aptamer for Targeting Staphylococcus Aureus, Streptococcus Agalactiae, and Escherichia Coli Bacteria. J. Genet. Eng. Biotechnol. 2022, 20, 95. [Google Scholar] [CrossRef]
- Ansari, N.; Ghazvini, K.; Ramezani, M.; Shahdordizadeh, M.; Yazdian-Robati, R.; Abnous, K.; Taghdisi, S.M. Selection of DNA Aptamers against Mycobacterium Tuberculosis Ag85A, and Its Application in a Graphene Oxide-Based Fluorometric Assay. Microchim. Acta 2018, 185, 21. [Google Scholar] [CrossRef]
- Zhong, Z.; Gao, X.; Gao, R.; Jia, L. Selective Capture and Sensitive Fluorometric Determination of Pseudomonas Aeruginosa by Using Aptamer Modified Magnetic Nanoparticles. Microchim. Acta 2018, 185, 377. [Google Scholar] [CrossRef]
- Wu, Z.; He, D.; Cui, B. A Fluorometric Assay for Staphylococcal Enterotoxin B by Making Use of Platinum Coated Gold Nanorods and of Upconversion Nanoparticles. Microchim. Acta 2018, 185, 516. [Google Scholar] [CrossRef]
- Yuan, J.; Wu, S.; Duan, N.; Ma, X.; Xia, Y.; Chen, J.; Ding, Z.; Wang, Z. A Sensitive Gold Nanoparticle-Based Colorimetric Aptasensor for Staphylococcus Aureus. Talanta 2014, 127, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Meng, X.; Sun, Y.; Li, Q.; Zhao, R.; Zhang, Y.; Wang, J.; Yi, Z. Aptamer-Based Fluorometric Assay for Direct Identification of Methicillin-Resistant Staphylococcus Aureus from Clinical Samples. J. Microbiol. Methods 2018, 153, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Phillips, T.; Montez, T.; Garcia, A.; Sivils, J.C.; Mayo, M.W.; Greis, A. Development of a Fluorescent Enzyme-Linked DNA Aptamer-Magnetic Bead Sandwich Assay and Portable Fluorometer for Sensitive and Rapid Listeria Detection. J. Fluoresc. 2015, 25, 173–183. [Google Scholar] [CrossRef]
- Wu, W.; Li, J.; Pan, D.; Li, J.; Song, S.; Rong, M.; Li, Z.; Gao, J.; Lu, J. Gold Nanoparticle-Based Enzyme-Linked Antibody-Aptamer Sandwich Assay for Detection of Salmonella Typhimurium. ACS Appl. Mater. Interfaces 2014, 6, 16974–16981. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yue, F.; Kong, Q.; Liu, Z.; Guo, Y.; Sun, X. Aptamers against Pathogenic Bacteria: Selection Strategies and Apta-Assay/Aptasensor Application for Food Safety. J. Agric. Food Chem. 2022, 70, 5477–5498. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wang, L.; Lu, W.; Zhao, J.; Zhang, H.; Chen, W. Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium Bifidum. Int. J. Mol. Sci. 2017, 18, 883. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Green, L.S.; Gold, L.; Janjic, N. Systematic Selection of Modified Aptamer Pairs for Diagnostic Sandwich Assays. Biotechniques 2014, 56, 125–133. [Google Scholar] [CrossRef]
- Kneißle, K.; Krämer, M.; Kissmann, A.-K.; Xing, H.; Müller, F.; Amann, V.; Noschka, R.; Gottschalk, K.-E.; Bozdogan, A.; Andersson, J.; et al. A Polyclonal SELEX Aptamer Library Allows Differentiation of Candida Albicans, C. Auris and C. Parapsilosis Cells from Human Dermal Fibroblasts. J. Fungi 2022, 8, 856. [Google Scholar] [CrossRef]
- Thevendran, R.; Navien, T.N.; Meng, X.; Wen, K.; Lin, Q.; Sarah, S.; Tang, T.-H.; Citartan, M. Mathematical Approaches in Estimating Aptamer-Target Binding Affinity. Anal. Biochem. 2020, 600, 113742. [Google Scholar] [CrossRef] [PubMed]
- Kubiczek, D.; Raber, H.; Bodenberger, N.; Oswald, T.; Sahan, M.; Mayer, D.; Wiese, S.; Stenger, S.; Weil, T.; Rosenau, F. The Diversity of a Polyclonal FluCell-SELEX Library Outperforms Individual Aptamers as Emerging Diagnostic Tools for the Identification of Carbapenem Resistant Pseudomonas Aeruginosa. Chem. A Eur. J. 2020, 26, 14536–14545. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Zhang, Y.; Krämer, M.; Kissmann, A.K.; Henkel, M.; Weil, T.; Knippschild, U.; Rosenau, F. A Polyclonal Selex Aptamer Library Directly Allows Specific Labelling of the Human Gut Bacterium Blautia Producta without Isolating Individual Aptamers. Molecules 2022, 27, 5693. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Kissmann, A.K.; Raber, H.F.; Krämer, M.; Amann, V.; Kohn, K.; Weil, T.; Rosenau, F. Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides Distasonis. Microorganisms 2021, 9, 2284. [Google Scholar] [CrossRef]
- Xing, H.; Zhang, Y.; Krämer, M.; Kissmann, A.K.; Amann, V.; Raber, H.F.; Weil, T.; Stieger, K.R.; Knippschild, U.; Henkel, M.; et al. A Polyclonal Aptamer Library for the Specific Binding of the Gut Bacterium Roseburia Intestinalis in Mixtures with Other Gut Microbiome Bacteria and Human Stool Samples. Int. J. Mol. Sci. 2022, 23, 7744. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xing, H.; Bolotnikov, G.; Krämer, M.; Gotzmann, N.; Knippschild, U.; Kissmann, A.-K.; Rosenau, F. Enriched Aptamer Libraries in Fluorescence-Based Assays for Rikenella Microfusus-Specific Gut Microbiome Analyses. Microorganisms 2023, 11, 2266. [Google Scholar] [CrossRef] [PubMed]
- Raber, H.F.; Kubiczek, D.H.; Bodenberger, N.; Kissmann, A.K.; D’souza, D.; Hu, X.; Mayer, D.; Xu, P.; Knippschild, U.; Spellerberg, B.; et al. Flucell-selex Aptamers as Specific Binding Molecules for Diagnostics of the Health Relevant Gut Bacterium Akkermansia Muciniphila. Int. J. Mol. Sci. 2021, 22, 10425. [Google Scholar] [CrossRef]
- Kissmann, A.K.; Wolf, D.; Krämer, M.; Müller, F.; Amann, V.; Xing, H.; Gottschalk, K.E.; Weil, T.; Eichmann, R.; Schäfer, P.; et al. Polyclonal Aptamer Libraries from a FluRoot-SELEX for the Specific Labeling of the Apical and Elongation/Differentiation Zones of Arabidopsis Thaliana Roots. Int. J. Mol. Sci. 2022, 23, 12220. [Google Scholar] [CrossRef]
- Kissmann, A.-K.; Andersson, J.; Bozdogan, A.; Amann, V.; Kraemer, M.; Xing, H.; Raber, H.; Kubiczek, D.H.; Aspermair, P.; Knoll, W.; et al. Polyclonal Aptamer Libraries as Binding Entities on a Graphene FET Based Biosensor for the Discrimination of Apo- and Holo- Retinol Binding Protein 4. Nanoscale Horiz. 2022, 7, 770–778. [Google Scholar] [CrossRef]
- Wan, Q.; Liu, X.; Zu, Y. Oligonucleotide Aptamers for Pathogen Detection and Infectious Disease Control. Theranostics 2021, 11, 9133–9161. [Google Scholar] [CrossRef]
- Lee, S.-H.; Ahn, J.-Y.; Lee, K.-A.; Um, H.-J.; Sekhon, S.S.; Sun Park, T.; Min, J.; Kim, Y.-H. Analytical Bioconjugates, Aptamers, Enable Specific Quantitative Detection of Listeria Monocytogenes. Biosens. Bioelectron. 2015, 68, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Trunzo, N.E.; Hong, K.L. Recent Progress in the Identification of Aptamers Against Bacterial Origins and Their Diagnostic Applications. Int. J. Mol. Sci. 2020, 21, 5074. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, X.; Duan, N.; Wu, S.; Wang, Z.; Wei, X.; Wang, Y. Selection and Characterization of DNA Aptamers against Staphylococcus Aureus Enterotoxin C1. Food Chem. 2015, 166, 623–629. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.-R.; Sekhon, S.S.; Kim, S.-G.; Rhee, S.J.; Yang, G.N.; Won, K.; Rhee, S.-K.; Ryu, H.; Kim, K.; Min, J.; et al. Aptamer-Based Pathogen Monitoring for Salmonella Enterica Ser. Typhimurium. J. Biomed. Nanotechnol. 2018, 14, 1992–2002. [Google Scholar] [CrossRef]
- Song, M.-S.; Sekhon, S.; Shin, W.-R.; Kim, H.; Min, J.; Ahn, J.-Y.; Kim, Y.-H. Detecting and Discriminating Shigella Sonnei Using an Aptamer-Based Fluorescent Biosensor Platform. Molecules 2017, 22, 825. [Google Scholar] [CrossRef]
- Chinnappan, R.; AlAmer, S.; Eissa, S.; Rahamn, A.A.; Abu Salah, K.M.; Zourob, M. Fluorometric Graphene Oxide-Based Detection of Salmonella Enteritis Using a Truncated DNA Aptamer. Microchim. Acta 2018, 185, 61. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).