The Chronic Effects of a Single Low-Intensity Blast Exposure on Phosphoproteome Networks and Cognitive Function Influenced by Mutant Tau Overexpression
Abstract
1. Introduction
1.1. Long Term Effects of Blast-Induced Neurotrauma
1.2. Utility of Phosphoproteomics
1.3. Linking Tau Hyperphosphorylation with Complex Behavioral Phenotypes and Synaptic Pathology Following BINT
1.4. Knowledge Gaps in Understanding the Role of Tau in Risk of BINT
1.5. Unveiling the Relationship between ADRD Genetic Markers and LIB-Induced Behavioral Deficits
1.6. Identification of Phosphopeptide Co-Expression Networks Related to Cognitive Changes
2. Results
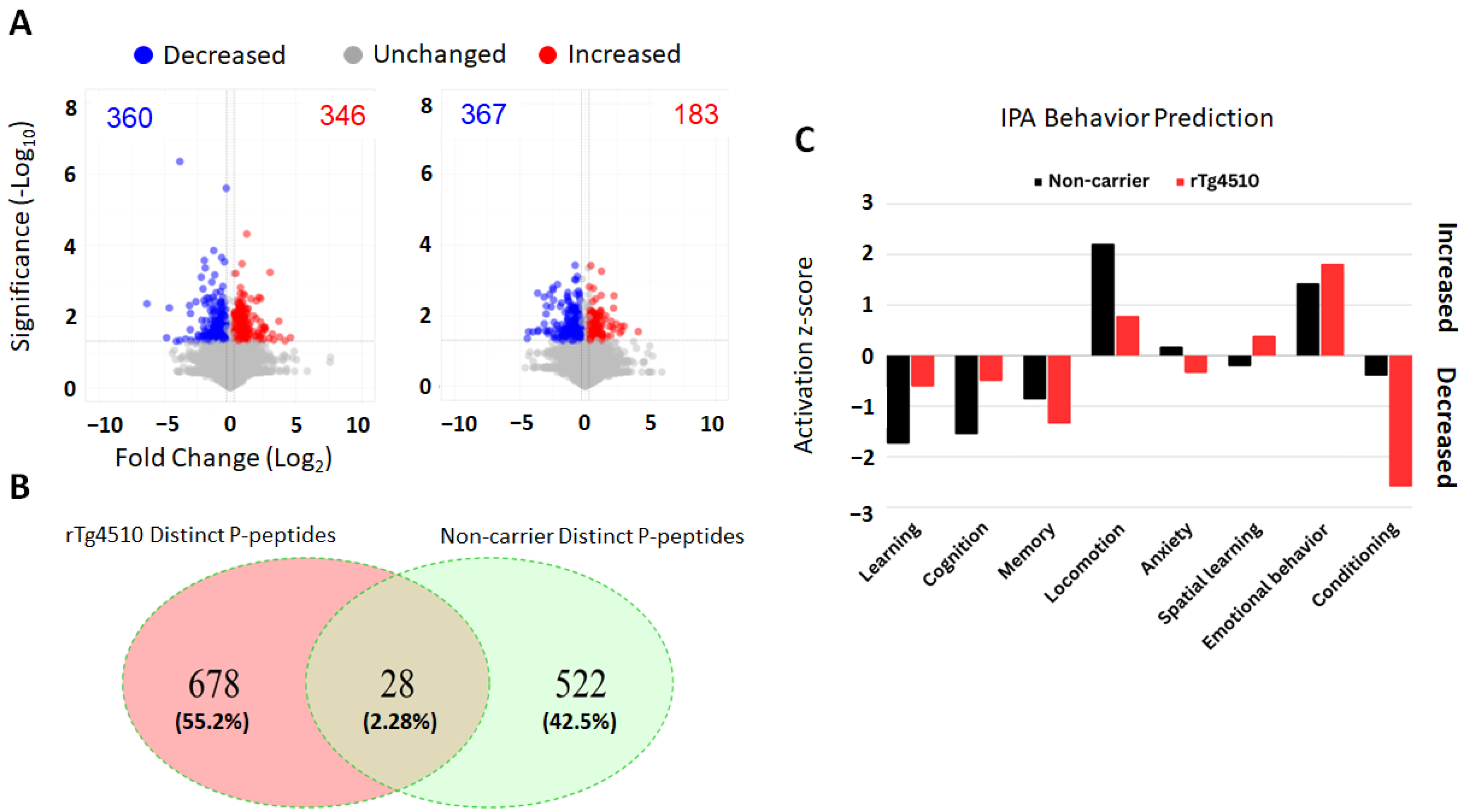
2.1. Quantitative Proteomics and Behavior Prediction by Ingenuity Pathway Analysis (IPA)
2.2. Results from Automated Home-Cage Monitoring System on the CognitionWall Experiment
2.3. Weighted Peptide Co-Expression Network Analysis (WpCNA): Network Construction and Topological Overlap Matrix
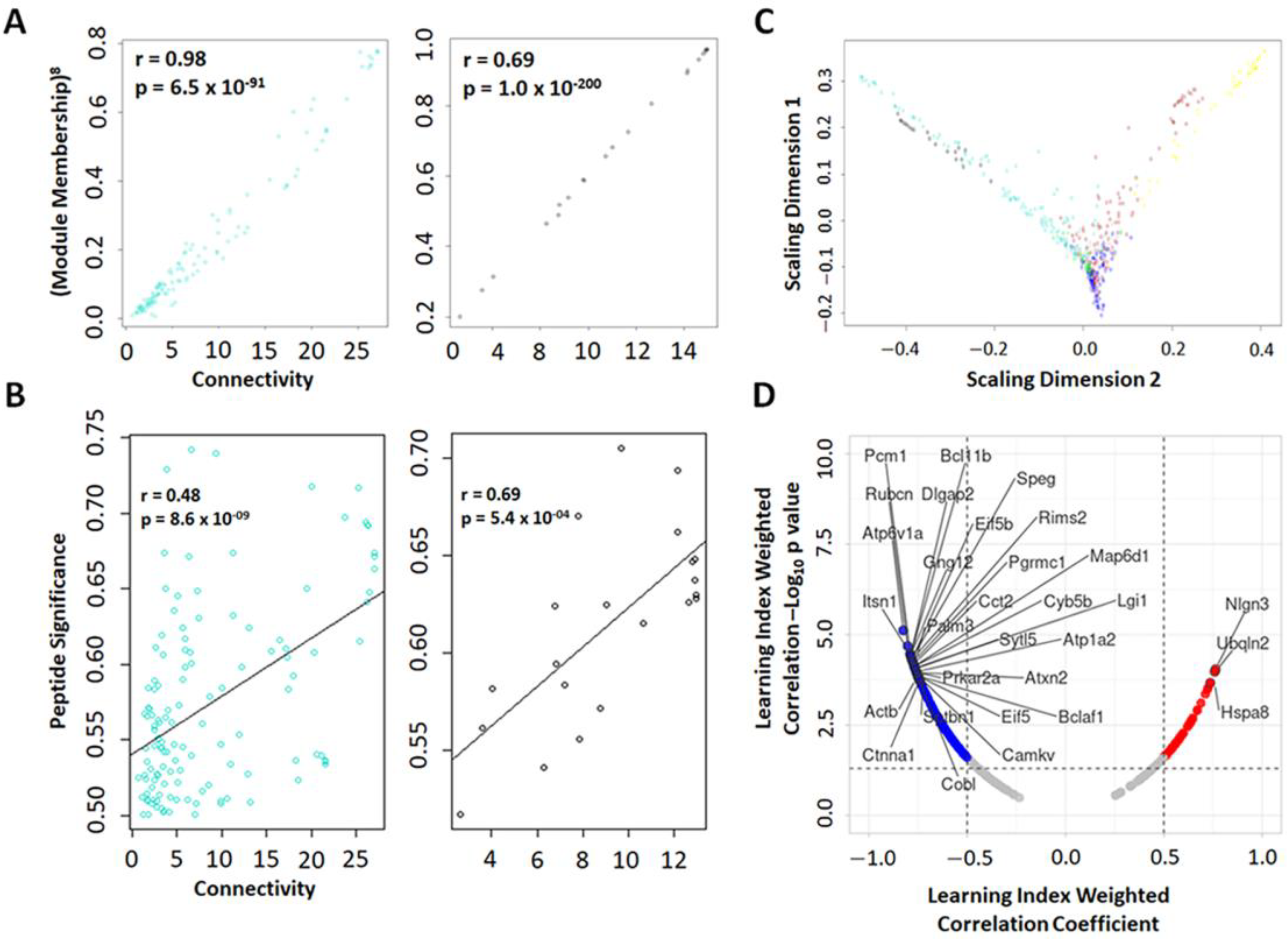
2.3.1. Weighted Peptide Co-Expression Network Analysis: Module–Module (Clustering and TOM Dissimilarity) and Module–Learning (Pearson Correlation) Relationship
2.3.2. Weighted Peptide Co-Expression Network Analysis: Phosphopeptide Module Connectivity Measure, Multidimensional Scaling, and Relationship with Learning Index
2.4. Weighted Peptide Co-Expression Network Analysis: Functional Characterization of Turquoise and Black Learning-Related Modules
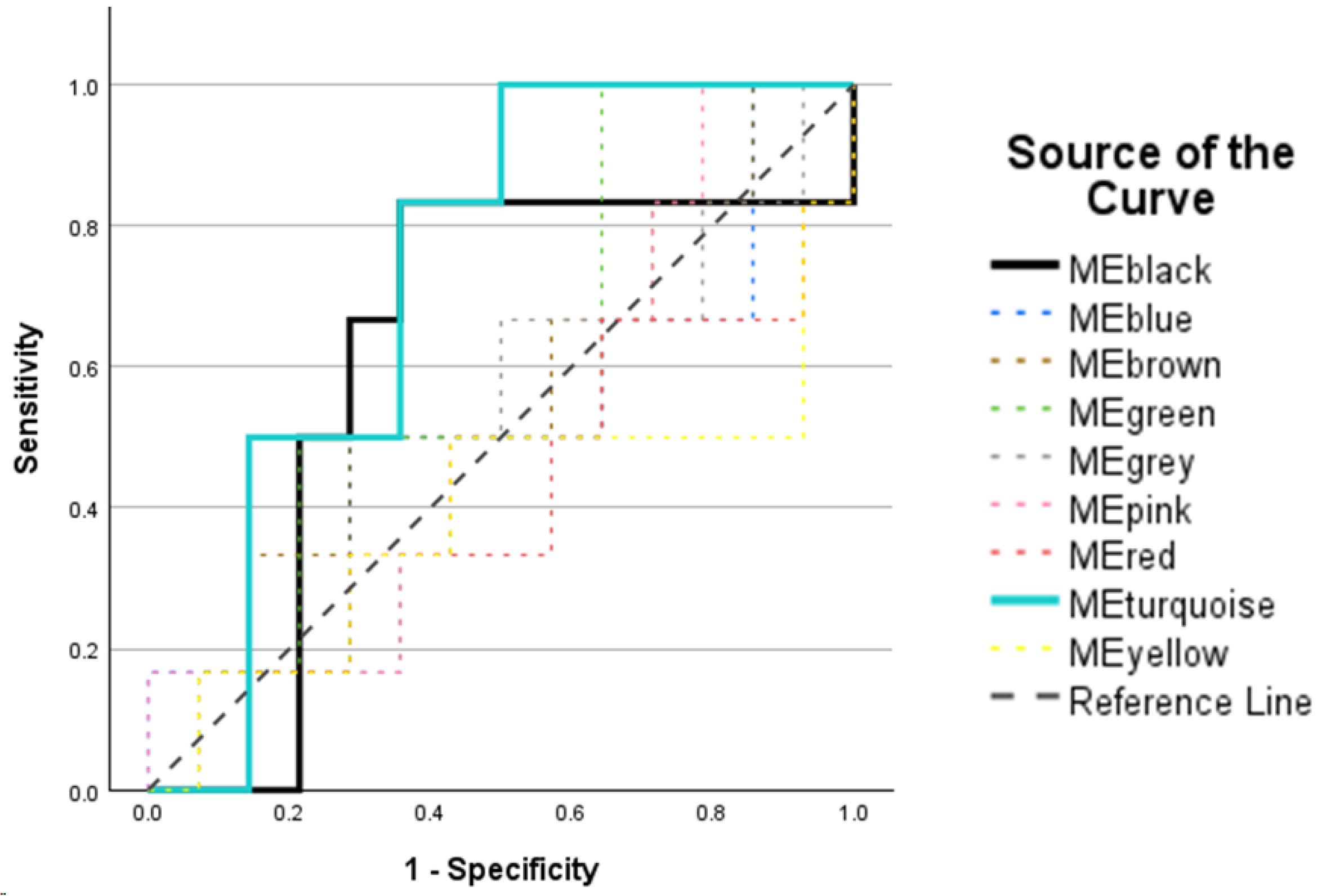
2.5. Post-Weighted Peptide Co-Expression Network Analysis: Receiver Operating Characteristic Analysis
2.6. Individual Level Analysis Identifies Phosphopeptides in Modules That Confer an Increased Risk for Learning Deficits Following a Single LIB Exposure
2.7. Group Level Analysis Using Protein Prioritization Identifies Blast-Relevant Phosphopeptides Related to Abnormal Behavorial and Molecular Phenotypes
3. Discussion
3.1. General Overview
3.2. Elevated Tau-ser262 Following LIB Exposure
3.3. WpCNA Module Eigenpeptides Predict and Correlate with Learning Behavior
3.4. WpCNA Individual-Level Analysis
3.4.1. Turquoise Module
3.4.2. Black Module
3.5. WpCNA Group-Level Analysis by Blast-Relevance Identified Phosphopeptides Related to Cognitive Decline, Synaptic Dysfunction, and Metabolic Disruption
3.6. Limitations
3.6.1. Other Genetic Risk Factors Related to BINT-Induced Cognitive Decline
3.6.2. Ongoing External Validation
3.6.3. CognitionWall Platform
3.6.4. WpCNA Approach
3.7. Future Studies
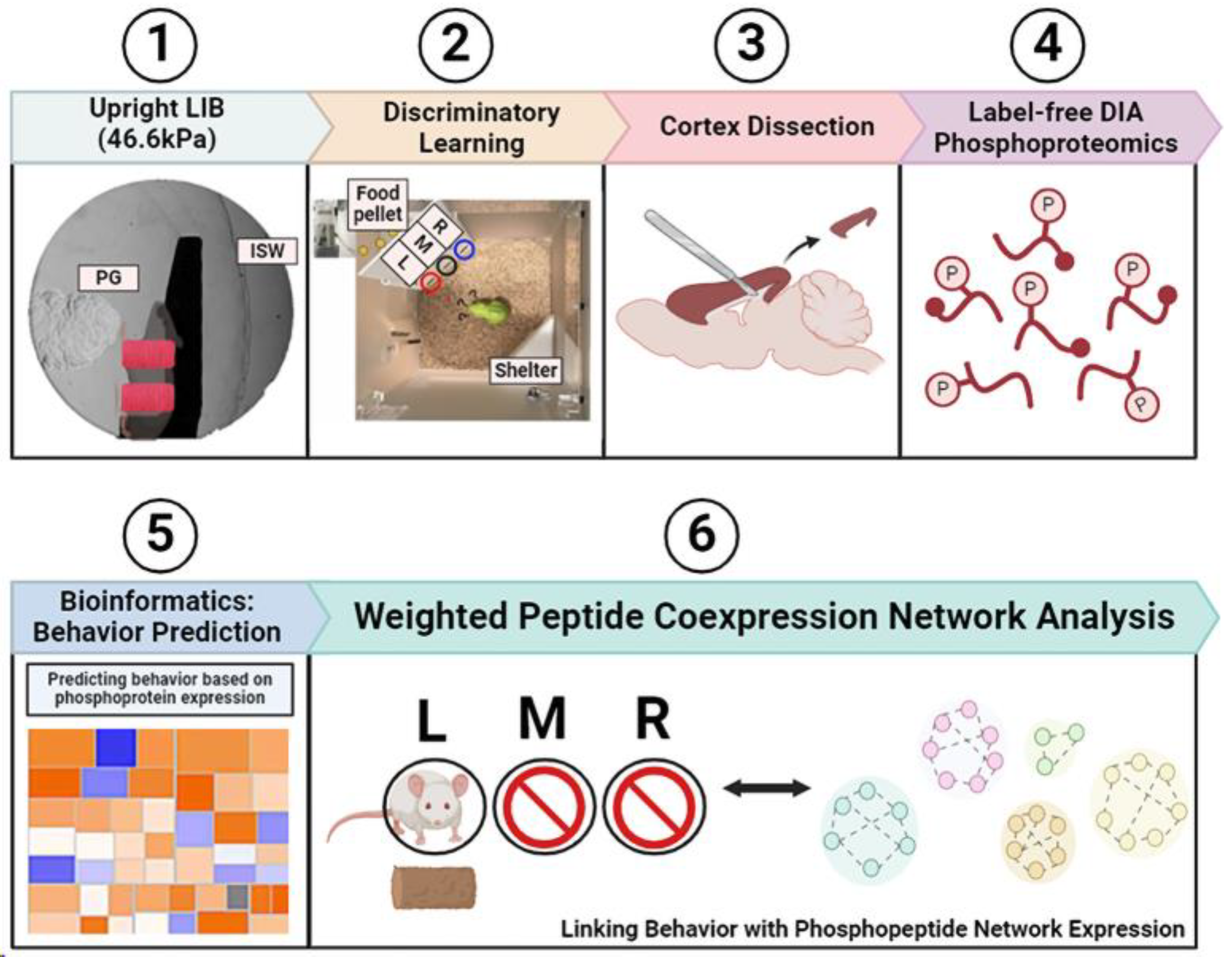
4. Materials and Methods
4.1. Open-Field Primary LIB in Mice
4.2. Automated Assessments of Learning Ability in a Home-Cage Environment
4.3. Tissue Collection and Protein Extraction
4.4. Protein Digestion for 4-D Tandem Mass Spectrometry
4.5. Spectral Library Generation, Phosphopeptide Identification, and Raw Data Processing
4.6. Processing of Expression (Phosphopeptidome) and Functional Outcome (Learning Index) Datasets for WpCNA
4.7. Adjacency Matrix Construction and Validation of Scale-Free Topology
4.8. Construction of Topological Overlap Matrix (TOM)
4.9. Detection of Phosphopeptide Module Membership
4.10. Relating Module Eigenpeptide to Learning Index
4.11. Statistical Methods
4.11.1. Behavioral Analysis
4.11.2. MS2 Quantitative Proteomics
4.11.3. ROC Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hoge, C.W.; Mc Gurk, D.; Thomas, J.L.; Cox, A.L.; Engel, C.C.; Castro, C.A. Mild Traumatic Brain Injury in U.S. Soldiers Returning from Iraq. N. Engl. J. Med. 2008, 358, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Phipps, H.; Mondello, S.; Wilson, A.; Dittmer, T.; Rohde, N.N.; Schroeder, P.J.; Nichols, J.; McGirt, C.; Hoffman, J.; Tanksley, K.; et al. Characteristics and Impact of U.S. Military Blast-Related Mild Traumatic Brain Injury: A Systematic Review. Front. Neurol. 2020, 11, 559318. [Google Scholar] [CrossRef] [PubMed]
- Cernak, I. Understanding Blast-Induced Neurotrauma: How Far Have We Come? Concussion 2017, 2, CNC42. [Google Scholar] [CrossRef] [PubMed]
- Siedhoff, H.R.; Chen, S.; Song, H.; Cui, J.; Cernak, I.; Cifu, D.X.; De Palma, R.G.; Gu, Z. Perspectives on Primary Blast Injury of the Brain: Translational Insights Into Non-inertial Low-Intensity Blast Injury. Front. Neurol. 2021, 12, 818169. [Google Scholar] [CrossRef]
- Sutar, S.; Ganpule, S. Investigation of Wave Propagation through Head Layers with Focus on Understanding Blast Wave Transmission. Biomech. Model Mechanobiol. 2020, 19, 875–892. [Google Scholar] [CrossRef]
- Arun, P.; Abu-Taleb, R.; Oguntayo, S.; Tanaka, M.; Wang, Y.; Valiyaveettil, M.; Long, J.B.; Zhang, Y.; Nambiar, M.P. Distinct Patterns of Expression of Traumatic Brain Injury Biomarkers after Blast Exposure: Role of Compromised Cell Membrane Integrity. Neurosci. Lett. 2013, 552, 87–91. [Google Scholar] [CrossRef]
- Li, C.; Chen, S.; Siedhoff, H.R.; Grant, D.; Liu, P.; Balderrama, A.; Jackson, M.; Zuckerman, A.; Greenlief, C.M.; Kobeissy, F.; et al. Low-Intensity Open-Field Blast Exposure Effects on Neurovascular Unit Ultrastructure in Mice. Acta Neuropathol. Commun. 2023, 11, 144. [Google Scholar] [CrossRef]
- Hayes, J.P.; Morey, R.A.; Tupler, L.A. A Case of Frontal Neuropsychological and Neuroimaging Signs Following Multiple Primary-Blast Exposure. Neurocase 2012, 18, 258–269. [Google Scholar] [CrossRef]
- Fleminger, S.; Oliver, D.L.; Lovestone, S.; Rabe-Hesketh, S.; Giora, A. Head Injury as a Risk Factor for Alzheimer’s Disease: The Evidence 10 Years on; a Partial Replication. J. Neurol. Neurosurg. Psychiatry 2003, 74, 857–862. [Google Scholar] [CrossRef]
- Englert, R.M.; Belding, J.N.; Thomsen, C.J. Self-Reported Symptoms in U.S. Marines Following Blast- and Impact-Related Concussion. Mil. Med. 2023, 188, e2118–e2125. [Google Scholar] [CrossRef]
- Chen, S.; Siedhoff, H.R.; Zhang, H.; Liu, P.; Balderrama, A.; Li, R.; Johnson, C.; Greenlief, C.M.; Koopmans, B.; Hoffman, T.; et al. Low-Intensity Blast Induces Acute Glutamatergic Hyperexcitability in Mouse Hippocampus Leading to Long-Term Learning Deficits and Altered Expression of Proteins Involved in Synaptic Plasticity and Serine Protease Inhibitors. Neurobiol. Dis. 2022, 165, 105634. [Google Scholar] [CrossRef]
- Chen, M.; Song, H.; Cui, J.; Johnson, C.E.; Hubler, G.K.; DePalma, R.G.; Gu, Z.; Xia, W. Proteomic Profiling of Mouse Brains Exposed to Blast-Induced Mild Traumatic Brain Injury Reveals Changes in Axonal Proteins and Phosphorylated Tau. J. Alzheimer’s Dis. 2018, 66, 751–773. [Google Scholar] [CrossRef]
- Konan, L.M.; Song, H.; Pentecost, G.; Fogwe, D.; Ndam, T.; Cui, J.; Johnson, C.E.; Grant, D.; White, T.; Chen, M.; et al. Multi-Focal Neuronal Ultrastructural Abnormalities and Synaptic Alterations in Mice after Low-Intensity Blast Exposure. J. Neurotrauma 2019, 36, 2117–2128. [Google Scholar] [CrossRef]
- Song, H.; Chen, M.; Chen, C.; Cui, J.; Johnson, C.E.; Cheng, J.; Wang, X.; Swerdlow, R.H.; DePalma, R.G.; Xia, W.; et al. Proteomic Analysis and Biochemical Correlates of Mitochondrial Dysfunction after Low-Intensity Primary Blast Exposure. J. Neurotrauma 2019, 36, 1591–1605. [Google Scholar] [CrossRef]
- Ruotolo, B.T. Collision Cross Sections for Native Proteomics: Challenges and Opportunities. J. Proteome Res. 2022, 21, 2–8. [Google Scholar] [CrossRef]
- Meier, F.; Brunner, A.D.; Koch, S.; Koch, H.; Lubeck, M.; Krause, M.; Goedecke, N.; Decker, J.; Kosinski, T.; Park, M.A.; et al. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell Proteomics 2018, 17, 2534–2545. [Google Scholar] [CrossRef] [PubMed]
- Li, K.W.; Gonzalez-Lozano, M.A.; Koopmans, F.; Smit, A.B. Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome. Front. Mol. Neurosci. 2020, 13, 564446. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Beck, S.; Grassl, N.; Lubeck, M.; Park, M.A.; Raether, O.; Mann, M. Parallel Accumulation-Serial Fragmentation (PASEF): Multiplying Sequencing Speed and Sensitivity by Synchronized Scans in a Trapped Ion Mobility Device. J. Proteome Res. 2015, 14, 5378–5387. [Google Scholar] [CrossRef] [PubMed]
- Possemato, A.P.; Paulo, J.A.; Mulhern, D.; Guo, A.; Gygi, S.P.; Beausoleil, S.A. Multiplexed Phosphoproteomic Profiling Using Titanium Dioxide and Immunoaffinity Enrichments Reveals Complementary Phosphorylation Events. J. Proteome Res. 2017, 16, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Gamache, J.; Benzow, K.; Forster, C.; Kemper, L.; Hlynialuk, C.; Furrow, E.; Ashe, K.H.; Koob, M.D. Factors Other than Htau Overexpression That Contribute to Tauopathy-Like Phenotype in rTg4510 Mice. Nat. Commun. 2019, 10, 2479. [Google Scholar] [CrossRef]
- Yanagisawa, D.; Hamezah, H.S.; Durani, L.W.; Taguchi, H.; Tooyama, I. Study of Tau Pathology in Male rTg4510 Mice Fed with a Curcumin Derivative Shiga-Y5. PLoS ONE 2018, 13, e0208440. [Google Scholar] [CrossRef]
- Blackmore, T.; Meftah, S.; Murray, T.K.; Craig, P.J.; Blockeel, A.; Phillips, K.; Eastwood, B.; O’Neill, M.J.; Marston, H.; Ahmed, Z.; et al. Tracking Progressive Pathological and Functional Decline in the rTg4510 Mouse Model of Tauopathy. Alzheimer’s Res. Ther. 2017, 9, 77. [Google Scholar] [CrossRef]
- Barabas, A.J.; Robbins, L.A.; Gaskill, B.N. Home Cage Measures of Alzheimer’s Disease in the rTg4510 Mouse Model. Genes Brain Behav. 2022, 21, e12795. [Google Scholar] [CrossRef]
- Camargo, L.C.; Honold, D.; Bauer, R.; Shah, N.J.; Langen, K.J.; Willbold, D.; Kutzsche, J.; Willuweit, A.; Schemmert, S. Sex-Related Motor Deficits in the Tau-P301L Mouse Model. Biomedicines 2021, 9, 1160. [Google Scholar] [CrossRef]
- Bailey, R.M.; Howard, J.; Knight, J.; Sahara, N.; Dickson, D.W.; Lewis, J. Effects of the C57BL/6 Strain Background on Tauopathy Progression in the rTg4510 Mouse Model. Mol. Neurodegener. 2014, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Gerson, J.; Castillo-Carranza, D.L.; Sengupta, U.; Bodani, R.; Prough, D.S.; DeWitt, D.S.; Hawkins, B.E.; Kayed, R. Tau Oligomers Derived from Traumatic Brain Injury Cause Cognitive Impairment and Accelerate Onset of Pathology in Htau Mice. J. Neurotrauma 2016, 33, 2034–2043. [Google Scholar] [CrossRef]
- Bressler, K.R.; Ross, J.A.; Ilnytskyy, S.; Vanden Dungen, K.; Taylor, K.; Patel, K.; Zovoilis, A.; Kovalchuk, I.; Thakor, N. Depletion of Eukaryotic Initiation Factor 5B (eIF5B) Reprograms the Cellular Transcriptome and Leads to Activation of Endoplasmic Reticulum (ER) Stress and c-Jun N-Terminal Kinase (JNK). Cell Stress Chaperones 2021, 26, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Tracy, T.E.; Madero-Perez, J.; Swaney, D.L.; Chang, T.S.; Moritz, M.; Konrad, C.; Ward, M.E.; Stevenson, E.; Huttenhain, R.; Kauwe, G.; et al. Tau Interactome Maps Synaptic and Mitochondrial Processes Associated with Neurodegeneration. Cell 2022, 185, 712–728.e14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, Y.; Ma, L.; Wei, Y.; Li, H. Possible Mechanisms of Tau Spread and Toxicity in Alzheimer’s Disease. Front. Cell Dev. Biol. 2021, 9, 707268. [Google Scholar] [CrossRef]
- Sahara, N.; Murayama, M.; Higuchi, M.; Suhara, T.; Takashima, A. Biochemical Distribution of Tau Protein in Synaptosomal Fraction of Transgenic Mice Expressing Human P301L Tau. Front. Neurol. 2014, 5, 26. [Google Scholar] [CrossRef]
- Tai, H.C.; Wang, B.Y.; Serrano-Pozo, A.; Frosch, M.P.; Spires-Jones, T.L.; Hyman, B.T. Frequent and Symmetric Deposition of Misfolded Tau Oligomers within Presynaptic and Postsynaptic Terminals in Alzheimer’s Disease. Acta Neuropathol. Commun. 2014, 2, 146. [Google Scholar] [CrossRef]
- Leiva-Salinas, C.; Singh, A.; Layfield, E.; Flors, L.; Patrie, J.T. Early Brain Amyloid Accumulation at PET in Military Instructors Exposed to Subconcussive Blast Injuries. Radiology 2023, 307, e221608. [Google Scholar] [CrossRef]
- Edwards, K.A.; Greer, K.; Leete, J.; Lai, C.; Devoto, C.; Qu, B.X.; Yarnell, A.M.; Polejaeva, E.; Dell, K.C.; LoPresti, M.L.; et al. Neuronally-Derived Tau is Increased in Experienced Breachers and Is Associated with Neurobehavioral Symptoms. Sci. Rep. 2021, 11, 19527. [Google Scholar] [CrossRef]
- Edwards, K.A.; Leete, J.J.; Tschiffely, A.E.; Moore, C.Y.; Dell, K.C.; Statz, J.K.; Carr, W.; Walker, P.B.; LoPresti, M.L.; Ahlers, S.T.; et al. Blast Exposure Results in Tau and Neurofilament Light Chain Changes in Peripheral Blood. Brain Inj. 2020, 34, 1213–1221. [Google Scholar] [CrossRef]
- Logsdon, A.F.; Schindler, A.G.; Meabon, J.S.; Yagi, M.; Herbert, M.J.; Banks, W.A.; Raskind, M.A.; Marshall, D.A.; Keene, C.D.; Perl, D.P.; et al. Nitric Oxide Synthase Mediates Cerebellar Dysfunction in Mice Exposed to Repetitive Blast-Induced Mild Traumatic Brain Injury. Sci. Rep. 2020, 10, 9420. [Google Scholar] [CrossRef]
- McKee, A.C.; Cantu, R.C.; Nowinski, C.J.; Hedley-Whyte, E.T.; Gavett, B.E.; Budson, A.E.; Santini, V.E.; Lee, H.S.; Kubilus, C.A.; Stern, R.A. Chronic Traumatic Encephalopathy in Athletes: Progressive Tauopathy after Repetitive Head Injury. J. Neuropathol. Exp. Neurol. 2009, 68, 709–735. [Google Scholar] [CrossRef]
- Turner, R.C.; Lucke-Wold, B.P.; Robson, M.J.; Lee, J.M.; Bailes, J.E. Alzheimer’s Disease and Chronic Traumatic Encephalopathy: Distinct but Possibly Overlapping Disease Entities. Brain Inj. 2016, 30, 1279–1292. [Google Scholar] [CrossRef]
- Hyman, B.T.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Carrillo, M.C.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; et al. National Institute on Aging-Alzheimer’s Association Guidelines for the Neuropathologic Assessment of Alzheimer’s Disease. Alzheimer’s Dement. 2012, 8, 1–13. [Google Scholar] [CrossRef]
- Banks, S.J.; Raman, R.; He, F.; Salmon, D.P.; Ferris, S.; Aisen, P.; Cummings, J. The Alzheimer’s disease Cooperative Study Prevention Instrument Project: Longitudinal Outcome of Behavioral Measures as Predictors of Cognitive Decline. Dement. Geriatr. Cogn. Dis. Extra 2014, 4, 509–516. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.; Chen, S.; Liu, P.; Langenderfer, M.; Li, C.; Siedhoff, H.R.; Balderrama, A.; Li, R.; Johnson, C.E.; Greenlief, C.M.; et al. Quantitative Proteomic Profiling in Brain Subregions of Mice Exposed to Open-Field Low-intensity Blast Reveals Position-Dependent Blast Effects. Shock Waves 2024. accepted. [Google Scholar]
- Elsworth, J.D.; Jentsch, J.D.; Groman, S.M.; Roth, R.H.; Redmond, E.D., Jr.; Leranth, C. Low Circulating Levels of Bisphenol-A Induce Cognitive Deficits and Loss of Asymmetric Spine Synapses in Dorsolateral Prefrontal Cortex and Hippocampus of Adult Male Monkeys. J. Comp. Neurol. 2015, 523, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Burke, S.N.; Barnes, C.A. Neural Plasticity in the Ageing Brain. Nat. Rev. Neurosci. 2006, 7, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Konan, L.M.; Cui, J.; Johnson, C.E.; Langenderfer, M.; Grant, D.; Ndam, T.; Simonyi, A.; White, T.; Demirci, U.; et al. Ultrastructural Brain Abnormalities and Associated Behavioral Changes in Mice after Low-Intensity Blast Exposure. Behav. Brain Res. 2018, 347, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Siedhoff, H.R.; Chen, S.; Balderrama, A.; Sun, G.Y.; Koopmans, B.; De Palma, R.G.; Cui, J.; Gu, Z. Long-Term Effects of Low-Intensity Blast Non-Inertial Brain Injury on Anxiety-Like Behaviors in Mice: Home-Cage Monitoring Assessments. Neurotrauma Rep. 2022, 3, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Aravind, A.; Kosty, J.; Chandra, N.; Pfister, B.J. Blast Exposure Predisposes the Brain to Increased Neurological Deficits in a Model of Blast Plus Blunt Traumatic Brain Injury. Exp. Neurol. 2020, 332, 113378. [Google Scholar] [CrossRef] [PubMed]
- Santacruz, K.; Lewis, J.; Spires, T.; Paulson, J.; Kotilinek, L.; Ingelsson, M.; Guimaraes, A.; DeTure, M.; Ramsden, M.; McGowan, E.; et al. Tau Suppression in a Neurodegenerative Mouse Model Improves Memory Function. Science 2005, 309, 476–481. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Presson, A.P.; Sobel, E.M.; Papp, J.C.; Suarez, C.J.; Whistler, T.; Rajeevan, M.S.; Vernon, S.D.; Horvath, S. Integrated Weighted Gene Co-Expression Network Analysis with an Application to Chronic Fatigue Syndrome. BMC Syst. Biol. 2008, 2, 95. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, D.; Qiu, W.; Shi, Y.; Yang, J.J.; Chen, S.; Wang, Q.; Pan, H. Application of Weighted Gene Co-expression Network Analysis for Data from Paired Design. Sci. Rep. 2018, 8, 622. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining Clusters from a Hierarchical Cluster Tree: The Dynamic Tree Cut package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef]
- Skelding, K.A.; Spratt, N.J.; Fluechter, L.; Dickson, P.W.; Rostas, J.A. alphaCaMKII is Differentially Regulated in Brain Regions that Exhibit Differing Sensitivities to Ischemia and Excitotoxicity. J. Cereb. Blood Flow. Metab. 2012, 32, 2181–2192. [Google Scholar] [CrossRef]
- Oka, M.; Fujisaki, N.; Maruko-Otake, A.; Ohtake, Y.; Shimizu, S.; Saito, T.; Hisanaga, S.I.; Iijima, K.M.; Ando, K. Ca2+/Calmodulin-Dependent Protein Kinase II Promotes Neurodegeneration Caused by Tau Phosphorylated at Ser262/356 in a Transgenic Drosophila Model of Tauopathy. J. Biochem. 2017, 162, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Iijima-Ando, K.; Sekiya, M.; Maruko-Otake, A.; Ohtake, Y.; Suzuki, E.; Lu, B.; Iijima, K.M. Loss of Axonal Mitochondria Promotes Tau-Mediated Neurodegeneration and Alzheimer’s Disease-Related Tau Phosphorylation via PAR-1. PLoS Genet. 2012, 8, e1002918. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; He, H.; Peng, T.; Lu, Y.; Yu, J. Identification of 9 Gene Signatures by WGCNA to Predict Prognosis for Colon Adenocarcinoma. Comput. Intell. Neurosci. 2022, 2022, 8598046. [Google Scholar] [CrossRef]
- Tang, X.; Xu, P.; Wang, B.; Luo, J.; Fu, R.; Huang, K.; Dai, L.; Lu, J.; Cao, G.; Peng, H.; et al. Identification of a Specific Gene Module for Predicting Prognosis in Glioblastoma Patients. Front. Oncol 2019, 9, 812. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Dai, Q.; Ding, Q.; Wei, M.; Kong, X. Identification of Key Genes in Hepatitis B Associated Hepatocellular Carcinoma Based on WGCNA. Infect. Agent. Cancer 2021, 16, 18. [Google Scholar] [CrossRef]
- Zhao, Z.J.; Wei, D.P.; Zheng, R.Z.; Peng, T.; Xiao, X.; Li, F.S. The Gene Coexpression Analysis Identifies Functional Modules Dynamically Changed after Traumatic Brain Injury. Comput. Math. Methods Med. 2021, 2021, 5511598. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Nakada, T.A.; Oshima, T.; Motoshima, T.; Kawaguchi, R.; Oda, S. Tau Protein as a Diagnostic Marker for Diffuse Axonal Injury. PLoS ONE 2019, 14, e0214381. [Google Scholar] [CrossRef]
- Platzer, K.; Sticht, H.; Edwards, S.L.; Allen, W.; Angione, K.M.; Bonati, M.T.; Brasington, C.; Cho, M.T.; Demmer, L.A.; Falik-Zaccai, T.; et al. De Novo Variants in MAPK8IP3 Cause Intellectual Disability with Variable Brain Anomalies. Am. J. Hum. Genet. 2019, 104, 203–212. [Google Scholar] [CrossRef]
- Koushika, S.P. “JIP”ing Along the Axon: The Complex Roles of JIPs in Axonal Transport. Bioessays 2008, 30, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Celestino, R.; Gama, J.B.; Castro-Rodrigues, A.F.; Barbosa, D.J.; Rocha, H.; d’Amico, E.A.; Musacchio, A.; Carvalho, A.X.; Morais-Cabral, J.H.; Gassmann, R. JIP3 Interacts with Dynein and Kinesin-1 to Regulate Bidirectional Organelle Transport. J. Cell Biol. 2022, 221, e202110057. [Google Scholar] [CrossRef] [PubMed]
- Rafiq, N.M.; Lyons, L.L.; Gowrishankar, S.; De Camilli, P.; Ferguson, S.M. JIP3 Links Lysosome Transport to Regulation of Multiple Components of the Axonal Cytoskeleton. Commun. Biol. 2022, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.; Nishitoh, H.; Takeda, K.; Matsuzawa, A.; Amagasa, T.; Ito, M.; Yoshioka, K.; Ichijo, H. Phosphorylation-Dependent Scaffolding Role of JSAP1/JIP3 in the ASK1-JNK Signaling pathWay. A New Mode of Regulation of the MAP Kinase Cascade. J. Biol. Chem. 2002, 277, 40703–40709. [Google Scholar] [CrossRef]
- Ye, Z.; Izadi, A.; Gurkoff, G.G.; Rickerl, K.; Sharp, F.R.; Ander, B.P.; Bauer, S.Z.; Lui, A.; Lyeth, B.G.; Liu, D. Combined Inhibition of Fyn and c-Src Protects Hippocampal Neurons and Improves Spatial Memory via ROCK after Traumatic Brain Injury. J. Neurotrauma 2022, 39, 520–529. [Google Scholar] [CrossRef]
- Ongusaha, P.P.; Qi, H.H.; Raj, L.; Kim, Y.B.; Aaronson, S.A.; Davis, R.J.; Shi, Y.; Liao, J.K.; Lee, S.W. Identification of ROCK1 as an Upstream Activator of the JIP-3 to JNK Signaling Axis in Response to UVB Damage. Sci. Signal. 2008, 1, ra14. [Google Scholar] [CrossRef]
- Kawasaki, A.; Okada, M.; Tamada, A.; Okuda, S.; Nozumi, M.; Ito, Y.; Kobayashi, D.; Yamasaki, T.; Yokoyama, R.; Shibata, T.; et al. Growth Cone Phosphoproteomics Reveals that GAP-43 Phosphorylated by JNK Is a Marker of Axon Growth and Regeneration. iScience 2018, 4, 190–203. [Google Scholar] [CrossRef]
- Ohrfelt, A.; Benedet, A.L.; Ashton, N.J.; Kvartsberg, H.; Vandijck, M.; Weiner, M.W.; Trojanowski, J.Q.; Shaw, L.M.; Zetterberg, H.; Blennow, K.; et al. Association of CSF GAP-43 With the Rate of Cognitive Decline and Progression to Dementia in Amyloid-Positive Individuals. Neurology 2023, 100, e275–e285. [Google Scholar] [CrossRef]
- Lu, N.N.; Tan, C.; Sun, N.H.; Shao, L.X.; Liu, X.X.; Gao, Y.P.; Tao, R.R.; Jiang, Q.; Wang, C.K.; Huang, J.Y.; et al. Cholinergic Grb2-Associated-Binding Protein 1 Regulates Cognitive Function. Cereb. Cortex 2018, 28, 2391–2404. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, T.; Hashimoto, R.; Sakoori, K.; Sugaya, Y.; Tanimura, A.; Hashimotodani, Y.; Ohi, K.; Yamamori, H.; Yasuda, Y.; Umeda-Yano, S.; et al. Emerging Roles of ARHGAP33 in Intracellular Trafficking of TrkB and Pathophysiology of Neuropsychiatric Disorders. Nat. Commun. 2016, 7, 10594. [Google Scholar] [CrossRef]
- Kim, Y.; Ha, C.M.; Chang, S. SNX26, a GTPase-Activating Protein for Cdc42, Interacts with PSD-95 Protein and Is Involved in Activity-Dependent Dendritic Spine Formation in Mature Neurons. J. Biol. Chem. 2013, 288, 29453–29466. [Google Scholar] [CrossRef]
- Bell, J.D.; Park, E.; Ai, J.; Baker, A.J. PICK1-Mediated GluR2 Endocytosis Contributes to Cellular Injury after Neuronal Trauma. Cell Death Differ. 2009, 16, 1665–1680. [Google Scholar] [CrossRef] [PubMed]
- Lucke-Wold, B.P.; Logsdon, A.F.; Smith, K.E.; Turner, R.C.; Alkon, D.L.; Tan, Z.; Naser, Z.J.; Knotts, C.M.; Huber, J.D.; Rosen, C.L. Bryostatin-1 Restores Blood Brain Barrier Integrity following Blast-Induced Traumatic Brain Injury. Mol. Neurobiol. 2015, 52, 1119–1134. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; Du, J.; Lu, S.; Li, X.; Chen, Y.; Yuan, C.; Hou, S.T.; Wang, Y. PKCalpha Phosphorylation of GLT-1 at Ser562/563 Induces Glutamate Excitotoxicity in Ischemia in Mice. Signal Transduct. Target. Ther. 2022, 7, 82. [Google Scholar] [CrossRef]
- Wang, P.; Joberty, G.; Buist, A.; Vanoosthuyse, A.; Stancu, I.C.; Vasconcelos, B.; Pierrot, N.; Faelth-Savitski, M.; Kienlen-Campard, P.; Octave, J.N.; et al. Tau Interactome Mapping Based Identification of Otub1 as Tau Deubiquitinase Involved in Accumulation of Pathological Tau forms In Vitro and In Vivo. Acta Neuropathol. 2017, 133, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Herhaus, L.; Perez-Oliva, A.B.; Cozza, G.; Gourlay, R.; Weidlich, S.; Campbell, D.G.; Pinna, L.A.; Sapkota, G.P. Casein Kinase 2 (CK2) Phosphorylates the Deubiquitylase OTUB1 at Ser16 to Trigger its Nuclear Localization. Sci. Signal. 2015, 8, ra35. [Google Scholar] [CrossRef]
- Kim, J.Y.; Oh, M.H.; Bernard, L.P.; Macara, I.G.; Zhang, H. The RhoG/ELMO1/Dock180 Signaling Module is Required for Spine Morphogenesis in Hippocampal Neurons. J. Biol. Chem. 2011, 286, 37615–37624. [Google Scholar] [CrossRef]
- Morikawa, M.; Tanaka, Y.; Cho, H.S.; Yoshihara, M.; Hirokawa, N. The Molecular Motor KIF21B Mediates Synaptic Plasticity and Fear Extinction by Terminating Rac1 Activation. Cell Rep. 2018, 23, 3864–3877. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Sherman, N.E.; Fox, J.W.; Ginsberg, M.H. Phosphorylation Sites in the Cerebral Cavernous Malformations Complex. J. Cell Sci. 2011, 124, 3929–3932. [Google Scholar] [CrossRef][Green Version]
- Bramswig, N.C.; Ludecke, H.J.; Pettersson, M.; Albrecht, B.; Bernier, R.A.; Cremer, K.; Eichler, E.E.; Falkenstein, D.; Gerdts, J.; Jansen, S.; et al. Identification of New TRIP12 Variants and Detailed Clinical Evaluation of Individuals with Non-Syndromic Intellectual Disability with or without Autism. Hum. Genet. 2017, 136, 179–192. [Google Scholar] [CrossRef]
- Edwards, K.A.; Motamedi, V.; Osier, N.D.; Kim, H.S.; Yun, S.; Cho, Y.E.; Lai, C.; Dell, K.C.; Carr, W.; Walker, P.; et al. A Moderate Blast Exposure Results in Dysregulated Gene Network Activity Related to Cell Death, Survival, Structure, and Metabolism. Front. Neurol. 2020, 11, 91. [Google Scholar] [CrossRef]
- Wu, X.; Zheng, Y.; Liu, M.; Li, Y.; Ma, S.; Tang, W.; Yan, W.; Cao, M.; Zheng, W.; Jiang, L.; et al. BNIP3L/NIX Degradation Leads to Mitophagy Deficiency in Ischemic Brains. Autophagy 2021, 17, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Gu, Y.; Shen, Y.; Feng, W. Identification of Immune-Associated Genes in Diagnosing Polycystic Ovary Syndrome with Metabolic Syndrome by Weighted Gene Co-expression Network Analysis. Res. Sq. 2023, 1–21. [Google Scholar] [CrossRef]
- Wooten, T.; Sullivan, D.R.; Logue, M.W.; Fonda, J.R.; Fortier, C.B.; DeGutis, J.; McGlinchey, R.; Milberg, W.; Esterman, M. Apolipoprotein E (APOE) epsilon4 Status Moderates the Relationship Between Close-Range Blast Exposure and Cognitive Functioning. J. Int. Neuropsychol. Soc. 2021, 27, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, D.R.; Logue, M.W.; Wolf, E.J.; Hayes, J.P.; Salat, D.H.; Fortier, C.B.; Fonda, J.R.; McGlinchey, R.E.; Milberg, W.P.; Miller, M.W. Close-Range Blast Exposure Is Associated with Altered White Matter Integrity in Apolipoprotein varepsilon4 Carriers. J. Neurotrauma 2019, 36, 3264–3273. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Konan, L.M.; Cui, J.; Johnson, C.E.; Hubler, G.K.; DePalma, R.G.; Gu, Z. Nanometer Ultrastructural Brain Damage Following Low Intensity Primary Blast Wave Exposure. Neural. Regen. Res. 2018, 13, 1516–1519. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, J.; Maroteaux, G.; Schut, D.; Loos, M.; Dubey, M.; Pitsch, J.; Remmelink, E.; Koopmans, B.; Crowley, J.; Cornelisse, L.N.; et al. Protein Instability, Haploinsufficiency, and Cortical Hyper-Excitability Underlie STXBP1 Encephalopathy. Brain 2018, 141, 1350–1374. [Google Scholar] [CrossRef] [PubMed]
- Logan, S.; Royce, G.H.; Owen, D.; Farley, J.; Ranjo-Bishop, M.; Sonntag, W.E.; Deepa, S.S. Accelerated Decline in Cognition in a Mouse Model of Increased Oxidative Stress. Geroscience 2019, 41, 591–607. [Google Scholar] [CrossRef]
- Remmelink, E.; Smit, A.B.; Verhage, M.; Loos, M. Measuring Discrimination- and Reversal Learning in Mouse Models within 4 Days and without Prior Food Deprivation. Learn Mem. 2016, 23, 660–667. [Google Scholar] [CrossRef]
- Song, H.; Cui, J.; Simonyi, A.; Johnson, C.E.; Hubler, G.K.; DePalma, R.G.; Gu, Z. Linking Blast Physics to Biological Outcomes in Mild Traumatic Brain Injury: Narrative review and preliminary report of an open-field blast model. Behav. Brain Res. 2018, 340, 147–158. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, K.; Fang, Z. Robust RNA-Seq Data Analysis Using an Integrated Method of ROC Curve and Kolmogorov-Smirnov Test. Commun. Stat. Simul. Comput. 2022, 51, 7444–7457. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jackson, M.; Chen, S.; Nguyen, T.T.; Siedhoff, H.R.; Balderrama, A.; Zuckerman, A.; Li, R.; Greenlief, C.M.; Cole, G.; Frautschy, S.A.; et al. The Chronic Effects of a Single Low-Intensity Blast Exposure on Phosphoproteome Networks and Cognitive Function Influenced by Mutant Tau Overexpression. Int. J. Mol. Sci. 2024, 25, 3338. https://doi.org/10.3390/ijms25063338
Jackson M, Chen S, Nguyen TT, Siedhoff HR, Balderrama A, Zuckerman A, Li R, Greenlief CM, Cole G, Frautschy SA, et al. The Chronic Effects of a Single Low-Intensity Blast Exposure on Phosphoproteome Networks and Cognitive Function Influenced by Mutant Tau Overexpression. International Journal of Molecular Sciences. 2024; 25(6):3338. https://doi.org/10.3390/ijms25063338
Chicago/Turabian StyleJackson, Marcus, Shanyan Chen, Thao Thi Nguyen, Heather R. Siedhoff, Ashley Balderrama, Amitai Zuckerman, Runting Li, C. Michael Greenlief, Gregory Cole, Sally A. Frautschy, and et al. 2024. "The Chronic Effects of a Single Low-Intensity Blast Exposure on Phosphoproteome Networks and Cognitive Function Influenced by Mutant Tau Overexpression" International Journal of Molecular Sciences 25, no. 6: 3338. https://doi.org/10.3390/ijms25063338
APA StyleJackson, M., Chen, S., Nguyen, T. T., Siedhoff, H. R., Balderrama, A., Zuckerman, A., Li, R., Greenlief, C. M., Cole, G., Frautschy, S. A., Cui, J., & Gu, Z. (2024). The Chronic Effects of a Single Low-Intensity Blast Exposure on Phosphoproteome Networks and Cognitive Function Influenced by Mutant Tau Overexpression. International Journal of Molecular Sciences, 25(6), 3338. https://doi.org/10.3390/ijms25063338







