The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins
Abstract
1. Introduction
2. Circulating Biomarkers: Is There an Ideal One?
- Clinical Relevance: The biomarker must offer meaningful insights that have a solid rationale for its application, reflecting significant measurements or variations in physiological or pathological states within a short timeframe.
- High Sensitivity and Specificity: Essential for evaluating treatment effects, these criteria ensure that the biomarker can accurately identify disease presence or absence and monitor therapeutic responses.
- Reliability: This encompasses the biomarker’s analytical measurement capabilities, highlighting the necessity for accurate detection with consistent accuracy, precision, robustness, and reproducibility.
- Practicality: Favoring non-invasive or minimally invasive methods reduces discomfort and inconvenience for individuals, making the biomarker patient-friendlier.
- Simplicity: The ease of use, affordability, and accessibility of necessary equipment are crucial for the biomarker’s adoption in drug development and clinical settings, promoting its widespread acceptance and implementation.
- Preclinical Testing: This initial stage involves assessing the biomarker using patient samples, with subsequent verification at both in vitro and in vivo levels to ensure preliminary efficacy and safety.
- Feasibility Analysis: This step aims to demonstrate the biomarker’s capability to distinguish between diseased and healthy individuals, establishing its potential diagnostic value.
- Validation Process: This vital stage verifies the accurate assay of the biomarker, ensuring that it meets all required standards for clinical application.
- Statistical Analysis: Performed to assess the biomarker’s discriminatory accuracy within a large patient cohort, this analysis evaluates its effectiveness in a broader clinical context.
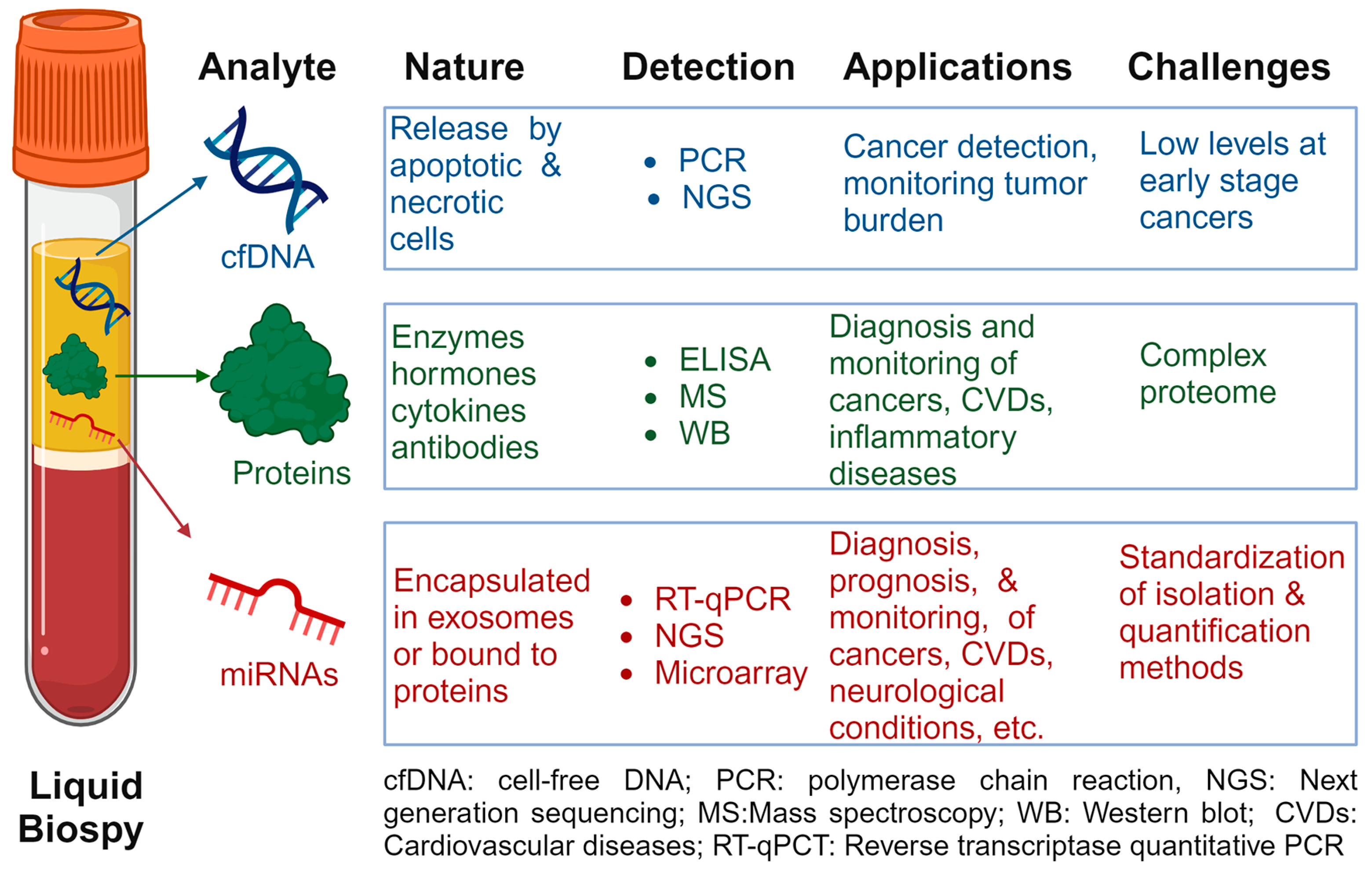
2.1. Cell-Free DNAs
2.2. Circulating Proteins
2.3. Circulating miRNAs
2.3.1. Advantages of C-miRNAs as Liquid Biopsies
- High Stability: C-miRNAs are stable in biofluids even under extreme conditions including pH, temperature changes, and freeze–thaw cycles.
- Non-Invasive: Detection in blood, urine, and other bodily fluids allows for minimally invasive testing compared to tissue biopsies.
- Early Detection: c-miRNAs can be indicative of disease before clinical symptoms appear, allowing for potentially earlier intervention.
- Tissue and Disease Specificity: Specific c-miRNAs are associated with specific tissues or diseases, aiding in targeted diagnosis and monitoring.
- Dynamic Response: Levels of c-miRNAs can change in response to disease progression or treatment, providing real-time monitoring capabilities.
2.3.2. Circulating miRNAs as Biomarkers: Challenges, Limitations, and Perspectives
- Complexity in Quantification: Precise quantification of c-miRNAs can be challenging due to their small size and the need for sensitive detection methods.
- Standardization Issues: Lack of standardized protocols for extraction, detection, and data analysis can lead to variability in results.
- Biological Function Uncertainties: The comprehensive roles and mechanisms of many c-miRNAs in various diseases are not fully understood.
- Interference by Endogenous Substances: Biological substances in samples may interfere with c-miRNA detection and analysis.
- Validation and Reproducibility: There is a need for large-scale studies to validate c-miRNAs as reliable biomarkers across different populations and conditions.
3. Combinatorial Potential of C-miRNAs with cfDNAs and Proteins as a Multi-Analyte Approach
4. Challenges and Limitations in Using a Multi-Analyte Biopsy Consisting of C-miRNAs, cfDNAs, and Circulating Proteins
- Lack of Standardization: There is a significant gap in standardized protocols for pre-analytical, analytical, and post-analytical procedures. This includes variability in biomarker source (e.g., blood fraction), types of blood collection tubes, methods of sample collection and handling, and techniques for biomarker isolation and detection.
- Influence of Individual Characteristics: The levels of biomarkers, including c-miRNAs, can be affected by various individual characteristics such as age, gender, physical activity, medication, and diet. This variability adds complexity to interpreting biomarker levels across different populations.
- Integration into Clinical Trials: Multi-analyte testing with more than two analytes has not yet been widely adopted in prospective interventional clinical trials. Demonstrating the clinical utility of this approach requires well-designed, large-scale, multicenter trials with rigorous conditions and standardized methodologies.
- Data Analysis and Interpretation: The combinatorial approach generates vast amounts of data, necessitating advanced bioinformatics and statistical analysis tools to identify meaningful correlations and potential causal links between biomarkers and disease states. The complexity of data interpretation is further compounded by the synergistic and orthogonal relationships between different types of biomarkers.
- Technological Limitations: Despite advancements in sequencing and proteomics technologies, detecting and quantifying low-abundance biomarkers with high sensitivity and specificity remain challenging. This is crucial for early disease detection and monitoring minimal residual disease.
- Clinical Translation: Translating the advantages of multi-analyte liquid biopsy testing into clinical practice faces hurdles in demonstrating improved diagnostic accuracy, prognostic value, and therapeutic decision-making over traditional single-analyte tests. This includes proving the approach’s cost-effectiveness and operational feasibility in a clinical setting.
- Regulatory and Ethical Considerations [192]: The introduction of new biomarker combinations into clinical practice involves navigating regulatory approvals and addressing ethical considerations related to patient consent, privacy, and data management.
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- AbdulRaheem, Y. Unveiling the significance and challenges of integrating prevention levels in healthcare practice. J. Prim. Care Community Health 2023, 14, 21501319231186500. [Google Scholar] [CrossRef] [PubMed]
- Alexander, K.E.; Brijnath, B.; Biezen, R.; Hampton, K.; Mazza, D. Preventive healthcare for young children: A systematic review of interventions in primary care. Prev. Med. 2017, 99, 236–250. [Google Scholar] [CrossRef]
- Kasoju, N.; Remya, N.S.; Sasi, R.; Sujesh, S.; Soman, B.; Kesavadas, C.; Muraleedharan, C.V.; Varma, P.R.H.; Behari, S. Digital health: Trends, opportunities and challenges in medical devices, pharma and bio-technology. CSI Trans. ICT 2023, 11, 11–30. [Google Scholar] [CrossRef]
- Fitzgerald, J.; Fenniri, H. Cutting edge methods for non-invasive disease diagnosis Uuing e-tongue and e-nose devices. Biosensors 2017, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Marchetti, D.; Lang, J.E. Liquid biopsy: From concept to clinical application. Sci. Rep. 2023, 13, 21685. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, A.; Imran, M.; Ahsan, H. Biomarkers as biomedical bioindicators: Approaches and techniques for the detection, analysis, and validation of novel biomarkers of diseases. Pharmaceutics 2023, 15, 1630. [Google Scholar] [CrossRef]
- Rho, J.H.; Bauman, A.J.; Boettger, H.G.; Yen, T.F. A search for porphyrin biomarkers in Nonesuch Shale and extraterrestrial samples. Space Life Sci. 1973, 4, 69–77. [Google Scholar] [CrossRef]
- Mundkur, B.D. Evidence excluding mutations, polysomy, and polyploidy as possible causes of non-Mendelian segregations in Saccharomyces. Ann. Missouri Bot. Gard. 1949, 36, 259–280. [Google Scholar] [CrossRef]
- Porter, K.A. Effect of homologous bone marrow injections in x-irradiated rabbits. Br. J. Exp. Pathol. 1957, 38, 401–412. [Google Scholar]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.B.; Siu, L.L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef]
- Aryutova, K.; Stoyanov, D.S.; Kandilarova, S.; Todeva-Radneva, A.; Kostianev, S.S. Clinical use of neurophysiological biomarkers and self-assessment scales to predict and monitor treatment response for psychotic and affective disorders. Curr. Pharm. Des. 2021, 27, 4039–4048. [Google Scholar] [CrossRef] [PubMed]
- Shama, A.; Soni, T.; Jawanda, I.K.; Upadhyay, G.; Sharma, A.; Prabha, V. The latest developments in using proteomic biomarkers from urine and serum for non-invasive disease diagnosis and prognosis. Biomark. Insights 2023, 18, 11772719231190218. [Google Scholar] [CrossRef]
- Allred, D.C. Commentary: Hormone receptor testing in breast cancer: A distress signal from Canada. Oncologist 2008, 13, 1134–1136. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer. 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Nishida, N.; Calin, G.A.; Pantel, K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat. Rev. Clin. Oncol. 2014, 11, 145–156. [Google Scholar] [CrossRef]
- Swarup, V.; Rajeswari, M.R. Circulating (cell-free) nucleic acids—A promising, non-invasive tool for early detection of several human diseases. FEBS Lett. 2007, 581, 795–799. [Google Scholar] [CrossRef]
- Takizawa, S.; Matsuzaki, J.; Ochiya, T. Circulating microRNAs: Challenges with their use as liquid biopsy biomarkers. Cancer Biomark. 2022, 35, 1–9. [Google Scholar] [CrossRef]
- Campos-Carrillo, A.; Weitzel, J.N.; Sahoo, P.; Rockne, R.; Mokhnatkin, J.V.; Murtaza, M.; Gray, S.W.; Goetz, L.; Goel, A.; Schork, N.; et al. Circulating tumor DNA as an early cancer detection tool. Pharmacol. Ther. 2020, 207, 107458. [Google Scholar] [CrossRef]
- Roberts, N.J.; Zhang, L.; Janku, F.; Collins, A.; Bai, R.Y.; Staedtke, V.; Rusk, A.W.; Tung, D.; Miller, M.; Roix, J.; et al. Intratumoral injection of Clostridium novyi-NT spores induces antitumor responses. Sci. Transl. Med. 2014, 6, 249ra111. [Google Scholar] [CrossRef] [PubMed]
- Adhyam, M.; Gupta, A.K. A Review on the clinical utility of PSA in cancer prostate. Indian J. Surg. Oncol. 2012, 3, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early detection of cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef]
- Harrow, J.; Nagy, A.; Reymond, A.; Alioto, T.; Patthy, L.; Antonarakis, S.E.; Guigó, R. Identifying protein-coding genes in genomic sequences. Genome Biol. 2009, 10, 201. [Google Scholar] [CrossRef]
- Vandevenne, M.; Delmarcelle, M.; Galleni, M. RNA regulatory networks as a control of stochasticity in biological systems. Front. Genet. 2019, 10, 403. [Google Scholar] [CrossRef]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function androle in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, A.; Tamanini, A.; Cabrini, G.; Dechecchi, M.C. Circulating microRNAs as emerging non-invasive biomarkers for gliomas. Ann. Transl. Med. 2017, 5, 277. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Pozniak, T.; Shcharbin, D.; Bryszewska, M. Circulating microRNAs in medicine. Int. J. Mol. Sci. 2022, 23, 3996. [Google Scholar] [CrossRef]
- Cardona, E.; Guyomar, C.; Desvignes, T.; Montfort, J.; Guendouz, S.; Postlethwait, J.H.; Skiba-Cassy, S.; Bobe, J. Circulating miRNA repertoire as a biomarker of metabolic and reproductive states in rainbow trout. BMC Biol. 2021, 19, 235. [Google Scholar] [CrossRef]
- Cheong, J.K.; Tang, Y.C.; Zhou, L.; Cheng, H.; Too, H.-P. Advances in quantifying circulatory microRNA for early disease detection. Curr. Opin. Biotechnol. 2022, 74, 256–262. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef]
- Keup, C.; Kimmig, R.; Kasimir-Bauer, S. Multimodality in liquid biopsy: Does a combination uncover insights undetectable in individual blood analytes? J. Lab. Med. 2022, 46, 255–264. [Google Scholar] [CrossRef]
- Hofmann, L.; Sallinger, K.; Haudum, C.; Smolle, M.; Heitzer, E.; Moser, T.; Novy, M.; Gesson, K.; Kroneis, T.; Bauernhofer, T.; et al. A multi-analyte approach for improved sensitivity of liquid biopsies in prostate cancer. Cancers 2020, 12, 2247. [Google Scholar] [CrossRef]
- Keup, C.; Kimmig, R.; Kasimir-Bauer, S. Combinatorial Power of cfDNA, CTCs and EVs in Oncology. Diagnostics 2022, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Shen, L.; Luo, M.; Zhang, K.; Li, J.; Yang, Q.; Zhu, F.; Zhou, D.; Zheng, S.; Chen, Y.; et al. Circulating tumor cells: Biology and clinical significance. Signal Transduct. Target Ther. 2021, 6, 404. [Google Scholar] [CrossRef] [PubMed]
- de Gramont, A.; Watson, S.; Ellis, L.M.; Rodón, J.; Tabernero, J.; de Gramont, A.; Hamilton, S.R. Pragmatic issues in biomarker evaluation for targeted therapies in cancer. Nat. Rev. Clin. Oncol. 2015, 12, 197–212. [Google Scholar] [CrossRef]
- Mert, D.G.; Terzi, H. Mean platelet volume in bipolar disorder: The search for an ideal biomarker. Neuropsychiatr. Dis. Treat. 2016, 12, 2057–2062. [Google Scholar] [CrossRef] [PubMed]
- Califf, R.M. Biomarker definitions and their applications. Exp. Biol. Med. 2018, 243, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Dongiovanni, P.; Meroni, M.; Casati, S.; Goldoni, R.; Thomaz, D.V.; Kehr, N.S.; Galimberti, D.; Del Fabbro, M.; Tartaglia, G.M. Salivary biomarkers: Novel noninvasive tools to diagnose chronic inflammation. Int. J. Oral Sci. 2023, 15, 27. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Prajzlerová, K.; Šenolt, L.; Filková, M. Is there a potential of circulating miRNAs as biomarkers in rheumatic diseases? Genes Dis. 2023, 10, 1263–1278. [Google Scholar] [CrossRef]
- Glaich, O.; Parikh, S.; Bell, R.E.; Mekahel, K.; Donyo, M.; Leader, Y.; Shayevitch, R.; Sheinboim, D.; Yannai, S.; Hollander, D.; et al. DNA methylation directs microRNA biogenesis in mammalian cells. Nat. Commun. 2019, 10, 5657. [Google Scholar] [CrossRef]
- Sallustio, F.; Gesualdo, L.; Gallone, A. New findings showing how DNA methylation influences diseases. World J. Biol. Chem. 2019, 10, 1–6. [Google Scholar] [CrossRef]
- Luo, H.; Wei, W.; Ye, Z.; Zheng, J.; Xu, R.-h. Liquid biopsy of methylation biomarkers in cell-free DNA. Trends. Mol. Med. 2021, 27, 482–500. [Google Scholar] [CrossRef] [PubMed]
- Saito, Y.; Saito, H.; Liang, G.; Friedman, J.M. Epigenetic alterations and microRNA misexpression in cancer and autoimmune diseases: A critical review. Clin. Rev. Allergy Immunol. 2014, 47, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, C.; Zhou, Y.; Yao, Y.; Liu, J.; Wu, M.; Su, J. Liquid biopsies for cancer: From bench to clinic. MedComm 2023, 4, e329. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, W.; Claret, F.X. Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 2017, 12, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Koch, A.; Krockenberger, K.; Grosshennig, A. Personalized medicine using DNA biomarkers: A review. Hum. Genet. 2012, 131, 1627–1638. [Google Scholar] [CrossRef] [PubMed]
- Kamel, H.F.M.; Al-Amodi, H. Exploitation of gene expression and cancer biomarkers in paving the path to era of personalized medicine. Genom. Proteom. Bioinform. 2017, 15, 220–235. [Google Scholar] [CrossRef]
- Ginsburg, G.S.; Willard, H.F. Genomic and personalized medicine: Foundations and applications. Transl. Res. 2009, 154, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Bartok, B.; Oler, E.; Liang, K.Y.H.; Budinski, Z.; Berjanskii, M.; Guo, A.; Cao, X.; Wilson, M. MarkerDB: An online database of molecular biomarkers. Nucleic Acids Res. 2021, 49, D1259–D1267. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.; Pan, D.; Wang, X.; Xu, Y.; Yan, J.; Wang, L.; Yang, X.; Yang, M.; Liu, G.P. Applications of multi-omics analysis in human diseases. MedComm 2023, 4, e315. [Google Scholar] [CrossRef]
- Al-Tashi, Q.; Saad, M.B.; Muneer, A.; Qureshi, R.; Mirjalili, S.; Sheshadri, A.; Le, X.; Vokes, N.I.; Zhang, J.; Wu, J. Machine learning models for the identification of prognostic and predictive cancer biomarkers: A systematic review. Int. J. Mol. Sci. 2023, 24, 7781. [Google Scholar] [CrossRef]
- Shegekar, T.; Vodithala, S.; Juganavar, A. The emerging role of liquid biopsies in revolutionising cancer diagnosis and therapy. Cureus 2023, 15, e43650. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, F.S.; Barauna, V.G.; Dos Santos, L.; Costa, G.; Vassallo, P.F.; Campos, L.C.G. Properties and application of cell-free DNA as a clinical biomarker. Int. J. Mol. Sci. 2021, 22, 9110. [Google Scholar] [CrossRef]
- Mandel, P.; Metais, P. Nuclear acids in humanblood plasma. C R Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Stroun, M.; Anker, P.; Lyautey, J.; Lederrey, C.; Maurice, P.A. Isolation and characterization of DNA from the plasma of cancer patients. Eur. J. Cancer Clin. Oncol. 1987, 23, 707–712. [Google Scholar] [CrossRef]
- Polina, I.A.; Ilatovskaya, D.V.; DeLeon-Pennell, K.Y. Cell free DNA as a diagnostic and prognostic marker for cardiovascular diseases. Clin. Chim. Acta. 2020, 503, 145–150. [Google Scholar] [CrossRef]
- Tsai, N.W.; Lin, T.K.; Chen, S.D.; Chang, W.N.; Wang, H.C.; Yang, T.M.; Lin, Y.J.; Jan, C.R.; Huang, C.R.; Liou, C.W.; et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin. Chim. Acta 2011, 412, 476–479. [Google Scholar] [CrossRef]
- Lou, X.; Hou, Y.; Liang, D.; Peng, L.; Chen, H.; Ma, S.; Zhang, L. A novel Alu-based real-time PCR method for the quantitative detection of plasma circulating cell-free DNA: Sensitivity and specificity for the diagnosis of myocardial infarction. Int. J. Mol. Med. 2015, 35, 72–80. [Google Scholar] [CrossRef]
- Lam, N.Y.; Rainer, T.H.; Chiu, R.W.; Joynt, G.M.; Lo, Y.M. Plasma mitochondrial DNA concentrations after trauma. Clin. Chem. 2004, 50, 213–216. [Google Scholar] [CrossRef]
- Kung, C.T.; Hsiao, S.Y.; Tsai, T.C.; Su, C.M.; Chang, W.N.; Huang, C.R.; Wang, H.C.; Lin, W.C.; Chang, H.W.; Lin, Y.J.; et al. Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J. Transl. Med. 2012, 10, 130. [Google Scholar] [CrossRef]
- Breitbach, S.; Tug, S.; Simon, P. Circulating cell-free DNA: An up-coming molecular marker in exercise physiology. Sports Med. 2012, 42, 565–586. [Google Scholar] [CrossRef]
- Teo, Y.V.; Capri, M.; Morsiani, C.; Pizza, G.; Faria, A.M.C.; Franceschi, C.; Neretti, N. Cell-free DNA as a biomarker of aging. Aging Cell 2019, 18, e12890. [Google Scholar] [CrossRef]
- Raptis, L.; Menard, H.A. Quantitation and characterization of plasma DNA in normals and patients with systemic lupus erythematosus. J. Clin. Investig. 1980, 66, 1391–1399. [Google Scholar] [CrossRef]
- Mosca, M.; Giuliano, T.; Cuomo, G.; Doveri, M.; Tani, C.; Curcio, M.; Abignano, G.; De Feo, F.; Bazzichi, L.; Della Rossa, A.; et al. Cell-free DNA in the plasma of patients with systemic sclerosis. Clin. Rheumatol. 2009, 28, 1437–1440. [Google Scholar] [CrossRef] [PubMed]
- Ou, H.L.; Schumacher, B. DNA damage responses and p53 in the aging process. Blood 2018, 131, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Lauková, L.; Konečná, B.; Vlková, B.; Mlynáriková, V.; Celec, P.; Šteňová, E. Anti-cytokine therapy and plasma DNA in patients with rheumatoid arthritis. Rheumatol. Int. 2018, 38, 1449–1454. [Google Scholar] [CrossRef] [PubMed]
- Yong, E. Cancer biomarkers: Written in blood. Nature 2014, 511, 524–526. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef]
- Hanibuchi, M.; Kanoh, A.; Kuramoto, T.; Saito, T.; Tobiume, M.; Saijo, A.; Kozai, H.; Kondo, M.; Morizumi, S.; Yoneda, H.; et al. Development, validation, and comparison of gene analysis methods for detecting EGFR mutation from non-small cell lung cancer patients-derived circulating free DNA. Oncotarget 2019, 10, 3654–3666. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Bronkhorst, A.J.; Ungerer, V.; Oberhofer, A.; Gabriel, S.; Polatoglou, E.; Randeu, H.; Uhlig, C.; Pfister, H.; Mayer, Z.; Holdenrieder, S. New Perspectives on the importance of cell-free DNA biology. Diagnostics 2022, 12, 2147. [Google Scholar] [CrossRef]
- Gao, Q.; Zeng, Q.; Wang, Z.; Li, C.; Xu, Y.; Cui, P.; Zhu, X.; Lu, H.; Wang, G.; Cai, S.; et al. Circulating cell-free DNA for cancer early detection. Innovation 2022, 3, 100259. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Sledge, G.W.; Jeffrey, S.S. Liquid biopsy enters the clinic—Implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021, 18, 297–312. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; Vettori, R.; Steffan, A.; Cannizzaro, R.; De Re, V. Circulating proteins as diagnostic markers in gastric cancer. Int. J. Mol. Sci. 2023, 24, 16931. [Google Scholar] [CrossRef] [PubMed]
- Scherl, A. Clinical protein mass spectrometry. Methods 2015, 81, 3–14. [Google Scholar] [CrossRef]
- Dhama, K.; Latheef, S.K.; Dadar, M.; Samad, H.A.; Munjal, A.; Khandia, R.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Bhatt, P.; et al. Biomarkers in stress related diseases/disorders: Diagnostic, prognostic, and therapeutic values. Front. Mol. Biosci. 2019, 6, 91. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Yang, F.; Jin, H.; Wang, X. The regulation of protein translation and its implications for cancer. Signal Transduct. Target Ther. 2021, 6, 68. [Google Scholar] [CrossRef]
- Zhang, N.; Hao, J.; Cai, Y.; Wang, M. Research advances of secretory proteins in malignant tumors. Chin. J. Cancer Res. 2021, 33, 115–132. [Google Scholar] [CrossRef]
- Veyssière, H.; Bidet, Y.; Penault-Llorca, F.; Radosevic-Robin, N.; Durando, X. Circulating proteins as predictive and prognostic biomarkers in breast cancer. Clin. Proteom. 2022, 19, 25. [Google Scholar] [CrossRef]
- Brandi, J.; Manfredi, M.; Speziali, G.; Gosetti, F.; Marengo, E.; Cecconi, D. Proteomic approaches to decipher cancer cell secretome. Semin. Cell Dev. Biol. 2018, 78, 93–101. [Google Scholar] [CrossRef]
- Lojanapiwat, B.; Anutrakulchai, W.; Chongruksut, W.; Udomphot, C. Correlation and diagnostic performance of the prostate-specific antigen level with the diagnosis, aggressiveness, and bone metastasis of prostate cancer in clinical practice. Prostate Int. 2014, 2, 133–139. [Google Scholar] [CrossRef]
- Scher, H.I.; Morris, M.J.; Larson, S.; Heller, G. Validation and clinical utility of prostate cancer biomarkers. Nat. Rev. Clin. Oncol. 2013, 10, 225–234. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Opstal-van Winden, A.W.; Krop, E.J.; Kåredal, M.H.; Gast, M.C.; Lindh, C.H.; Jeppsson, M.C.; Jönsson, B.A.; Grobbee, D.E.; Peeters, P.H.; Beijnen, J.H.; et al. Searching for early breast cancer biomarkers by serum protein profiling of pre-diagnostic serum; a nested case-control study. BMC Cancer 2011, 11, 381. [Google Scholar] [CrossRef]
- Fredolini, C.; Pathak, K.V.; Paris, L.; Chapple, K.M.; Tsantilas, K.A.; Rosenow, M.; Tegeler, T.J.; Garcia-Mansfield, K.; Tamburro, D.; Zhou, W.; et al. Shotgun proteomics coupled to nanoparticle-based biomarker enrichment reveals a novel panel of extracellular matrix proteins as candidate serum protein biomarkers for early-stage breast cancer detection. Breast Cancer Res. 2020, 22, 135. [Google Scholar] [CrossRef]
- Rifai, N.; Gillette, M.A.; Carr, S.A. Protein biomarker discovery and validation: The long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. [Google Scholar] [CrossRef]
- Westermann, D.; Neumann, J.T.; Sörensen, N.A.; Blankenberg, S. High-sensitivity assays for troponin in patients with cardiac disease. Nat. Rev. Cardiol. 2017, 14, 472–483. [Google Scholar] [CrossRef]
- Park, K.C.; Gaze, D.C.; Collinson, P.O.; Marber, M.S. Cardiac troponins: From myocardial infarction to chronic disease. Cardiovasc. Res. 2017, 113, 1708–1718. [Google Scholar] [CrossRef]
- Twerenbold, R.; Boeddinghaus, J.; Nestelberger, T.; Wildi, K.; Rubini Gimenez, M.; Badertscher, P.; Mueller, C. Clinical use of high-sensitivity cardiac troponin in patients with suspected myocardial infarction. J. Am. Coll. Cardiol. 2017, 70, 996–1012. [Google Scholar] [CrossRef] [PubMed]
- Dayon, L.; Cominetti, O.; Affolter, M. Proteomics of human biological fluids for biomarker discoveries: Technical advances and recent applications. Expert Rev. Proteom. 2022, 19, 131–151. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.; Rudnick, P.A.; Martinez, M.Y.; Cheek, K.L.; Stein, S.E.; Slebos, R.J.; Liebler, D.C. Depletion of abundant plasma proteins and limitations of plasma proteomics. J. Proteome Res. 2010, 9, 4982–4991. [Google Scholar] [CrossRef] [PubMed]
- Boschetti, E.; Righetti, P.G. Optimized sample treatment protocol by solid-phase peptide libraries to enrich for protein traces. Amino Acids 2013, 45, 1431–1442. [Google Scholar] [CrossRef] [PubMed]
- Hartl, J.; Kurth, F.; Kappert, K.; Horst, D.; Mülleder, M.; Hartmann, G.; Ralser, M. Quantitative protein biomarker panels: A path to improved clinical practice through proteomics. EMBO Mol. Med. 2023, 15, e16061. [Google Scholar] [CrossRef] [PubMed]
- Curtis, J.R.; van der Helm-van Mil, A.H.; Knevel, R.; Huizinga, T.W.; Haney, D.J.; Shen, Y.; Ramanujan, S.; Cavet, G.; Centola, M.; Hesterberg, L.K.; et al. Validation of a novel multibiomarker test to assess rheumatoid arthritis disease activity. Arthritis Care Res. 2012, 64, 1794–1803. [Google Scholar] [CrossRef] [PubMed]
- Meznerics, F.A.; Kemény, L.V.; Gunther, E.; Bakó, E.; Dembrovszky, F.; Szabó, B.; Ascsillán, A.; Lutz, E.; Csupor, D.; Hegyi, P.; et al. Multibiomarker disease activity score: An objective tool for monitoring rheumatoid arthritis? A systematic review and meta-analysis. Rheumatology 2023, 62, 2048–2059. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Valinezhad Orang, A.; Safaralizadeh, R.; Kazemzadeh-Bavili, M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int. J. Genom. 2014, 2014, 970607. [Google Scholar] [CrossRef]
- Bracken, C.P.; Scott, H.S.; Goodall, G.J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 2016, 17, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef]
- Arif, K.M.T.; Elliott, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory mechanisms of epigenetic miRNA relationships in human cancer and potential as therapeutic targets. Cancers 2020, 12, 2922. [Google Scholar] [CrossRef]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From mechanism to organism. Front. Cell Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef]
- Luo, B.; Zhou, K.; Liufu, Y.; Huang, X.; Zeng, H.; Zhang, Z. Novel insight into miRNA biology and its role in the pathogenesis of systemic lupus erythematosus. Front. Immunol. 2022, 13, 1059887. [Google Scholar] [CrossRef]
- Ying, S.Y.; Chang, D.C.; Lin, S.L. The microRNA (miRNA): Overview of the RNA genes that modulate gene function. Mol. Biotechnol. 2008, 38, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, Q.; Zhang, R.; Dai, X.; Chen, W.; Xing, D. Circulating microRNAs: Biomarkers of disease. Clin. Chim. Acta 2021, 516, 46–54. [Google Scholar] [CrossRef]
- Conti, I.; Varano, G.; Simioni, C.; Laface, I.; Milani, D.; Rimondi, E.; Neri, L.M. miRNAs as influencers of cell-cell communication in tumor microenvironment. Cells 2020, 9, 220. [Google Scholar] [CrossRef]
- Ortiz-Quintero, B. Extracellular microRNAs as intercellular mediators and noninvasive biomarkers of cancer. Cancers 2020, 12, 3455. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Quintero, B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016, 49, 281–303. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenetics 2018, 10, 59. [Google Scholar] [CrossRef]
- Gayosso-Gómez, L.V.; Ortiz-Quintero, B. Circulating microRNAs in blood and other body fluids as biomarkers for diagnosis, prognosis, and therapy response in lung cancer. Diagnostics 2021, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Liao, J.; Guarnera, M.A.; Fang, H.; Cai, L.; Stass, S.A.; Jiang, F. Analysis of microRNAs in sputum to improve computed tomography for lung cancer diagnosis. J. Thorac. Oncol. 2014, 9, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Roman-Canal, B.; Moiola, C.P.; Gatius, S.; Bonnin, S.; Ruiz-Miró, M.; González, E.; Ojanguren, A.; Recuero, J.L.; Gil-Moreno, A.; Falcón-Pérez, J.M.; et al. EV-associated miRNAs from pleural lavage as potential diagnostic biomarkers in lung cancer. Sci. Rep. 2019, 9, 15057. [Google Scholar] [CrossRef] [PubMed]
- Lan, H.; Lu, H.; Wang, X.; Jin, H. MicroRNAs as potential biomarkers in cancer: Opportunities and challenges. Biomed. Res. Int. 2015, 2015, 125094. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Zhang, Q.; Chen, H.; He, P.; Li, Y.; Si, M.; Jiao, X. Circulating microRNAs as a biomarker to predict therapy efficacy in hepatitis C patients with different genotypes. Microb. Pathog. 2017, 112, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Solé, C.; Moliné, T.; Vidal, M.; Ordi-Ros, J.; Cortés-Hernández, J. An exosomal urinary miRNA signature for early diagnosis of renal fibrosis in lupus nephritis. Cells 2019, 8, 773. [Google Scholar] [CrossRef]
- Allegra, A.; Alonci, A.; Campo, S.; Penna, G.; Petrungaro, A.; Gerace, D.; Musolino, C. Circulating microRNAs: New biomarkers in diagnosis, prognosis and treatment of cancer. Int. J. Oncol. 2012, 41, 1897–1912. [Google Scholar] [CrossRef]
- Zhou, X.; Wen, W.; Zhu, J.; Huang, Z.; Zhang, L.; Zhang, H.; Qi, L.W.; Shan, X.; Wang, T.; Cheng, W.; et al. A six-microRNA signature in plasma was identified as a potential biomarker in diagnosis of esophageal squamous cell carcinoma. Oncotarget 2017, 8, 34468–34480. [Google Scholar] [CrossRef]
- Kanaoka, R.; Iinuma, H.; Dejima, H.; Sakai, T.; Uehara, H.; Matsutani, N.; Kawamura, M. Usefulness of plasma eExosomal microRNA-451a as a noninvasive biomarker for early prediction of recurrence and prognosis of non-small cell lung cancer. Oncology 2018, 94, 311–323. [Google Scholar] [CrossRef]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol. Ther. Nucleic Acids 2020, 20, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Moshiri, F.; Salvi, A.; Gramantieri, L.; Sangiovanni, A.; Guerriero, P.; De Petro, G.; Bassi, C.; Lupini, L.; Sattari, A.; Cheung, D.; et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget 2018, 9, 15350–15364. [Google Scholar] [CrossRef] [PubMed]
- Suehiro, T.; Miyaaki, H.; Kanda, Y.; Shibata, H.; Honda, T.; Ozawa, E.; Miuma, S.; Taura, N.; Nakao, K. Serum exosomal microRNA-122 and microRNA-21 as predictive biomarkers in transarterial chemoembolization-treated hepatocellular carcinoma patients. Oncol. Lett. 2018, 16, 3267–3273. [Google Scholar] [CrossRef] [PubMed]
- Hortmann, M.; Walter, J.E.; Benning, L.; Follo, M.; Mayr, R.M.; Honegger, U.; Robinson, S.; Stallmann, D.; Duerschmied, D.; Twerenbold, R.; et al. Droplet digital PCR of serum miR-499, miR-21 and miR-208a for the detection of functionally relevant coronary artery disease. Int. J. Cardiol. 2019, 275, 129–135. [Google Scholar] [CrossRef]
- Li, Z.; Wu, J.; Wei, W.; Cai, X.; Yan, J.; Song, J.; Wang, C.; Wang, J. Association of serum miR-186-5p with the prognosis of acute coronary syndrome patients after percutaneous coronary intervention. Front. Physiol. 2019, 10, 686. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Zhao, X.; Liu, Y.Z.; Zeng, Q.T.; Mao, X.B.; Li, S.N.; Zhang, M.; Jiang, C.; Zhou, Y.; Qian, C.; et al. Circulating miR-19b-3p, miR-134-5p and miR-186-5p are promising novel biomarkers for early diagnosis of acute myocardial infarction. Cell Physiol. Biochem. 2016, 38, 1015–1029. [Google Scholar] [CrossRef] [PubMed]
- Velle-Forbord, T.; Eidlaug, M.; Debik, J.; Sæther, J.C.; Follestad, T.; Nauman, J.; Gigante, B.; Røsjø, H.; Omland, T.; Langaas, M.; et al. Circulating microRNAs as predictive biomarkers of myocardial infarction: Evidence from the HUNT study. Atherosclerosis 2019, 289, 1–7. [Google Scholar] [CrossRef]
- Dong, H.; Li, J.; Huang, L.; Chen, X.; Li, D.; Wang, T.; Hu, C.; Xu, J.; Zhang, C.; Zen, K.; et al. Serum microRNA profiles serve as novel biomarkers for the diagnosis of Alzheimer’s disease. Dis. Markers 2015, 2015, 625659. [Google Scholar] [CrossRef]
- Ding, H.; Huang, Z.; Chen, M.; Wang, C.; Chen, X.; Chen, J.; Zhang, J. Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Park. Relat. Disord. 2016, 22, 68–73. [Google Scholar] [CrossRef]
- Liu, X.; Ni, S.; Li, C.; Xu, N.; Chen, W.; Wu, M.; van Wijnen, A.J.; Wang, Y. Circulating microRNA-23b as a new biomarker for rheumatoid arthritis. Gene 2019, 712, 143911. [Google Scholar] [CrossRef]
- Zeng, L.; Wu, J.L.; Liu, L.M.; Jiang, J.Q.; Wu, H.J.; Zhao, M.; Lu, Q.J. Serum miRNA-371b-5p and miRNA-5100 act as biomarkers for systemic lupus erythematosus. Clin. Immunol. 2018, 196, 103–109. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Si, H.; Li, X.; Ding, X.; Sheng, Q.; Chen, P.; Zhang, H. Serum miR-23a, a potential biomarker for diagnosis of pre-diabetes and type 2 diabetes. Acta Diabetol. 2014, 51, 823–831. [Google Scholar] [CrossRef]
- Gahlawat, A.W.; Witte, T.; Sinn, P.; Schott, S. Circulating cf-miRNA as a more appropriate surrogate liquid biopsy marker than cfDNA for ovarian cancer. Sci. Rep. 2023, 13, 5503. [Google Scholar] [CrossRef] [PubMed]
- Lewandowski, P.; Goławski, M.; Baron, M.; Reichman-Warmusz, E.; Wojnicz, R. A systematic review of miRNA and cfDNA as potential biomarkers for liquid biopsy in myocarditis and inflammatory dilated cardiomyopathy. Biomolecules 2022, 12, 1476. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, A.P.; Ohlsson, R.; Henikoff, S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006, 7, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef]
- Galvão-Lima, L.J.; Morais, A.H.F.; Valentim, R.A.M.; Barreto, E.J.S.S. miRNAs as biomarkers for early cancer detection and their application in the development of new diagnostic tools. Biomed. Eng. Online 2021, 20, 21. [Google Scholar] [CrossRef] [PubMed]
- Chiam, K.; Wang, T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating serum exosomal miRNAs as Potential biomarkers for esophageal adenocarcinoma. J. Gastrointest. Surg. 2015, 19, 1208–1215. [Google Scholar] [CrossRef]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Kato, K.; et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019, 110, 408–419. [Google Scholar] [CrossRef]
- Felekkis, K.; Papaneophytou, C. Challenges in using circulating micro-RNAs as biomarkers for cardiovascular diseases. Int. J. Mol. Sci. 2020, 21, 561. [Google Scholar] [CrossRef]
- Xu, J.; Wu, C.; Che, X.; Wang, L.; Yu, D.; Zhang, T.; Huang, L.; Li, H.; Tan, W.; Wang, C.; et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol. Carcinog. 2011, 50, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Boutz, D.R.; Collins, P.J.; Suresh, U.; Lu, M.; Ramírez, C.M.; Fernández-Hernando, C.; Huang, Y.; Abreu Rde, S.; Le, S.Y.; Shapiro, B.A.; et al. Two-tiered approach identifies a network of cancer and liver disease-related genes regulated by miR-122. J. Biol. Chem. 2011, 286, 18066–18078. [Google Scholar] [CrossRef]
- Felekkis, K.; Pieri, M.; Papaneophytou, C. Variability in the levels of exosomal miRNAs among human subjects could be explained by differential interactions of exosomes with the endothelium. IUBMB Life 2021, 73, 1400–1405. [Google Scholar] [CrossRef] [PubMed]
- Chorley, B.N.; Atabakhsh, E.; Doran, G.; Gautier, J.C.; Ellinger-Ziegelbauer, H.; Jackson, D.; Sharapova, T.; Yuen, P.S.T.; Church, R.J.; Couttet, P.; et al. Methodological considerations for measuring biofluid-based microRNA biomarkers. Crit. Rev. Toxicol. 2021, 51, 264–282. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Pieri, M.; Papaneophytou, C. Exploring the feasibility of circulating miRNAs as diagnostic and prognostic biomarkers in osteoarthritis: Challenges and opportunities. Int. J. Mol. Sci. 2023, 24, 13144. [Google Scholar] [CrossRef]
- Bottani, M.; Banfi, G.; Lombardi, G. The clinical potential of circulating miRNAs as biomarkers: Present and future applications for diagnosis and prognosis of age-associated bone diseases. Biomolecules 2020, 10, 589. [Google Scholar] [CrossRef]
- Wang, K.; Yuan, Y.; Cho, J.-H.; McClarty, S.; Baxter, D.; Galas, D.J. Comparing the microRNA spectrum between serum and plasma. PLoS ONE 2012, 7, e41561. [Google Scholar] [CrossRef]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as biomarkers and therapeutic targets for cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef]
- Ricci, C.; Salvemini, A.; Dalmiglio, C.; Castagna, M.G.; Cantara, S. From circulating tumor eclls to mirna: New challenges in the diagnosis and prognosis of medullary thyroid cancer. Cancers 2023, 15, 4009. [Google Scholar] [CrossRef]
- Bortolini Silveira, A.; Bidard, F.C.; Tanguy, M.L.; Girard, E.; Trédan, O.; Dubot, C.; Jacot, W.; Goncalves, A.; Debled, M.; Levy, C.; et al. Multimodal liquid biopsy for early monitoring and outcome prediction of chemotherapy in metastatic breast cancer. NPJ Breast Cancer 2021, 7, 115. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.E.; Vuppalapaty, M.; Wilkerson, C.; Renier, C.; Chiu, M.; Lemaire, C.; Che, J.; Matsumoto, M.; Carroll, J.; Crouse, S.; et al. Detection of EGFR mutations in cfDNA and CTCs, and comparison to tumor tissue in non-small-cell-lung-cancer (NSCLC) patients. Front. Oncol. 2020, 10, 572895. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Loginov, V.I.; Pronina, I.V.; Burdennyy, A.M.; Filippova, E.A.; Kazubskaya, T.P.; Kushlinsky, D.N.; Utkin, D.O.; Khodyrev, D.S.; Kushlinskii, N.E.; Dmitriev, A.A.; et al. Novel miRNA genes deregulated by aberrant methylation in ovarian carcinoma are involved in metastasis. Gene 2018, 662, 28–36. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, Y.; Mouliere, F. Toward the early detection of cancer by decoding the epigenetic and environmental fingerprints of cell-free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef]
- Ward Gahlawat, A.; Lenhardt, J.; Witte, T.; Keitel, D.; Kaufhold, A.; Maass, K.K.; Pajtler, K.W.; Sohn, C.; Schott, S. Evaluation of storage tubes for combined analysis of circulating nucleic acids in liquid biopsies. Int. J. Mol. Sci. 2019, 20, 704. [Google Scholar] [CrossRef]
- Otandault, A.; Anker, P.; Al Amir Dache, Z.; Guillaumon, V.; Meddeb, R.; Pastor, B.; Pisareva, E.; Sanchez, C.; Tanos, R.; Tousch, G.; et al. Recent advances in circulating nucleic acids in oncology. Ann. Oncol. 2019, 30, 374–384. [Google Scholar] [CrossRef]
- Albitar, M.; Zhang, H.; Charifa, A.; Ip, A.; Ma, W.; McCloskey, J.; Donato, M.; Siegel, D.; Waintraub, S.; Gutierrez, M.; et al. Combining cell-free RNA with cell-free DNA in liquid biopsy for hematologic and solid tumors. Heliyon 2023, 9, e16261. [Google Scholar] [CrossRef]
- Ibrahim, M.R.K.; Waly, N.G.F.M.; Moness, H.; Ahmed, S.S.; Ibrahem, R. Serum miRNA-21, miRNA-146a and plasma cell free DNA as novel biomarkers for assessing systemic lupus erythematosus activity. Mol. Biol. Rep. 2023, 50, 10025–10036. [Google Scholar] [CrossRef]
- Peng, C.; Wang, J.; Gao, W.; Huang, L.; Liu, Y.; Li, X.; Li, Z.; Yu, X. Meta-analysis of the diagnostic performance of circulating microRNAs for pancreatic cancer. Int. J. Health Sci. 2021, 18, 660–671. [Google Scholar] [CrossRef]
- Ahmed, S.I.; Sarwar, S.; Verma, H.; Verma, R.; Lakkireddy, C.; Adil, M.A.M.; Khan, A.A. Implication of circulating microRNA-21 as a potential non-invasive diagnostic predictor of prostate cancer patients. Hum. Gene 2022, 34, 201081. [Google Scholar] [CrossRef]
- Souza, M.F.d.; Kuasne, H.; Barros-Filho, M.d.C.; Cilião, H.L.; Marchi, F.A.; Fuganti, P.E.; Paschoal, A.R.; Rogatto, S.R.; Cólus, I.M.d.S. Circulating mRNAs and miRNAs as candidate markers for the diagnosis and prognosis of prostate cancer. PLoS ONE 2017, 12, e0184094. [Google Scholar] [CrossRef]
- Cirillo, P.D.R.; Margiotti, K.; Fabiani, M.; Barros-Filho, M.C.; Sparacino, D.; Cima, A.; Longo, S.A.; Cupellaro, M.; Mesoraca, A.; Giorlandino, C. Multi-analytical test based on serum miRNAs and proteins quantification for ovarian cancer early detection. PLoS ONE 2021, 16, e0255804. [Google Scholar] [CrossRef]
- Radwan, E.; Shaltout, A.S.; Mansor, S.G.; Shafik, E.A.; Abbas, W.A.; Shehata, M.R.; Ali, M. Evaluation of circulating microRNAs-211 and 25 as diagnostic biomarkers of colorectal cancer. Mol. Biol. Rep. 2021, 48, 4601–4610. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Barwari, T.; Joshi, A.; Theofilatos, K.; Zampetaki, A.; Barallobre-Barreiro, J.; Singh, B.; Sorensen, N.A.; Neumann, J.T.; Zeller, T.; et al. Comparative analysis of circulating noncoding RNAs versus protein biomarkers in the detection of myocardial injury. Circ. Res. 2019, 125, 328–340. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Han, R.; Su, J.; Chen, H.; Li, D. Multi-marker diagnosis method for early Hepatocellular Carcinoma based on surface plasmon resonance. Clin. Chim. Acta 2020, 502, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Tomeva, E.; Switzeny, O.J.; Heitzinger, C.; Hippe, B.; Haslberger, A.G. Comprehensive approach to distinguish patients with solid tumors from healthy controls by combining androgen receptor mutation p.H875Y with cell-free DNA methylation and circulating miRNAs. Cancers 2022, 14, 462. [Google Scholar] [CrossRef]
- Chan, A.K.; Lockhart, D.C.; von Bernstorff, W.; Spanjaard, R.A.; Joo, H.G.; Eberlein, T.J.; Goedegebuure, P.S. Soluble MUC1 secreted by human epithelial cancer cells mediates immune suppression by blocking T-cell activation. Int. J. Cancer 1999, 82, 721–726. [Google Scholar] [CrossRef]
- Kirmiz, C.; Li, B.; An, H.J.; Clowers, B.H.; Chew, H.K.; Lam, K.S.; Ferrige, A.; Alecio, R.; Borowsky, A.D.; Sulaimon, S.; et al. A serum glycomics approach to breast cancer biomarkers. Mol. Cell Proteom. 2007, 6, 43–55. [Google Scholar] [CrossRef]
- Gendler, S.J.; Spicer, A.P.; Lalani, E.N.; Duhig, T.; Peat, N.; Burchell, J.; Pemberton, L.; Boshell, M.; Taylor-Papadimitriou, J. Structure and biology of a carcinoma-associated mucin, MUC1. Am. Rev. Respir. Dis. 1991, 144, S42–S47. [Google Scholar] [CrossRef] [PubMed]
- Stout, R.L.; Fulks, M.; Dolan, V.F.; Magee, M.E.; Suarez, L. Increased mortality associated with elevated carcinoembryonic antigen in insurance applicants. J. Insur. Med. 2007, 39, 251–258. [Google Scholar]
- Anoop, T.M.; Joseph, P.R.; Soman, S.; Chacko, S.; Mathew, M. Significance of serum carcinoembryonic antigen in metastatic breast cancer patients: A prospective study. World J. Clin. Oncol. 2022, 13, 529–539. [Google Scholar] [CrossRef]
- Bast, R.C., Jr. Status of tumor markers in ovarian cancer screening. J. Clin. Oncol. 2003, 21, 200s–205s. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, Y.; Xu, F.; Berchuck, A.; van Haaften-Day, C.; Havrilesky, L.J.; de Bruijn, H.W.; van der Zee, A.G.; Woolas, R.P.; Jacobs, I.J.; et al. Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol. Oncol. 2007, 107, 526–531. [Google Scholar] [CrossRef]
- Bast, R.C., Jr.; Xu, F.J.; Yu, Y.H.; Barnhill, S.; Zhang, Z.; Mills, G.B. CA 125: The past and the future. Int. J. Biol. Markers 1998, 13, 179–187. [Google Scholar] [CrossRef]
- Mazouni, C.; Baggerly, K.; Hawke, D.; Tsavachidis, S.; André, F.; Buzdar, A.U.; Martin, P.M.; Kobayashi, R.; Pusztai, L. Evaluation of changes in serum protein profiles during neoadjuvant chemotherapy in HER2-positive breast cancer using an LC-MALDI-TOF/MS procedure. Proteomics 2010, 10, 3525–3532. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Tang, W.; Xie, Y.; Wang, S.; Chen, Y.; Qi, J.; Qiao, Y.; Ma, J. New combined microRNA and protein plasmatic biomarker panel for pancreatic cancer. Oncotarget 2016, 7, 80033–80045. [Google Scholar] [CrossRef]
- Cai, Z.; Poulos, R.C.; Liu, J.; Zhong, Q. Machine learning for multi-omics data integration in cancer. iScience 2022, 25, 103798. [Google Scholar] [CrossRef] [PubMed]
- Adhit, K.K.; Wanjari, A.; Menon, S.; Siddhaarth, K. Liquid biopsy: An evolving paradigm for non-invasive disease diagnosis and monitoring in medicine. Cureus 2023, 15, e50176. [Google Scholar] [CrossRef]
- Jung, K.; Fleischhacker, M.; Rabien, A. Cell-free DNA in the blood as a solid tumor biomarker—A critical appraisal of the literature. Clin. Chim. Acta 2010, 411, 1611–1624. [Google Scholar] [CrossRef] [PubMed]
- Batool, S.M.; Yekula, A.; Khanna, P.; Hsia, T.; Gamblin, A.S.; Ekanayake, E.; Escobedo, A.K.; You, D.G.; Castro, C.M.; Im, H.; et al. The Liquid Biopsy Consortium: Challenges and opportunities for early cancer detection and monitoring. Cell Rep. Med. 2023, 4, 101198. [Google Scholar] [CrossRef] [PubMed]
- Armakolas, A.; Kotsari, M.; Koskinas, J. Liquid biopsies, novel approaches and future directions. Cancers 2023, 15, 1579. [Google Scholar] [CrossRef] [PubMed]
- Sandau, U.S.; Wiedrick, J.T.; McFarland, T.J.; Galasko, D.R.; Fanning, Z.; Quinn, J.F.; Saugstad, J.A. Analysis of the longitudinal stability of human plasma miRNAs and implications for disease biomarkers. Sci. Rep. 2024, 14, 2148. [Google Scholar] [CrossRef] [PubMed]
- Kappel, A.; Keller, A. miRNA assays in the clinical laboratory: Workflow, detection technologies and automation aspects. Clin. Chem. Lab. Med. 2017, 55, 636–647. [Google Scholar] [CrossRef]
- Ginghina, O.; Hudita, A.; Zamfir, M.; Spanu, A.; Mardare, M.; Bondoc, I.; Buburuzan, L.; Georgescu, S.E.; Costache, M.; Negrei, C.; et al. Liquid biopsy and artificial intelligence as tools to detect signatures of colorectal malignancies: A modern approach in patient’s stratification. Front. Oncol. 2022, 12, 856575. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Petinrin, O.O.; Zhang, W.; Rahaman, S.; Tang, Z.-R.; Wong, K.-C. Machine learning protocols in early cancer detection based on liquid biopsy: A survey. Life 2021, 11, 638. [Google Scholar] [CrossRef]
- Choi, G.H.; Yun, J.; Choi, J.; Lee, D.; Shim, J.H.; Lee, H.C.; Chung, Y.H.; Lee, Y.S.; Park, B.; Kim, N.; et al. Development of machine learning-based clinical decision support system for hepatocellular carcinoma. Sci. Rep. 2020, 10, 14855. [Google Scholar] [CrossRef]


| Advantages | Disadvantages |
|---|---|
| Non-invasive: cfDNAs can be obtained from blood samples and other biological fluids. | Sensitivity: The sensitivity of cfDNA to detect certain mutations or low-abundance genetic alterations can be lower compared to tissue biopsies. |
| Applicability: cfDNAs can be used as prognostic, diagnostic biomarkers and for monitoring disease progression. | Heterogeneity: The presence of cfDNA from other sources can complicate the interpretation of results. |
| Specificity: They can be used for various diseases such as cancer, CVDs 2, and autoimmune disorders. | Standardization: The lack of standardized methods for cfDNA isolation, quantification, and analysis can lead to variability in results. |
| Rapid turnover: cfDNAs have a short half-life in biofluids, enabling timely reflection of the current disease state [78]. | Quantitative limitations: The absolute quantity of cfDNA can be low, especially in the early stages of the disease, which may hinder detection and analysis. |
| Comprehensive detection: cfDNA analysis can detect several genetic alterations, offering a comprehensive view of the genetic landscape of diseases. | Cost and accessibility: The costs associated with cfDNA analysis and the need for specialized equipment is a drawback. |
| Advantages | Disadvantages |
|---|---|
| Improved Diagnosis: Multiple protein markers can significantly enhance the prediction accuracy of diagnoses. | Low Concentration: Often, disease-related protein markers are present at concentrations too low for detection by current proteomics techniques. |
| Discovery of Disease Signatures: Specific proteins associated with metabolic diseases, offering insights into the molecular underpinnings of these conditions. | Cost and Time: Discovering new protein biomarkers is both time-consuming and expensive, hindered by the complex structure of proteins and the difficulty in finding accurate detection methods. |
| Early Detection: Detection of disease-specific proteins is essential for early-stage diagnosis, where treatment can be more effective. | Methodological Limitations: Traditional proteomics methodologies struggle to detect low-abundance proteins due to the masking effects of high-abundance species. |
| Deregulated c-miR | Blood Fraction | Disease | Function | Ref. |
|---|---|---|---|---|
| miR-429, miR-205, miR-200b, miR-203, miR-125b, miR-34b | Plasma | Lung Cancer | Diagnostic | [130] |
| miR-451a, miR-21 | Plasma | Lung cancer | Prognostic | [131] [132] |
| miR-106b-3p, miR-101-3p, miR-1246 | Plasma | Liver cancer | Diagnostic | [133] |
| miR-122, miR-21 | Serum | Liver cancer | Prognostic | [134] |
| miR-499, miR-21, miR-208a | Serum |
Coronary artery disease | Diagnostic | [135] |
| miR-186-5p | Serum |
Coronary artery disease | Prognostic | [136] |
| miR-19b-3p, miR-134-5p, miR-186-5p | Plasma |
Acute myocardial infarction | Diagnostic | [137] |
| miR-21-5p, miR-26a-5p, miR-29c-3p, miR-144-3p, miR-151a-5p | Serum | Myocardial infarction | Prognostic | [138] |
| miR-31, miR-93, miR-143, miR-146a | Serum | Alzheimer’s disease | Diagnostic | [139] |
| miR-195, miR-185, miR-15b, miR-221, miR-181a | Serum | Parkinson’s disease | Diagnostic | [140] |
| miR-23b | Plasma | Rheumatoid arthritis | Monitoring disease progression | [141] |
| miR-371b-5p, miR-5100 | Serum |
Systemic Lupus Erythematosus | Diagnostic | [142] |
| miR-23a | Serum | Diabetes | Diagnostic | [143] |
| Biomarker Panel | Disease | Outcome | Ref. |
|---|---|---|---|
|
Serum miR-21 and miR-146a, with plasma cfDNA | Systemic Lupus Erythematosus (SLE) Activity | Significant correlations with SLE activity indicators | [169] |
|
Serum miR-96 and miR-200, with CA19-9 2 | Pancreatic Cancer Diagnosis | Enhanced diagnostic accuracy | [170] |
| Serum miR-21 with serum PSA 3 | Prostate Cancer |
Sensitivity: 71.05%, Specificity: 77.35% | [171] |
|
Plasma miR-200b and miR-200c with plasma mRNAs (SIM2 and OR51E2) | Prostate Cancer | Differential expression between normal and tumor samples | [172] |
|
Serum miR-320b and miR-141-3p, with serum CA-125 4 and HE4 5 | Ovarian Cancer Diagnosis | High specificity and sensitivity in differentiation | [173] |
|
Plasma miR-211 and miR-25, with plasma TGF-β1 6 | Colorectal Cancer Diagnosis | Correlation with lymph node metastasis, diagnostic utility | [174] |
|
Serum or Plasma: Muscle-enriched miR-1 and miR-133a with cMyBP-C 8 and cardiac troponins | Acute Myocardial Infarction | Enhanced diagnostic accuracy, highest AUC 7 values | [175] |
| Serum AFP 9 with serum miR-125b | Early Hepatocellular Carcinoma | Low detection limit for both markers, enhanced sensitivity and specificity | [176] |
|
Plasma: cfDNA mutations: COSM10758, COSM18561 cfDNA methylation markers: MLH1, MDR1, GATA5, SFN miR-17-5p, -20a-5p, -21-5p, -26a-5p, -27a-3p, -29c-3p, -92a-3p, -101-3p, -133a-3p, 148b-3p, -155-5p195-5p |
Various cancer types: Bladder, brain, breast, colorectal, lung, ovarian, pancreas, prostate, stomach |
Simultaneous detection of several cancer types: Specificity: 80% Sensitivity: 97.7% Accuracy 95.4% | [177] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felekkis, K.; Papaneophytou, C. The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins. Int. J. Mol. Sci. 2024, 25, 3403. https://doi.org/10.3390/ijms25063403
Felekkis K, Papaneophytou C. The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins. International Journal of Molecular Sciences. 2024; 25(6):3403. https://doi.org/10.3390/ijms25063403
Chicago/Turabian StyleFelekkis, Kyriacos, and Christos Papaneophytou. 2024. "The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins" International Journal of Molecular Sciences 25, no. 6: 3403. https://doi.org/10.3390/ijms25063403
APA StyleFelekkis, K., & Papaneophytou, C. (2024). The Circulating Biomarkers League: Combining miRNAs with Cell-Free DNAs and Proteins. International Journal of Molecular Sciences, 25(6), 3403. https://doi.org/10.3390/ijms25063403







