Mapping of Prion Structures in the Yeast Rnq1
Abstract
1. Introduction
2. Results
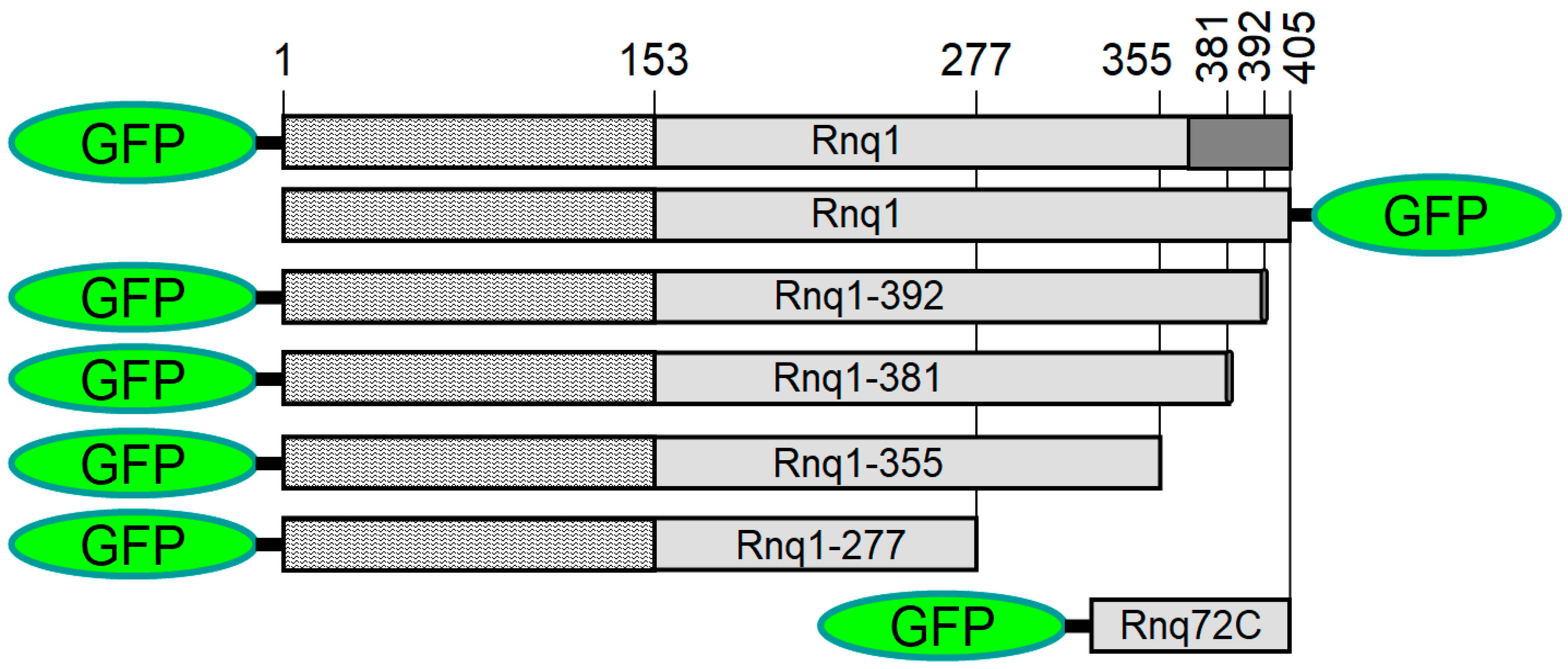
2.1. Rnq1 Constructs Used
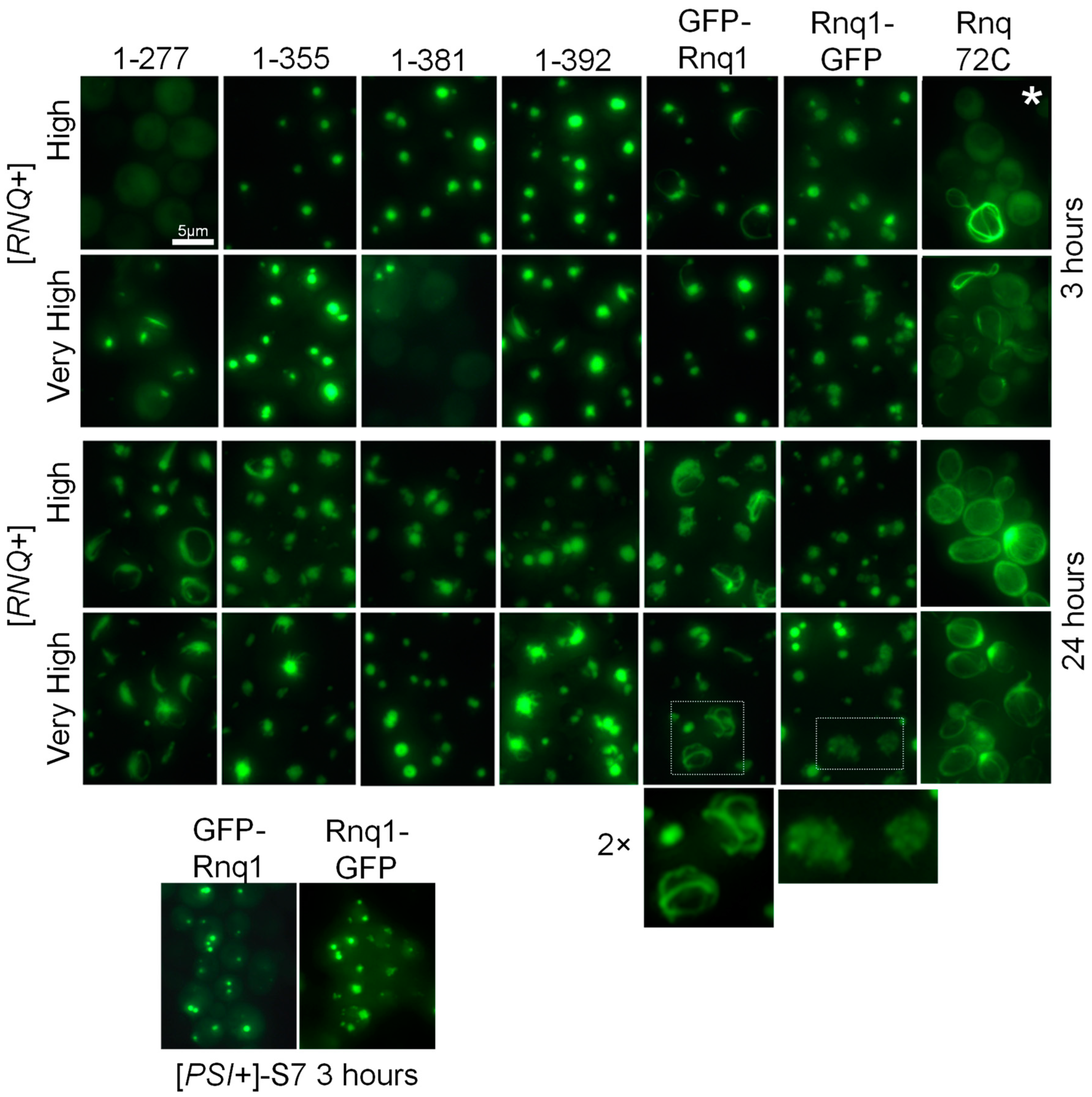
2.2. Aggregation Patterns of GFP-Labeled Rnq1 Proteins
2.3. Chromosomal Alterations of RNQ1
2.4. Structural Maps of the Rnq1 Protein Constructs
2.4.1. Structures of Full-Sized Rnq1 and Rnq1-GFP

2.4.2. Structures Observed in C-Terminally Deleted Rnq1 Variants
3. Discussion
3.1. On the Protease Mapping Procedure
3.2. Rnq1 Prion Structures
3.3. General Considerations
3.4. Problems to Be Solved
3.5. Concluding Remarks
- The actual and potential prion structures of Rnq1 were mapped, and their location better correlates with high QN content than with predictions of some computer algorithms.
- Prion structures significantly rearrange when transferring from native Rnq1 to its truncated versions or to Rnq1-GFP. This implies that in the cases when such a transfer is successful, one most likely obtains a different prion fold.
- The reluctance of GFP-Rnq72C to aggregate was unexpected, since the “mirror” Sup35 protein Sup35(1-61)-GFP readily decorates different Sup35 prion variants. Thus, some important details of how prion co-aggregation occurs are not well understood and require further investigation.
- The Rnq1 prion appears to differ from Swi1 and Sup35 in that the key Rnq1 prion structure is not restricted to a single terminal prion core.
- While the Rnq1 prion readily induces the prion state of Sup35, the opposite is not true: the Sup35 prion seeds Rnq1 very inefficiently, if at all.
- Prion transition from Rnq1 to Rnq1-GFP does not occur when the Rnq1 protein is changed rapidly through chromosomal replacement of the RNQ1 gene but occurs when the change proceeds during many cellular generations through plasmid shuffle. The C-terminal Rnq1 prion core disappears in Rnq1-GFP overproduced in both studied [RNQ+] prion variants. The microscopic aggregation patterns of Rnq1-GFP differ significantly from that of GFP-Rnq1. Thus, Rnq1-GFP does not appear to be an adequate instrument for studying Rnq1 prion properties. In general, GFP or other tags should not be fused to the same side of a protein as its PFD.
4. Materials and Methods
4.1. Yeast Strains and Media
4.2. Plasmids and DNA Manipulations
4.3. Genetic Manipulations
4.4. Microscopy
4.5. Determination of the Rnq1 Prion State
4.6. Isolation of Prion Aggregates
4.7. PK Digestion, Mass Spectrometry, and Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Galkin, A.P. Functional amyloids of eukaryotes: Criteria, classification, and biological significance. Curr. Genet. 2020, 66, 849–866. [Google Scholar] [CrossRef]
- Roberts, B.T.; Wickner, R.B. Heritable activity: A prion that propagates by covalent autoactivation. Genes Dev. 2003, 17, 2083–2087. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.C.S.; Lindquist, S. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 2009, 23, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Chakrabortee, S.; Byers, J.S.; Jones, S.; Garcia, D.M.; Bhullar, B.; Chang, A.; She, R.; Lee, L.; Fremin, B.; Lindquist, S.; et al. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell 2016, 167, 369–381.e12. [Google Scholar] [CrossRef]
- Zhouravleva, G.A.; Bondarev, S.A.; Trubitsina, N.P. How Big Is the Yeast Prion Universe? Int. J. Mol. Sci. 2023, 24, 11651. [Google Scholar] [CrossRef]
- Halfmann, R.; Wright, J.R.; Alberti, S.; Lindquist, S.; Rexach, M. Prion formation by a yeast GLFG nucleoporin. Prion 2012, 6, 391–399. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: The story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, G.; Shimazu, N.; Tanaka, M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 2012, 336, 355–359. [Google Scholar] [CrossRef]
- Goncharoff, D.K.; Cabral, R.; Applebey, S.V.; Pagadala, M.; Du, Z.; Li, L. Defining Key Residues of the Swi1 Prion Domain in Prion Formation and Maintenance. Mol. Cell. Biol. 2021, 41, e0004421. [Google Scholar] [CrossRef]
- Dergalev, A.A.; Alexandrov, A.I.; Ivannikov, R.I.; Ter-Avanesyan, M.D.; Kushnirov, V. V Yeast Sup35 Prion Structure: Two Types, Four Parts, Many Variants. Int. J. Mol. Sci. 2019, 20, 2633. [Google Scholar] [CrossRef]
- Chang, H.-Y.; Lin, J.-Y.; Lee, H.-C.; Wang, H.-L.; King, C.-Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc. Natl. Acad. Sci. USA 2008, 105, 13345–13350. [Google Scholar] [CrossRef] [PubMed]
- Depace, A.H.; Santoso, A.; Hillner, P.; Weissman, J.S. A Critical Role for Amino-Terminal Glutamine/Asparagine Repeats in the Formation and Propagation of a Yeast Prion. Cell 1998, 93, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Vitrenko, Y.A.; Pavon, M.E.; Stone, S.I.; Liebman, S.W. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr. Genet. 2007, 51, 309–319. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kadnar, M.L.; Articov, G.; Derkatch, I.L. Distinct Type of Transmission Barrier Revealed by Study of Multiple Prion Determinants of Rnq1. PLOS Genet. 2010, 6, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Dagkesamanskaia, A.R.; Kushnirov, V.V.; Paushkin, S.V.; Ter-Avanesian, M.D. Fusion of glutathione S-transferase with the N-terminus of yeast Sup35p protein inhibits its prion-like properties. Genetika 1997, 33, 610–615. [Google Scholar] [PubMed]
- Dergalev, A.A.; Urakov, V.N.; Agaphonov, M.O.; Alexandrov, A.I.; Kushnirov, V. V Dangerous Stops: Nonsense Mutations Can Dramatically Increase Frequency of Prion Conversion. Int. J. Mol. Sci. 2021, 22, 1542. [Google Scholar] [CrossRef]
- Huh, W.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [PSI+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef]
- Bradley, M.E.; Edskes, H.K.; Hong, J.Y.; Wickner, R.B.; Liebman, S.W. Interactions among prions and prion “strains” in yeast. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. 4), 16392–16399. [Google Scholar] [CrossRef]
- Zhou, P.; Derkatch, I.L.; Liebman, S.W. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)]. Mol. Microbiol. 2001, 39, 37–46. [Google Scholar] [CrossRef]
- Tyedmers, J.; Treusch, S.; Dong, J.; McCaffery, J.M.; Bevis, B.; Lindquist, S. Prion induction involves an ancient system for the sequestration of aggregated proteins and heritable changes in prion fragmentation. Proc. Natl. Acad. Sci. USA 2010, 107, 8633–8638. [Google Scholar] [CrossRef]
- Huang, Y.-W.; King, C.-Y. A complete catalog of wild-type Sup35 prion variants and their protein-only propagation. Curr. Genet. 2020, 66, 97–122. [Google Scholar] [CrossRef]
- Stein, K.C.; True, H.L. Extensive Diversity of Prion Strains Is Defined by Differential Chaperone Interactions and Distinct Amyloidogenic Regions. PLoS Genet. 2014, 10, e1004337. [Google Scholar] [CrossRef]
- Manogaran, A.L.; Fajardo, V.M.; Reid, R.J.D.; Rothstein, R.; Liebman, S.W. Most, but not all, yeast strains in the deletion library contain the [PIN(+)] prion. Yeast 2010, 27, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Salnikova, A.B.; Kryndushkin, D.S.; Smirnov, V.N.; Kushnirov, V.V.; Ter-Avanesyan, M.D. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J. Biol. Chem. 2005, 280, 8808–8812. [Google Scholar] [CrossRef] [PubMed]
- Alieva, M.K.; Nikishina, S.B.; Kireev, I.I.; Golyshev, S.A.; Tyurin-Kuzmin, P.A.; Ivanova, L.V.; Alexandrov, A.I.; Kushnirov, V.V.; Dergalev, A.A. A liquid-to-solid phase transition of biomolecular condensates drives in vivo formation of yeast amyloids and prions. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Ter-Avanesyan, M.D. Structure and replication of yeast prions. Cell 1998, 94, 13–16. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Shen, H.C.-H.; Komi, Y.; Sugiyama, S.; Kurinomaru, T.; Tomabechi, Y.; Krayukhina, E.; Okamoto, K.; Yokoyama, T.; Shirouzu, M.; et al. Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system. Nat. Chem. Biol. 2022, 18, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Khan, T.; Kandola, T.S.; Wu, J.; Venkatesan, S.; Ketter, E.; Lange, J.J.; Rodríguez Gama, A.; Box, A.; Unruh, J.R.; Cook, M.; et al. Quantifying Nucleation In Vivo Reveals the Physical Basis of Prion-like Phase Behavior. Mol. Cell 2018, 71, 155–168.e7. [Google Scholar] [CrossRef]
- Subedi, S.; Sasidharan, S.; Nag, N.; Saudagar, P.; Tripathi, T. Amyloid Cross-Seeding: Mechanism, Implication, and Inhibition. Molecules 2022, 27, 1776. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Lancaster, A.K.; Nutter-Upham, A.; Lindquist, S.; King, O.D. PLAAC: A web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics 2014, 30, 2501–2502. [Google Scholar] [CrossRef] [PubMed]
- Prilusky, J.; Felder, C.E.; Zeev-Ben-Mordehai, T.; Rydberg, E.H.; Man, O.; Beckmann, J.S.; Silman, I.; Sussman, J.L. FoldIndex: A simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics 2005, 21, 3435–3438. [Google Scholar] [CrossRef] [PubMed]
- Toombs, J.A.; McCarty, B.R.; Ross, E.D. Compositional Determinants of Prion Formation in Yeast. Mol. Cell. Biol. 2010, 30, 319–332. [Google Scholar] [CrossRef]
- Bailleul, P.A.; Newnam, G.P.; Steenbergen, J.N.; Chernoff, Y.O. Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics 1999, 153, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Shaw, W. Quick and Easy CRISPR Engineering in Saccharomyces Cerevisiae. Available online: https://benchling.com/pub/ellis-crispr-tools (accessed on 7 March 2024).
- Alexandrov, A.I.; Dergalev, A.A. Increasing throughput of manual microscopy of cell suspensions using solid medium pads. MethodsX 2019, 6, 329–332. [Google Scholar] [CrossRef] [PubMed]
- Kroschwald, S.; Maharana, S.; Simon, A. Hexanediol: A chemical probe to investigate the material properties of membrane-less compartments. Matters 2017, 3, e201702000010. [Google Scholar] [CrossRef]
- Burnette, W.N. “Western blotting”: Electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 1981, 112, 195–203. [Google Scholar] [CrossRef]

| Primer | Sequence | Description |
|---|---|---|
| sGFP-Rnq-Df | gatgaactgtacaaaggccaaagatctgaaATGgatacgga | sGFP to RNQ1 junction |
| Yes-Rnq-Rf | tacatgatgcggccctctagaatcatcgtTCAgtagcggt | RNQ1 to pYes2 junction |
| Yes-Rnq277x-Rf | catgatgcggccctctagaTTAgtaagattgagccatggag | RNQ1-277 to pYes2 junction |
| Yes-Rnq355x-Rf | catgatgcggccctctagaTTAatactcattagcctgttgctg | RNQ1-355 to pYes2 junction |
| Yes-Rnq381-Rf | catgatgcggccctctagaTCAgtagcgattggagttctggtttccgcc | RNQ1-381 to pYes2 junction |
| Yes-Rnq392-Rf | catgatgcggccctctagaTCAgtagcggttgccagaaaaattgaagga | RNQ1-392 to pYes2 junction |
| sGfp-Rnq72cDf | ctgtacaaaggccaaagatcctacctgggcaataactcc | sGFP to RNQ72C junction |
| Rnq1-CrC-D | gacttacggaagaccgcaatacgg | CRISPR spacer for pWS172 plasmid |
| Rnq1-CrC-R | aaacccgtattgcggtcttccgta | CRISPR spacer for pWS172 |
| Rnq1-CR2-D | gactactgaatcatcgttcagtag | CRISPR spacer for pWS172 |
| Rnq1-CR2-R | aaacctactgaacgatgattcagt | CRISPR spacer for pWS172 |
| Yes-Rnq1-Df | cggatcggactactagcagcaaagatctgaaATGgatacg | pYes2 to RNQ1 junction |
| Yes-GFP65-Rf | atgatgcggccctctagacgcgccctatttgtatagtt | pYes2 to GFP junction |
| Rnq1-Din | gggagccaaagtatgggtg | RNQ1 inside ORF |
| Rnq1-R1 | gggcatcctgcagagataca | RNQ1 terminator |
| YesXba-Rnq3′D | tctagagggccgcatcatgcgatgattcagttcgccttc | PCR of RNQ1 3′ region |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galliamov, A.A.; Malukhina, A.D.; Kushnirov, V.V. Mapping of Prion Structures in the Yeast Rnq1. Int. J. Mol. Sci. 2024, 25, 3397. https://doi.org/10.3390/ijms25063397
Galliamov AA, Malukhina AD, Kushnirov VV. Mapping of Prion Structures in the Yeast Rnq1. International Journal of Molecular Sciences. 2024; 25(6):3397. https://doi.org/10.3390/ijms25063397
Chicago/Turabian StyleGalliamov, Arthur A., Alena D. Malukhina, and Vitaly V. Kushnirov. 2024. "Mapping of Prion Structures in the Yeast Rnq1" International Journal of Molecular Sciences 25, no. 6: 3397. https://doi.org/10.3390/ijms25063397
APA StyleGalliamov, A. A., Malukhina, A. D., & Kushnirov, V. V. (2024). Mapping of Prion Structures in the Yeast Rnq1. International Journal of Molecular Sciences, 25(6), 3397. https://doi.org/10.3390/ijms25063397






