Applying Proteomics and Computational Approaches to Identify Novel Targets in Blast-Associated Post-Traumatic Epilepsy
Abstract
1. Introduction
2. Results
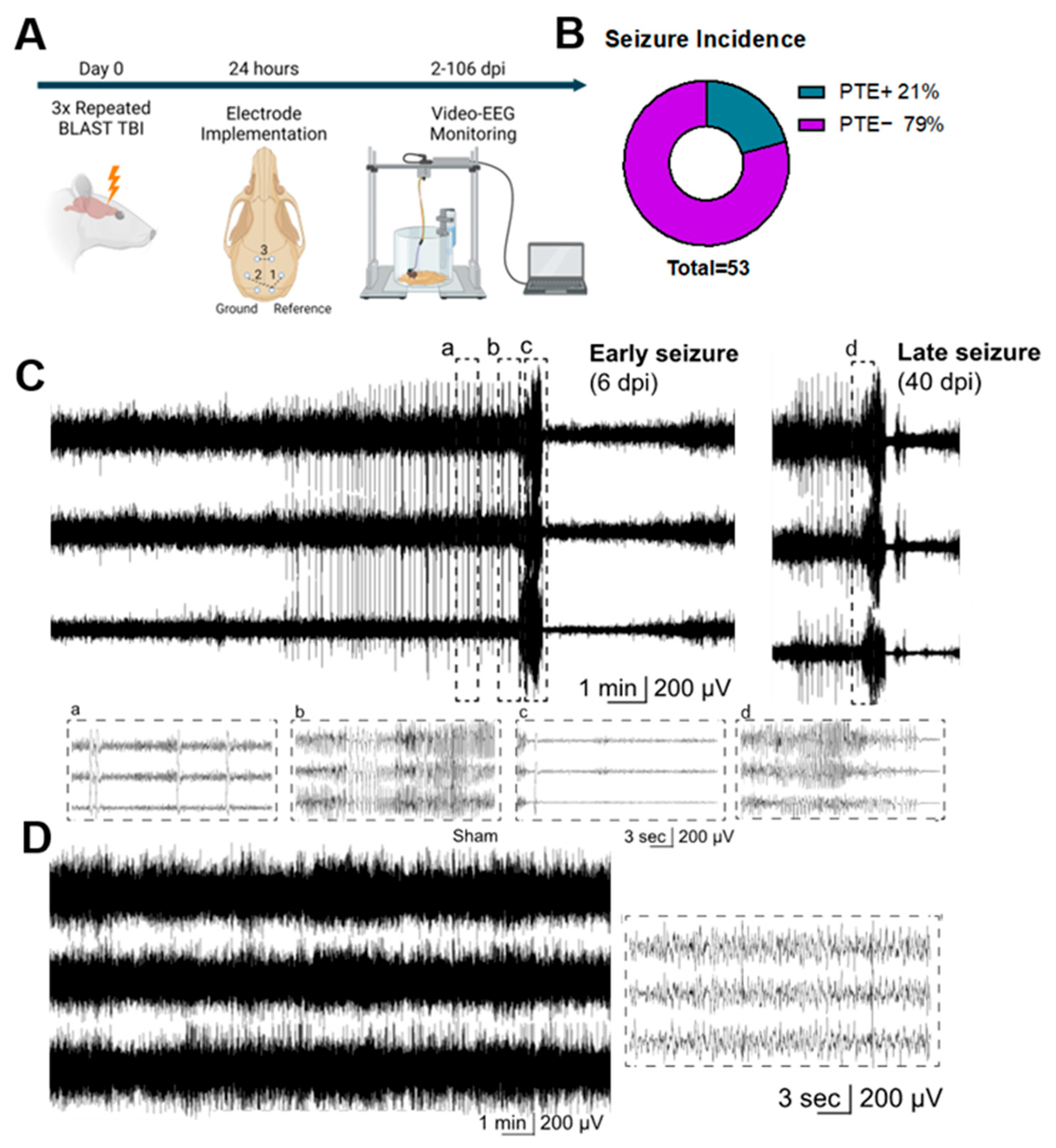
2.1. Repeated Blast TBI Caused Spontaneous, Unprovoked, Recurrent Seizures
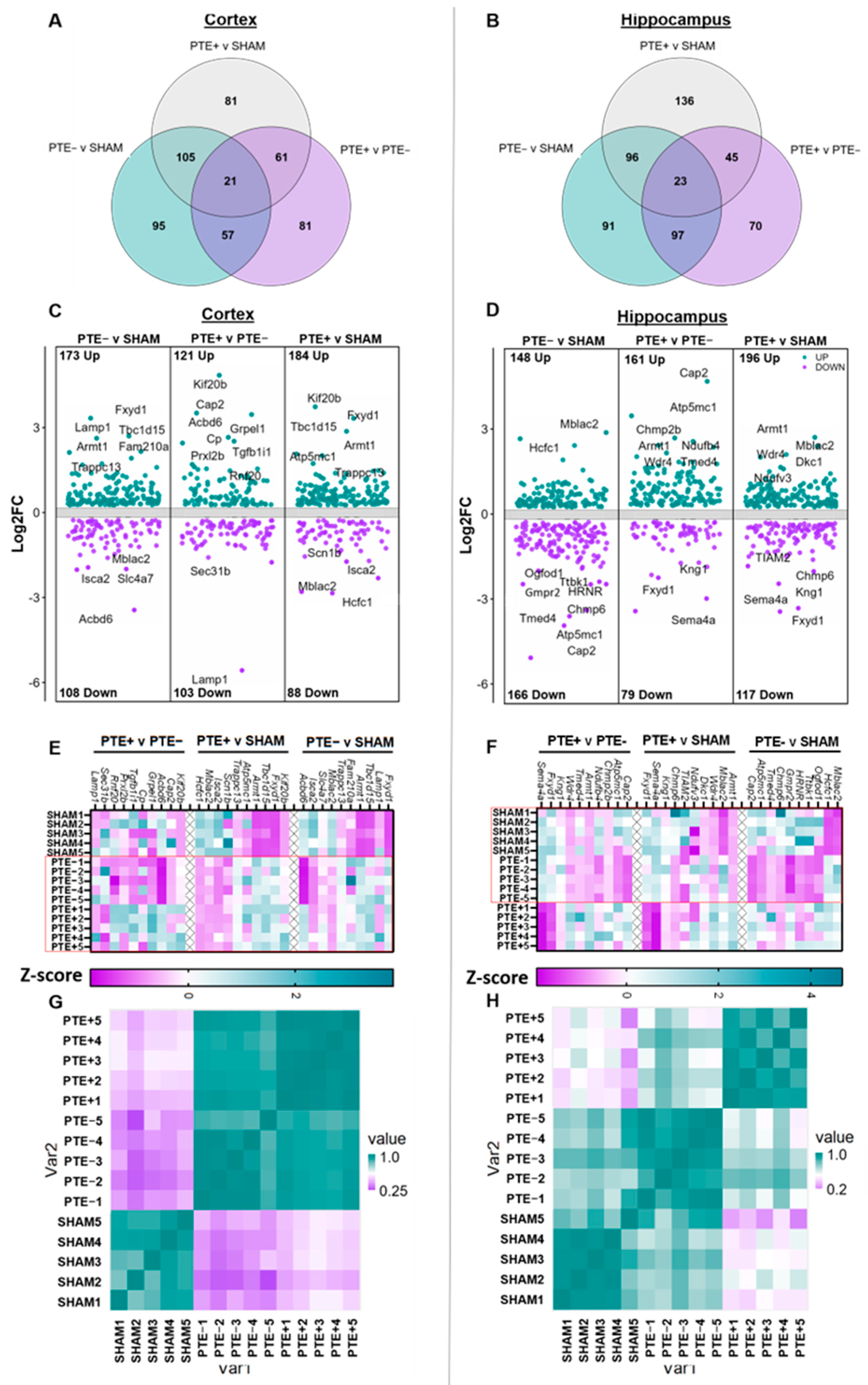
2.2. Chronic Protein Dysregulation in the Cortex and Hippocampus following rbTBI
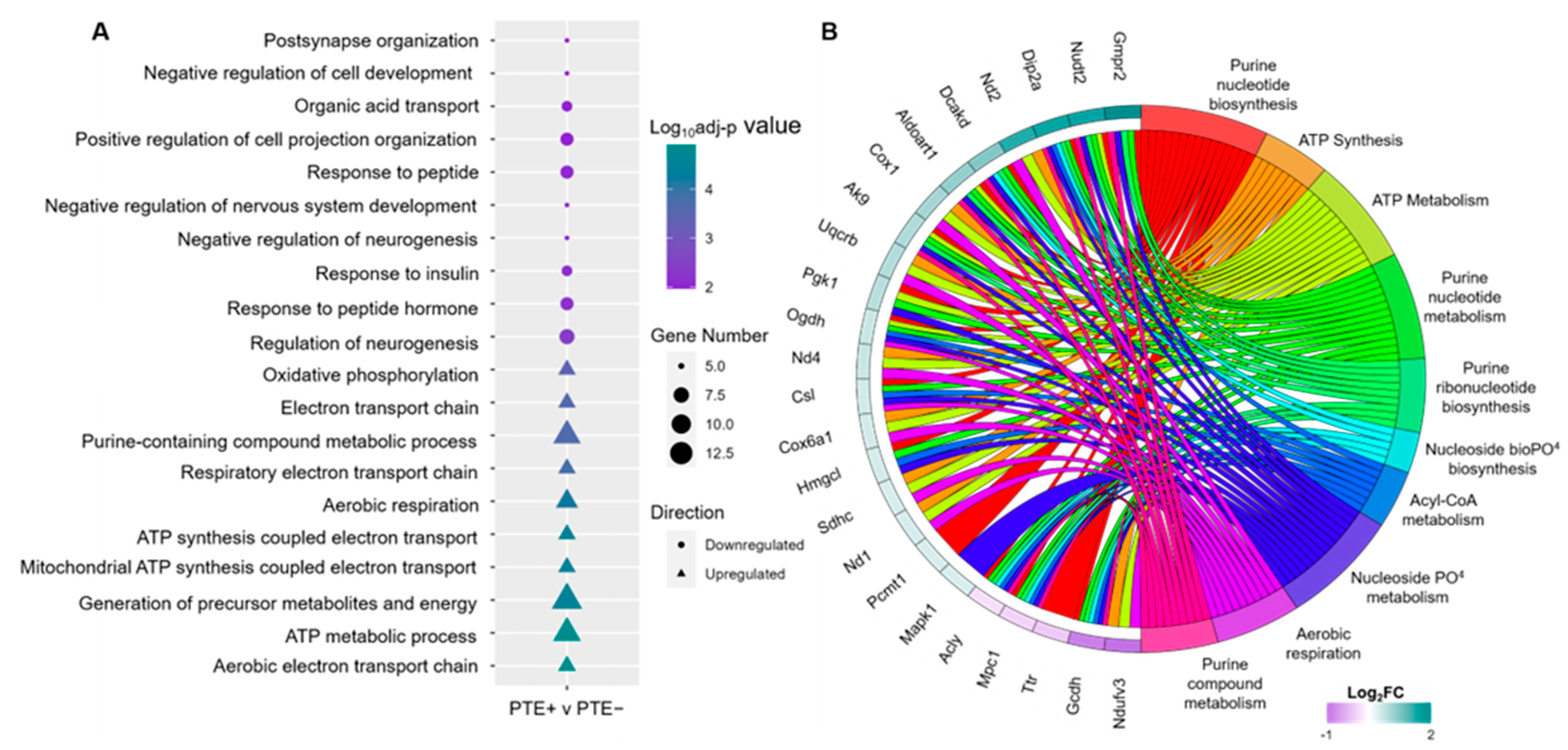
2.3. Functional Enrichment Analysis of Dysregulated DEPs in rbTBI
2.4. Predicted Alterations in Flux Reactions Caused by DEPs in PTE+ rbTBI
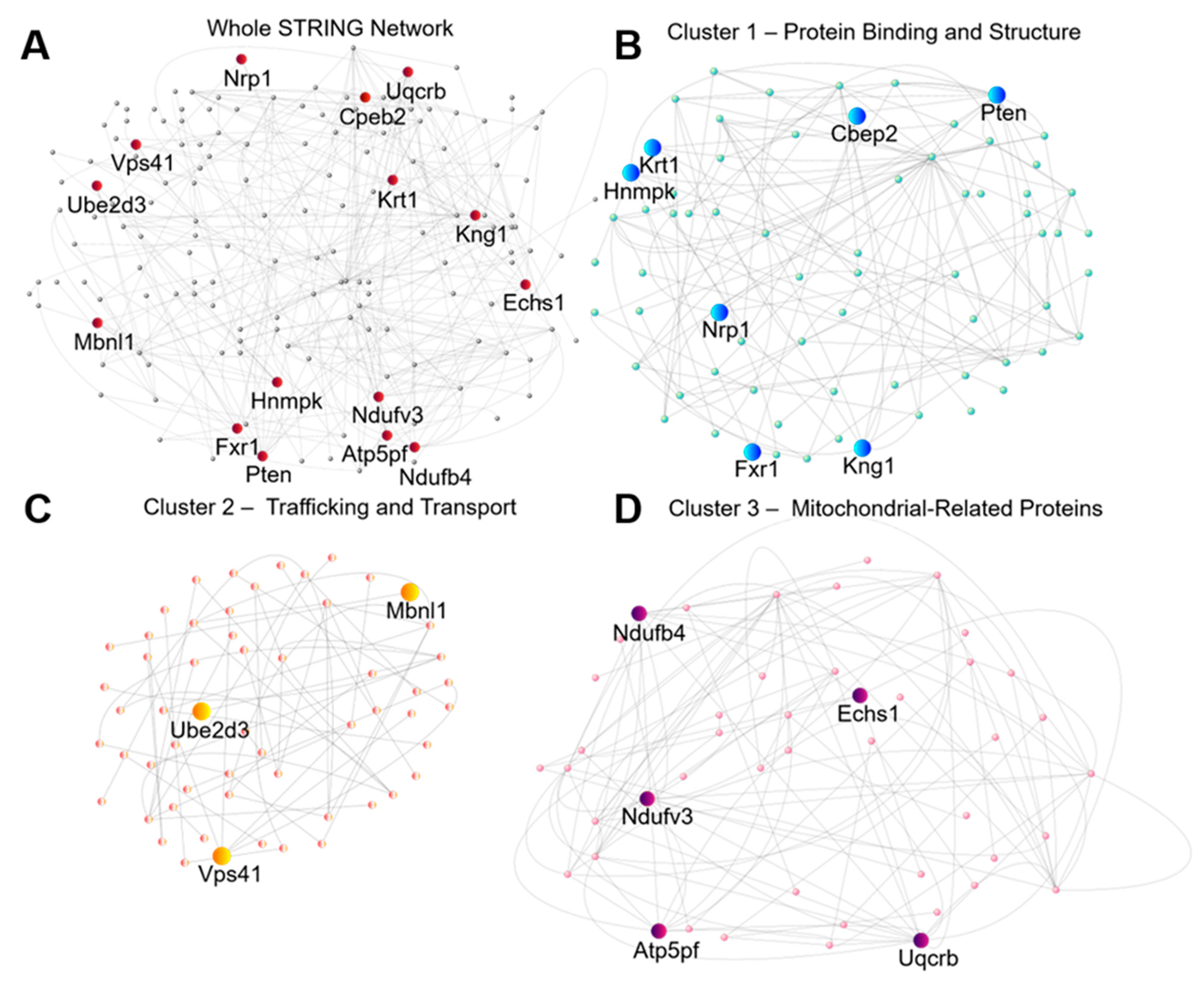
2.5. Identification of High Confidence Targets for Drug Intervention in Seizure Prevention
3. Discussion
4. Materials and Methods
4.1. Animal Experimentation
4.2. EEG Electrode Placement and Video-EEG Data Acquisition
4.3. Seizure Detection
4.4. Cortical and Hippocampal Dissection and Dissociation
4.5. Trypsin Digestion and Untargeted LC–MS/MS Proteomic Acquisition
4.6. Untargeted Proteomic Data Analysis
4.7. Methodological Approach for Predicting Cellular Metabolism
4.8. Protein Network Centrality Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zack, M.M.; Kobau, R. National and State Estimates of the Numbers of Adults and Children with Active Epilepsy—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, P.L.; Smith, G.M.; Wannamaker, B.B.; Thurman, D.J.; Pickelsimer, E.E.; Selassie, A.W. A population-based study of risk of epilepsy after hospitalization for traumatic brain injury. Epilepsia 2010, 51, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Wat, R.; Mammi, M.; Paredes, J.; Haines, J.; Alasmari, M.; Liew, A.; Lu, V.M.; Arnaout, O.; Smith, T.R.; Gormley, W.B.; et al. The Effectiveness of Antiepileptic Medications as Prophylaxis of Early Seizure in Patients with Traumatic Brain Injury Compared with Placebo or No Treatment: A Systematic Review and Meta-Analysis. World Neurosurg. 2019, 122, 433–440. [Google Scholar] [CrossRef]
- Yu, W.; Ravelo, A.; Wagner, T.H.; Phibbs, C.S.; Bhandari, A.; Chen, S.; Barnett, P.G. Prevalence and costs of chronic conditions in the VA health care system. Med. Care Res. Rev. 2003, 60, 146S–167S. [Google Scholar] [CrossRef] [PubMed]
- Hainsworth, J.B.; Johnson, A.; Godfred-Cato, S.; Smolinski, G.J.; Jorgensen-Wagers, K. The 8 January 2020 theatre ballistic missile attack on US soldiers stationed at Al Asad Air Base, Iraq: Case series using a concussion subtypes framework to approach a real-world, chaotic blast-related TBI mass casualty event. BMJ Neurol. Open 2023, 5, e000343. [Google Scholar] [CrossRef]
- Saar-Ashkenazy, R.; Naparstek, S.; Dizitzer, Y.; Zimhoni, N.; Friedman, A.; Shelef, I.; Cohen, H.; Shalev, H.; Oxman, L.; Novack, V.; et al. Neuro-psychiatric symptoms in directly and indirectly blast exposed civilian survivors of urban missile attacks. BMC Psychiatry 2023, 23, 423. [Google Scholar] [CrossRef]
- Menon, D.K.; Schwab, K.; Wright, D.W.; Maas, A.I.; The Demographics and Clinical Assessment Working Group of the International and Interagency Initiative toward Common Data Elements for Research on Traumatic Brain Injury and Psychological Health. Position statement: Definition of traumatic brain injury. Arch. Phys. Med. Rehabil. 2010, 91, 1637–1640. [Google Scholar] [CrossRef]
- Stone, J.R.; Avants, B.B.; Tustison, N.J.; Wassermann, E.M.; Gill, J.; Polejaeva, E.; Dell, K.C.; Carr, W.; Yarnell, A.M.; LoPresti, M.L.; et al. Functional and Structural Neuroimaging Correlates of Repetitive Low-Level Blast Exposure in Career Breachers. J. Neurotrauma 2020, 37, 2468–2481. [Google Scholar] [CrossRef]
- Chen, L.L.; Baca, C.B.; Choe, J.; Chen, J.W.; Ayad, M.E.; Cheng, E.M. Posttraumatic epilepsy in Operation Enduring Freedom/Operation Iraqi Freedom veterans. Mil. Med. 2014, 179, 492–496. [Google Scholar] [CrossRef][Green Version]
- Rehman, R.; Kelly, P.R.; Husain, A.M.; Tran, T.T. Characteristics of Veterans diagnosed with seizures within Veterans Health Administration. J. Rehabil. Res. Dev. 2015, 52, 751–762. [Google Scholar] [CrossRef]
- Frey, L.C. Epidemiology of posttraumatic epilepsy: A critical review. Epilepsia 2003, 44, 11–17. [Google Scholar] [CrossRef]
- Lowenstein, D.H. Epilepsy after head injury: An overview. Epilepsia 2009, 50 (Suppl. S2), 4–9. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, M.; Patel, M.Y.; Wilder, D.M.; Long, J.B.; Arun, P. Blast Exposure Dysregulates Nighttime Melatonin Synthesis and Signaling in the Pineal Gland: A Potential Mechanism of Blast-Induced Sleep Disruptions. Brain Sci. 2022, 12, 1340. [Google Scholar] [CrossRef] [PubMed]
- Agoston, D.V. Modeling the Long-Term Consequences of Repeated Blast-Induced Mild Traumatic Brain Injuries. J. Neurotrauma 2017, 34, S44–S52. [Google Scholar] [CrossRef]
- Dickerson, M.R.; Murphy, S.F.; Urban, M.J.; White, Z.; VandeVord, P.J. Chronic Anxiety- and Depression-Like Behaviors Are Associated With Glial-Driven Pathology Following Repeated Blast Induced Neurotrauma. Front. Behav. Neurosci. 2021, 15, 787475. [Google Scholar] [CrossRef]
- Bugay, V.; Bozdemir, E.; Vigil, F.A.; Chun, S.H.; Holstein, D.M.; Elliott, W.R.; Sprague, C.J.; Cavazos, J.E.; Zamora, D.O.; Rule, G.; et al. A Mouse Model of Repetitive Blast Traumatic Brain Injury Reveals Post-Trauma Seizures and Increased Neuronal Excitability. J. Neurotrauma 2020, 37, 248–261. [Google Scholar] [CrossRef] [PubMed]
- Pitkanen, A.; Immonen, R. Epilepsy related to traumatic brain injury. Neurotherapeutics 2014, 11, 286–296. [Google Scholar] [CrossRef]
- Shandra, O.; Winemiller, A.R.; Heithoff, B.P.; Munoz-Ballester, C.; George, K.K.; Benko, M.J.; Zuidhoek, I.A.; Besser, M.N.; Curley, D.E.; Edwards, G.F., 3rd; et al. Repetitive Diffuse Mild Traumatic Brain Injury Causes an Atypical Astrocyte Response and Spontaneous Recurrent Seizures. J. Neurosci. 2019, 39, 1944–1963. [Google Scholar] [CrossRef]
- Rust, M.B.; Marcello, E. Disease association of cyclase-associated protein (CAP): Lessons from gene-targeted mice and human genetic studies. Eur. J. Cell Biol. 2022, 101, 151207. [Google Scholar] [CrossRef]
- Scheffer, I.E.; Harkin, L.A.; Grinton, B.E.; Dibbens, L.M.; Turner, S.J.; Zielinski, M.A.; Xu, R.; Jackson, G.; Adams, J.; Connellan, M.; et al. Temporal lobe epilepsy and GEFS+ phenotypes associated with SCN1B mutations. Brain 2007, 130, 100–109. [Google Scholar] [CrossRef]
- Al-Hassnan, Z.N.; Kaya, N. ISCA2-Related Mitochondrial Disorder. In GeneReviews((R)); Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Braun, D.A.; Shril, S.; Sinha, A.; Schneider, R.; Tan, W.; Ashraf, S.; Hermle, T.; Jobst-Schwan, T.; Widmeier, E.; Majmundar, A.J.; et al. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med. Genet. A 2018, 176, 2460–2465. [Google Scholar] [CrossRef]
- Wang, J.; Lin, Z.J.; Liu, L.; Xu, H.Q.; Shi, Y.W.; Yi, Y.H.; He, N.; Liao, W.P. Epilepsy-associated genes. Seizure 2017, 44, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, W.B.; Vekaria, H.J.; Velmurugan, G.V.; Kalimon, O.J.; Prajapati, P.; Brown, E.; Geisler, J.G.; Sullivan, P.G. Mitochondrial Dysfunction After Repeated Mild Blast Traumatic Brain Injury Is Attenuated by a Mild Mitochondrial Uncoupling Prodrug. J. Neurotrauma 2023, 40, 2396–2409. [Google Scholar] [CrossRef]
- Schmitt, R.; Qayum, S.; Pliss, A.; Kuzmin, A.N.; Muthaiah, V.P.K.; Kaliyappan, K.; Prasad, P.N.; Mahajan, S.D. Mitochondrial Dysfunction and Apoptosis in Brain Microvascular Endothelial Cells Following Blast Traumatic Brain Injury. Cell Mol. Neurobiol. 2023, 43, 3639–3651. [Google Scholar] [CrossRef] [PubMed]
- Guilhaume-Correa, F.; Pickrell, A.M.; VandeVord, P.J. The Imbalance of Astrocytic Mitochondrial Dynamics Following Blast-Induced Traumatic Brain Injury. Biomedicines 2023, 11, 329. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Chen, M.; Chen, C.; Cui, J.; Johnson, C.E.; Cheng, J.; Wang, X.; Swerdlow, R.H.; DePalma, R.G.; Xia, W.; et al. Proteomic Analysis and Biochemical Correlates of Mitochondrial Dysfunction after Low-Intensity Primary Blast Exposure. J. Neurotrauma 2019, 36, 1591–1605. [Google Scholar] [CrossRef] [PubMed]
- King, Z.A.; Lu, J.; Drager, A.; Miller, P.; Federowicz, S.; Lerman, J.A.; Ebrahim, A.; Palsson, B.O.; Lewis, N.E. BiGG Models: A platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res. 2016, 44, D515–D522. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.S.; Mendez, R.; Fernandez, M.; Richter, J.D. CPEB and translational control by cytoplasmic polyadenylation: Impact on synaptic plasticity, learning, and memory. Mol. Psychiatry 2023, 28, 2728–2736. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Qu, Y.; Mu, D. The Regulatory Network of METTL3 in the Nervous System: Diagnostic Biomarkers and Therapeutic Targets. Biomolecules 2023, 13, 664. [Google Scholar] [CrossRef]
- Hu, T.; Chen, X.; Lu, S.; Zeng, H.; Guo, L.; Han, Y. Biological Role and Mechanism of Lipid Metabolism Reprogramming Related Gene ECHS1 in Cancer. Technol. Cancer Res. Treat. 2022, 21, 15330338221140655. [Google Scholar] [CrossRef]
- Gowthami, N.; Pursotham, N.; Dey, G.; Ghose, V.; Sathe, G.; Pruthi, N.; Shukla, D.; Gayathri, N.; Santhoshkumar, R.; Padmanabhan, B.; et al. Neuroanatomical zones of human traumatic brain injury reveal significant differences in protein profile and protein oxidation: Implications for secondary injury events. J. Neurochem. 2023, 167, 218–247. [Google Scholar] [CrossRef]
- Shen, M.; Guo, Y.; Dong, Q.; Gao, Y.; Stockton, M.E.; Li, M.; Kannan, S.; Korabelnikov, T.; Schoeller, K.A.; Sirois, C.L.; et al. FXR1 regulation of parvalbumin interneurons in the prefrontal cortex is critical for schizophrenia-like behaviors. Mol. Psychiatry 2021, 26, 6845–6867. [Google Scholar] [CrossRef]
- Knight, H.M.; Demirbugen Oz, M.; PerezGrovas-Saltijeral, A. Dysregulation of RNA modification systems in clinical populations with neurocognitive disorders. Neural Regen. Res. 2024, 19, 1256–1261. [Google Scholar] [CrossRef]
- Do Canto, A.M.; Donatti, A.; Geraldis, J.C.; Godoi, A.B.; da Rosa, D.C.; Lopes-Cendes, I. Neuroproteomics in Epilepsy: What Do We Know so Far? Front. Mol. Neurosci. 2020, 13, 604158. [Google Scholar] [CrossRef] [PubMed]
- Montes-Cano, M.A. Heterogeneous Nuclear Ribonucleoprotein K Autoantibodies in Patients who Suffered Severe Traumatic Brain Injury. SOJ Immunol. 2014, 2, 1–6. [Google Scholar] [CrossRef]
- Malik, I.; Kelley, C.P.; Wang, E.T.; Todd, P.K. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat. Rev. Mol. Cell Biol. 2021, 22, 589–607. [Google Scholar] [CrossRef] [PubMed]
- Dorsett, C.R.; McGuire, J.L.; Niedzielko, T.L.; DePasquale, E.A.; Meller, J.; Floyd, C.L.; McCullumsmith, R.E. Traumatic Brain Injury Induces Alterations in Cortical Glutamate Uptake without a Reduction in Glutamate Transporter-1 Protein Expression. J. Neurotrauma 2017, 34, 220–234. [Google Scholar] [CrossRef]
- Shevlyakov, A.D.; Kolesnikova, T.O.; de Abreu, M.S.; Petersen, E.V.; Yenkoyan, K.B.; Demin, K.A.; Kalueff, A.V. Forward Genetics-Based Approaches to Understanding the Systems Biology and Molecular Mechanisms of Epilepsy. Int. J. Mol. Sci. 2023, 24, 5280. [Google Scholar] [CrossRef]
- Wen, M.; Jin, Y.; Zhang, H.; Sun, X.; Kuai, Y.; Tan, W. Proteomic Analysis of Rat Cerebral Cortex in the Subacute to Long-Term Phases of Focal Cerebral Ischemia-Reperfusion Injury. J. Proteome Res. 2019, 18, 3099–3118. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, C.; Cao, H.; Zhang, L.; Wang, X.; Chen, S. Identification of target genes in neuroinflammation and neurodegeneration after traumatic brain injury in rats. PeerJ 2019, 7, e8324. [Google Scholar] [CrossRef]
- Kim, J.E.; Lee, D.S.; Park, H.; Kang, T.C. Src/CK2/PTEN-Mediated GluN2B and CREB Dephosphorylations Regulate the Responsiveness to AMPA Receptor Antagonists in Chronic Epilepsy Rats. Int. J. Mol. Sci. 2020, 21, 9633. [Google Scholar] [CrossRef]
- White, A.R.; Tiwari, D.; MacLeod, M.C.; Danzer, S.C.; Gross, C. PI3K isoform-selective inhibition in neuron-specific PTEN-deficient mice rescues molecular defects and reduces epilepsy-associated phenotypes. Neurobiol. Dis. 2020, 144, 105026. [Google Scholar] [CrossRef]
- Zhu, H.; Xu, H.; Ma, H.; Luo, L.; Yang, L.; Chen, F.; Qu, X.; Liu, H.; Zhang, R. LncRNA CASC2 inhibits astrocytic activation and adenosine metabolism by regulating PTEN in pentylenetetrazol-induced epilepsy model. J. Chem. Neuroanat. 2020, 105, 101749. [Google Scholar] [CrossRef]
- Walker, C.L.; Wu, X.; Liu, N.K.; Xu, X.M. Bisperoxovanadium Mediates Neuronal Protection through Inhibition of PTEN and Activation of PI3K/AKT-mTOR Signaling after Traumatic Spinal Injuries. J. Neurotrauma 2019, 36, 2676–2687. [Google Scholar] [CrossRef]
- Xiong, Y.; Cao, F.; Hu, L.; Yan, C.; Chen, L.; Panayi, A.C.; Sun, Y.; Zhou, W.; Zhang, P.; Wu, Q.; et al. miRNA-26a-5p Accelerates Healing via Downregulation of PTEN in Fracture Patients with Traumatic Brain Injury. Mol. Ther. Nucleic Acids 2019, 17, 223–234. [Google Scholar] [CrossRef]
- Gao, X.; Xiong, Y.; Li, Q.; Han, M.; Shan, D.; Yang, G.; Zhang, S.; Xin, D.; Zhao, R.; Wang, Z.; et al. Extracellular vesicle-mediated transfer of miR-21-5p from mesenchymal stromal cells to neurons alleviates early brain injury to improve cognitive function via the PTEN/Akt pathway after subarachnoid hemorrhage. Cell Death Dis. 2020, 11, 363. [Google Scholar] [CrossRef] [PubMed]
- Pearson, A.; Ortiz, C.; Eisenbaum, M.; Arrate, C.; Browning, M.; Mullan, M.; Bachmeier, C.; Crawford, F.; Ojo, J.O. Deletion of PTEN in microglia ameliorates chronic neuroinflammation following repetitive mTBI. Mol. Cell Neurosci. 2023, 125, 103855. [Google Scholar] [CrossRef] [PubMed]
- Bolte, A.C.; Shapiro, D.A.; Dutta, A.B.; Ma, W.F.; Bruch, K.R.; Kovacs, M.A.; Royo Marco, A.; Ennerfelt, H.E.; Lukens, J.R. The meningeal transcriptional response to traumatic brain injury and aging. eLife 2023, 12, e81154. [Google Scholar] [CrossRef] [PubMed]
- Bando, S.Y.; Bertonha, F.B.; Pimentel-Silva, L.R.; de Oliveira, J.G.M.; Carneiro, M.A.D.; Oku, M.H.M.; Wen, H.T.; Castro, L.H.M.; Moreira-Filho, C.A. Hippocampal CA3 transcriptional modules associated with granule cell alterations and cognitive impairment in refractory mesial temporal lobe epilepsy patients. Sci. Rep. 2021, 11, 10257. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Vizarra, E.; Zeviani, M. Mitochondrial disorders of the OXPHOS system. FEBS Lett. 2021, 595, 1062–1106. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; He, Y.; Ji, J.; Shi, Y. A Machine Learning Method for Identifying Critical Interactions Between Gene Pairs in Alzheimer’s Disease Prediction. Front. Neurol. 2019, 10, 1162. [Google Scholar] [CrossRef]
- Zhang, S.N.; Li, H.M.; Liu, Q.; Li, X.Z.; Yang, W.D.; Zhou, Y. Eucommiae Folium and Active Compounds Protect Against Mitochondrial Dysfunction-Calcium Overload in Epileptic Hippocampal Neurons Through the Hypertrophic Cardiomyopathy Pathway. Neurochem. Res. 2023, 48, 2674–2686. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, J.; Cui, H.; Li, P.; Li, H.; Wang, Y.; Tang, T. Quantitative proteomic analysis of intracerebral hemorrhage in rats with a focus on brain energy metabolism. Brain Behav. 2018, 8, e01130. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Tong, F.; Zhang, L.; Zhu, L.; Li, W.; Huang, W.; Zhao, S.; He, G.; Zhou, Y. iTRAQ-based proteomic analysis discovers potential biomarkers of diffuse axonal injury in rats. Brain Res. Bull. 2019, 153, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, L.E.; Lanko, K.; Alsagob, M.; Almass, R.; Al-Ahmadi, N.; Najafi, M.; Al-Muhaizea, M.A.; Alzaidan, H.; AlDhalaan, H.; Perenthaler, E.; et al. Bi-allelic variants in HOPS complex subunit VPS41 cause cerebellar ataxia and abnormal membrane trafficking. Brain 2021, 144, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Sheils, T.K.; Mathias, S.L.; Kelleher, K.J.; Siramshetty, V.B.; Nguyen, D.T.; Bologa, C.G.; Jensen, L.J.; Vidovic, D.; Koleti, A.; Schurer, S.C.; et al. TCRD and Pharos 2021: Mining the human proteome for disease biology. Nucleic Acids Res 2021, 49, D1334–D1346. [Google Scholar] [CrossRef] [PubMed]
- Vigil, F.A.; Belchior, H.; Bugay, V.; Bazaldua, I.I.; Stoja, A.; Dantas, D.C.; Chun, S.H.; Farmer, A.; Bozdemir, E.; Holstein, D.M.; et al. Acute Treatment with the M-Channel (K(v)7, KCNQ) Opener Retigabine Reduces the Long-Term Effects of Repetitive Blast Traumatic Brain Injuries. Neurotherapeutics 2023, 20, 853–869. [Google Scholar] [CrossRef] [PubMed]
- Arun, P.; Abu-Taleb, R.; Oguntayo, S.; Wang, Y.; Valiyaveettil, M.; Long, J.B.; Nambiar, M.P. Acute mitochondrial dysfunction after blast exposure: Potential role of mitochondrial glutamate oxaloacetate transaminase. J. Neurotrauma 2013, 30, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Arun, P.; Wei, Y.; Oguntayo, S.; Gharavi, R.; Valiyaveettil, M.; Nambiar, M.P.; Long, J.B. Repeated blast exposures cause brain DNA fragmentation in mice. J. Neurotrauma 2014, 31, 498–504. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, C.; Mitkin, V.V.; Lankford, M.F.; Li, J.; Zuo, Z.; Meyer, C.H.; Goyne, C.P.; Ahlers, S.T.; Stone, J.R.; et al. Comprehensive Characterization of Cerebrovascular Dysfunction in Blast Traumatic Brain Injury Using Photoacoustic Microscopy. J. Neurotrauma 2019, 36, 1526–1534. [Google Scholar] [CrossRef]
- Gama Sosa, M.A.; De Gasperi, R.; Pryor, D.; Perez Garcia, G.S.; Perez, G.M.; Abutarboush, R.; Kawoos, U.; Hogg, S.; Ache, B.; Janssen, W.G.; et al. Low-level blast exposure induces chronic vascular remodeling, perivascular astrocytic degeneration and vascular-associated neuroinflammation. Acta Neuropathol. Commun. 2021, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Gama Sosa, M.A.; De Gasperi, R.; Pryor, D.; Perez Garcia, G.S.; Perez, G.M.; Abutarboush, R.; Kawoos, U.; Hogg, S.; Ache, B.; Sowa, A.; et al. Late chronic local inflammation, synaptic alterations, vascular remodeling and arteriovenous malformations in the brains of male rats exposed to repetitive low-level blast overpressures. Acta Neuropathol. Commun. 2023, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Kaur, C.; Singh, J.; Lim, M.K.; Ng, B.L.; Yap, E.P.; Ling, E.A. Ultrastructural changes of macroglial cells in the rat brain following an exposure to a non-penetrative blast. Ann. Acad. Med. Singap. 1997, 26, 27–29. [Google Scholar] [PubMed]
- Robinson, J.L.; Kocabas, P.; Wang, H.; Cholley, P.E.; Cook, D.; Nilsson, A.; Anton, M.; Ferreira, R.; Domenzain, I.; Billa, V.; et al. An atlas of human metabolism. Sci. Signal 2020, 13, eaaz1482. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.S.; Bai, Y.M.; Hsu, J.W.; Huang, K.L.; Ko, N.Y.; Tsai, C.K.; Yeh, T.C.; Chu, H.T.; Tsai, S.J.; Chen, T.J.; et al. The Risk of Epilepsy after Long-term Proton Pump Inhibitor Therapy. Seizure 2021, 87, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Poulos, R.C.; Cai, Z.; Robinson, P.J.; Reddel, R.R.; Zhong, Q. Opportunities for pharmacoproteomics in biomarker discovery. Proteomics 2023, 23, e2200031. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Sajja, V.S.; Vandevord, P.J.; Lee, Y.W. Blast induces oxidative stress, inflammation, neuronal loss and subsequent short-term memory impairment in rats. Neuroscience 2013, 253, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Shandra, O.; Robel, S. Inducing Post-Traumatic Epilepsy in a Mouse Model of Repetitive Diffuse Traumatic Brain Injury. J. Vis. Exp. JoVE 2020, e60360. [Google Scholar] [CrossRef]
- Ng, R.H.; Lee, J.W.; Baloni, P.; Diener, C.; Heath, J.R.; Su, Y. Constraint-Based Reconstruction and Analyses of Metabolic Models: Open-Source Python Tools and Applications to Cancer. Front. Oncol. 2022, 12, 914594. [Google Scholar] [CrossRef]
- Ivan, G.; Grolmusz, V. When the Web meets the cell: Using personalized PageRank for analyzing protein interaction networks. Bioinformatics 2011, 27, 405–407. [Google Scholar] [CrossRef]
| # | Reaction ID | Optimized Flux Equation (mol hr-1gDw-1) | Dysregulated Flux Equation (mol hr-1gDw-1) |
|---|---|---|---|
| 1 | CYOO3mi | 7.92Hm (−3.187) + O2m (0.7967) + 4FocytCm (−3.187) ⇌ 1.96 H2Om (1.561) + 4.0FicytCm (3.187) + 0.02O2sm (0.015) + 4Hi (3.187) | 7.92Hm (−13.940) + O2m (−1.761) + 4.0FocytCm (−7.043) ⇌ +1.96H2Om (3.451) + 4.0FicytCm (7.043) + 0.02O2sm (0.035) + 4.0Hi (0.917) |
| 2 | CYOR_u10mi | 2.0Hm (−0.907) + 2.0FicytCm (−0.907) +q10H2m (−0.453) ⇌ 2.0FocytCm (0.907) + q10m (0.453) + 4.0Hi (1.813) | 2.0Hm (−1.360) + 2.0FicytCm (−1.360) + q10H2m (-0.680) ⇌ 2.0FocytCm (1.360) + q10m (0.680) + 4.0Hi (1.152) |
| 3 | SUCD1m | FADm (−0.578) + Succm (−0.578) ⇌FADH2m (0.578) + Fumm (0.578) | FADm (−0.752) + Succm (−0.752) ⇌ FADH2m (0.752) + Fumm (0.752) |
| 4 | ECOAH9m | H2Om (0.791) + 2mb2CoAm (0.791) ⇌ 3hmbCoAm (−0.791) | H2Om (1.107) + 2mb2CoAm (1.107) ⇌ 3hmbCoAm (−1.107) |
| 5 | AKDGm | Akgm (−0.366) + CoAm (−0.366) + NADm (−0.366) ⇌ CO2m (0.366) + NADHm (0.366) + SucCoAm (0.366) | Akgm (−0.513) + CoAm (−0.513) + NADm (−0.513) ⇌ CO2m (0.513) + NADHm (0.513) + SucCoAm (0.513) |
| 6 | GMPR | GMPc (0.027) + 2.0Hc (0.054) + NADPHc (0.027) ⇌ impc (-0.027) + NADPc (−0.027) + NH4c (−0.027) | GMPc (0.109) + 2.0Hc (0.218) + NADPHc (0.109) ⇌ impc (−0.109) + NADPc (−0.109) + NH4c (−0.109) |
| 7 | PGK | 3pgc (0.449) + ATPc (0.449) ⇌ 13dpgc (−0.449) + ADPc (−0.449) | 3pgc (0.674) + ATPc (0.674) ⇌ 13dpgc (−0.674) + ADPc (−0.674) |
| Gene Symbol | Degree # | Betweenness Centrality $ | Closeness Centrality % | Stress Centrality & | Adj p Value | Protein Function * |
|---|---|---|---|---|---|---|
| Cpeb2 | 2 | 0.166667 | 0.292683 | 26 | 8.78 × 10−2 | mRNA binding, regulating cytoplasmic polyadenylation of mRNA |
| Echs1 | 2 | 0.054054 | 0.355769 | 180 | 4.43 × 10−6 | Catalyzes CoA intermediates to L-3-hydroxyacyl-CoAs in mitochondrial fatty acid beta-oxidation pathway |
| Fxr1 | 2 | 0.30303 | 0.375 | 48 | 2.67 × 10−16 | RNA binding protein which shuttles between the nucleus and cytoplasm to bind to polyribosomes |
| Hnrnpk | 4 | 0.712121 | 0.48 | 118 | 2.28 × 10−6 | RNA binding protein which complexes with heterogeneous nuclear RNA and influence pre-mRNA processing and metabolism |
| Mbnl1 | 2 | 0.166667 | 0.352941 | 26 | 1.93 × 10−2 | C3H-Zinc finger binding protein which modulates external splicing of pre-mRNAs |
| Ndufb4 | 18 | 0.001661 | 0.506849 | 32 | 2.67 × 10−16 | Non-catalytic subunit of multisubunit NADH:oxidoreductase |
| Ndufv3 | 15 | 0.000374 | 0.474359 | 8 | 2.55 × 10−6 | Subunit of multisubunit NADH:oxidoreductase; function unknown |
| Pten | 3 | 0.7 | 0.625 | 14 | 1.17 × 10−3 | Tumor suppressor which negatively regulates AKT/PKB signaling. Longer isoform may play a role in energy metabolism in the mitochondria |
| Ube2d3 | 3 | 0.166667 | 0.315789 | 38 | 3.04 × 10−2 | Member of the E2 ubiquitin conjugating enzyme family, which functions in ubiquitination tumor suppressor protein p53 |
| Uqcrb | 19 | 0.006285 | 0.569231 | 114 | 2.07 × 10−3 | Binds ubiquinone and participates in electron transfer while bound to ubiquinone |
| Gene Symbol | Protein Function * |
|---|---|
| Atp5pf | F6 subunit of the F0 complex, required for F1 and F0 interactions |
| Hnrnpk # | RNA binding protein which complexes with heterogeneous nuclear RNA and influence pre-mRNA processing and metabolism |
| Kng1 | Uses alternative splicing to generate high and low molecular weight kininogens |
| Krt1 | Member of the keratin family, which are expressed during simple and stratified epithelial cell differentiation |
| Ndufb4 # | Non-catalytic subunit of multisubunit NADH:oxidoreductase |
| Nrp1 | Cell surface receptor involved in the development of the cardiovascular system, in angiogenesis, in the formation of certain neuronal circuits |
| Pten # | Tumor supressor which negatively regulates AKT/PKB signaling. Longer isoform may play a role in energy metabolism in the mitochondria |
| Ube2d3 # | Member of the E2 ubiquitin conjugating enzyme family, which functions in ubiquitination tumor suppressor protein p53 |
| Uqcrb # | Binds ubiquinone and participates in electron transfer while bound to ubiquinone |
| Vps41 | Plays a role in transport and fusion of vacuoles from the Golgi |
| Gene Symbol | Supporting Epilepsy Literature | Supporting TBI Literature | Biomarker Validation Database | |||||
|---|---|---|---|---|---|---|---|---|
| DisGenNet | * CTD | ** Other | ||||||
| Cpeb2 | [29] | [29,30] | n | n | y | n | n | n |
| Echs1 | [31] | [32] | n | n | y | n | y | n |
| Fxr1 | [33] | [34] | n | n | y | y | n | n |
| Hnrnpk | [35] | [36] | n | n | y | n | n | n |
| Mbnl1 | [37] | [38] | n | n | y | n | n | n |
| Ndufb4 | [39] | [32] | n | n | y | n | n | n |
| Ndufv3 | [39] | [40,41] | n | n | n | n | n | n |
| Pten | [39,42,43,44] | [45,46,47,48] | y | n | y | y | y | n |
| Ube2d3 | [49] | n | n | y | n | n | n | |
| Uqcrb | [50,51] | [32,52] | n | n | n | n | n | |
| Additional nodes weighted through PageRank | ||||||||
| Atp5pf | [53] | [32] | n | N | y | n | n | n |
| Kng1 | [54] | [32] | y | N | y | n | n | n |
| Krt1 | [55] | n | N | n | n | n | n | |
| Nrp1 | [39] | n | N | y | n | y | n | |
| Vps41 | [56] | [56] | n | N | y | n | n | n |
| Gene Symbol | Drug/Chemical | Drug/Chemical, Group | Database | Species | Protein Details | |||
|---|---|---|---|---|---|---|---|---|
| HU | MO | pPTMs | TDL | DTO Fam | ||||
| Cpeb2 | Bisphenol A | Experimental | CTD | y | y | p, me, ac, gl, ub, hy | Tbio | Nab |
| Echs1 | Hexanoyl-CoA | Experimental | DrugBank | y | y | p, gl, ub, ac, | Tbio | E |
| Fxr1 | Valproic Acid | Experimental | CTD | y | y | p, ac, pc, su, ub, gl, me, pa | Tbio | Nab |
| Hnrnpk | Phenethyl isothiocyanate, Artenimol | Investigational, Approved | DrugBank | y | y | gl, ub, ac, p, pc, hy, me, su | Tbio | E |
| Mbnl1 | Bisphenol A | Experimental | CTD | y | y | pc, gl, p, ub, ac, hy, pa | Tbio | Nab |
| Ndufb4 | Metformin | Approved | Pharos | y | y | me, su, p, gl, ub, ac | Tclin | E |
| Ndufv3 | Metformin | Approved | Pharos | y | y | p, ub, gl, hy, su, ac | Tclin | na |
| Pten | Bisphenol A | Experimental | CTD | y | y | ac, ub, me, su, gl | Tbio | E |
| Ube2d3 | Bisphenol A | Experimental | CTD | y | y | me, p, gl | Tchem | E |
| Uqcrb | Azoxystrobin | Experimental | DrugBank | y | y | ac, ub, p | Tbio | E |
| Additional nodes weighted through PageRank | ||||||||
| Atp5pf | Bisphenol A | Experimental | CTD | y | y | p, ub, ac, pc, su | Tbio | T |
| Kng1 | Copper | Approved | DrugBank | y | y | pc, p, gl, ub, hy, me, ac | Tbio | EM |
| Krt1 | Copper | Approved | DrugBank | y | y | p, pc, pa, me, gl, ac, ub, su | Tbio | E |
| Nrp1 | PEGAPTANIB | Approved | CTD | y | y | su, p, ub, ac, gl, me, pc, hy, | Tchem | na |
| Vps41 | Valproic Acid | Experimental | CTD | y | y | pc, p, su, gl, ub, ac, hy, | Tbio | T |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Browning, J.L.; Wilson, K.A.; Shandra, O.; Wei, X.; Mahmutovic, D.; Maharathi, B.; Robel, S.; VandeVord, P.J.; Olsen, M.L. Applying Proteomics and Computational Approaches to Identify Novel Targets in Blast-Associated Post-Traumatic Epilepsy. Int. J. Mol. Sci. 2024, 25, 2880. https://doi.org/10.3390/ijms25052880
Browning JL, Wilson KA, Shandra O, Wei X, Mahmutovic D, Maharathi B, Robel S, VandeVord PJ, Olsen ML. Applying Proteomics and Computational Approaches to Identify Novel Targets in Blast-Associated Post-Traumatic Epilepsy. International Journal of Molecular Sciences. 2024; 25(5):2880. https://doi.org/10.3390/ijms25052880
Chicago/Turabian StyleBrowning, Jack L., Kelsey A. Wilson, Oleksii Shandra, Xiaoran Wei, Dzenis Mahmutovic, Biswajit Maharathi, Stefanie Robel, Pamela J. VandeVord, and Michelle L. Olsen. 2024. "Applying Proteomics and Computational Approaches to Identify Novel Targets in Blast-Associated Post-Traumatic Epilepsy" International Journal of Molecular Sciences 25, no. 5: 2880. https://doi.org/10.3390/ijms25052880
APA StyleBrowning, J. L., Wilson, K. A., Shandra, O., Wei, X., Mahmutovic, D., Maharathi, B., Robel, S., VandeVord, P. J., & Olsen, M. L. (2024). Applying Proteomics and Computational Approaches to Identify Novel Targets in Blast-Associated Post-Traumatic Epilepsy. International Journal of Molecular Sciences, 25(5), 2880. https://doi.org/10.3390/ijms25052880






