Upregulation of Notch Signaling and Cell-Differentiation Inhibitory Transcription Factors in Stable Chronic Obstructive Pulmonary Disease Patients
Abstract
1. Introduction
2. Results
2.1. Clinical Characteristics of Subjects Providing Bronchial Biopsies and Lung Parenchyma
2.2. Immunohistochemistry for Notch Signaling Proteins in the Bronchial Epithelium of Bronchial Biopsies
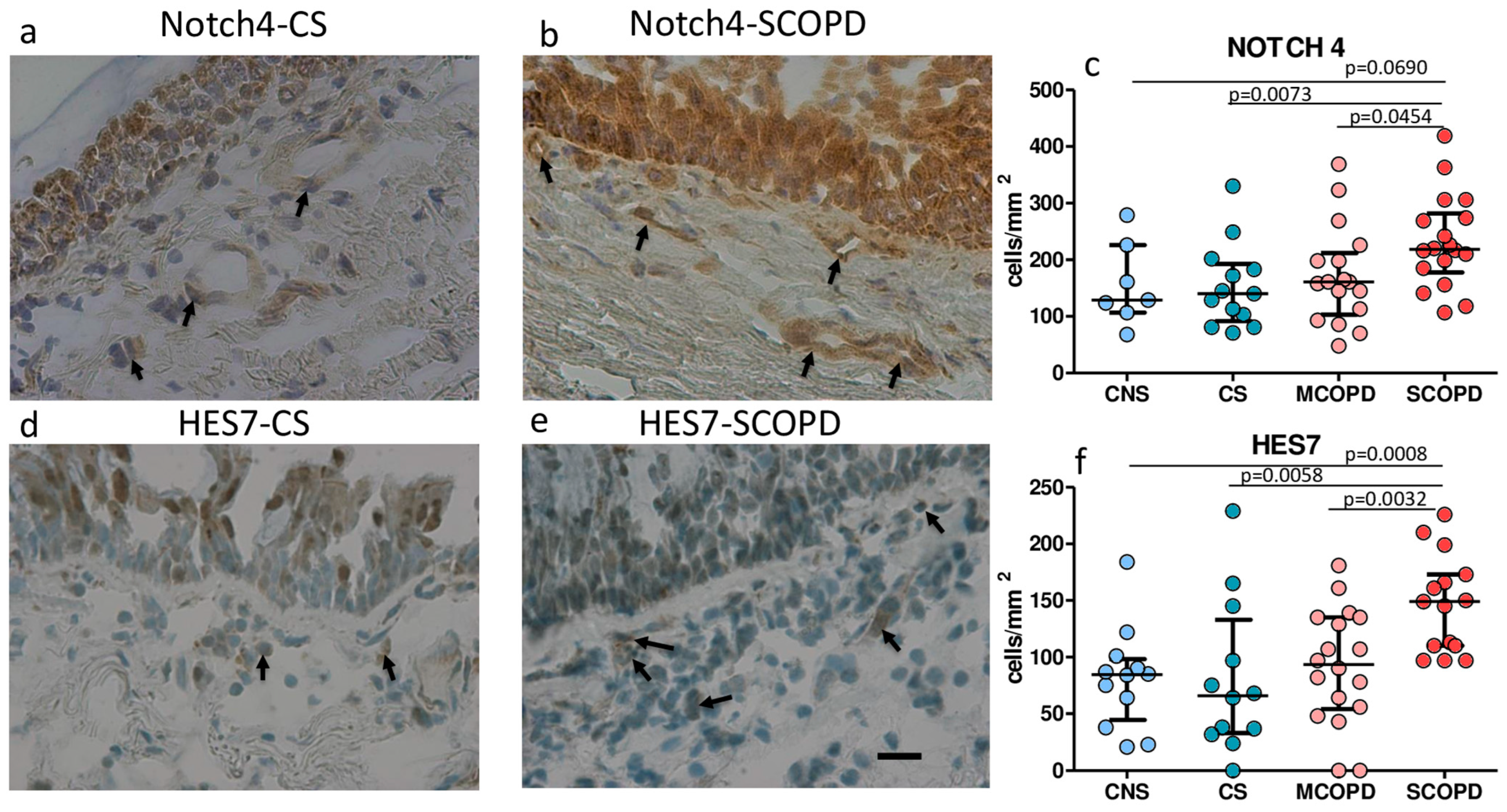
2.3. Immunohistochemistry for Notch Signaling Proteins in the Bronchial Lamina Propria of Bronchial Biopsies
2.4. Immunohistochemistry for Notch Signaling Proteins in the Peripheral Airways and Lung Parenchyma
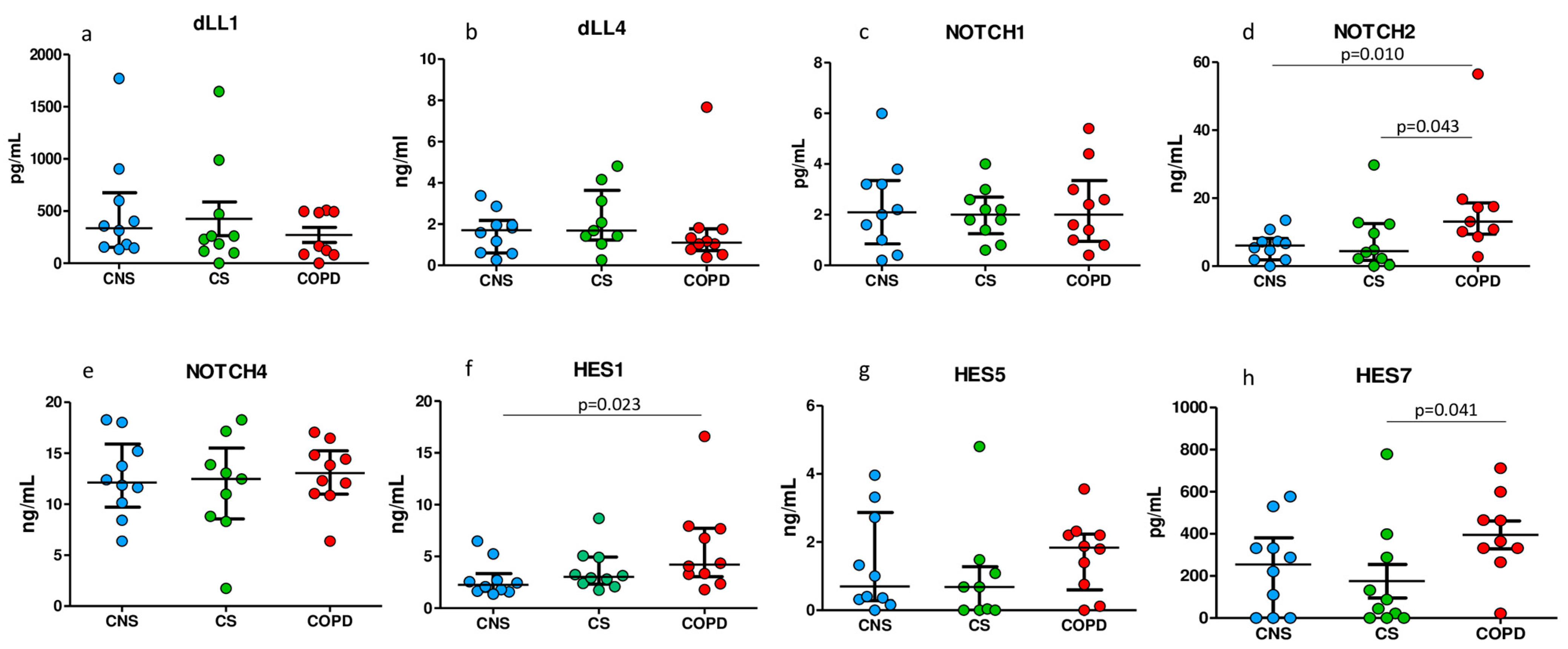
2.5. ELISA Tests for Notch Signaling Proteins in Homogenized Peripheral Lung Tissue
2.6. Gene Expression Level of Notch Signaling Molecules in Bronchial Rings and Lung Parenchyma
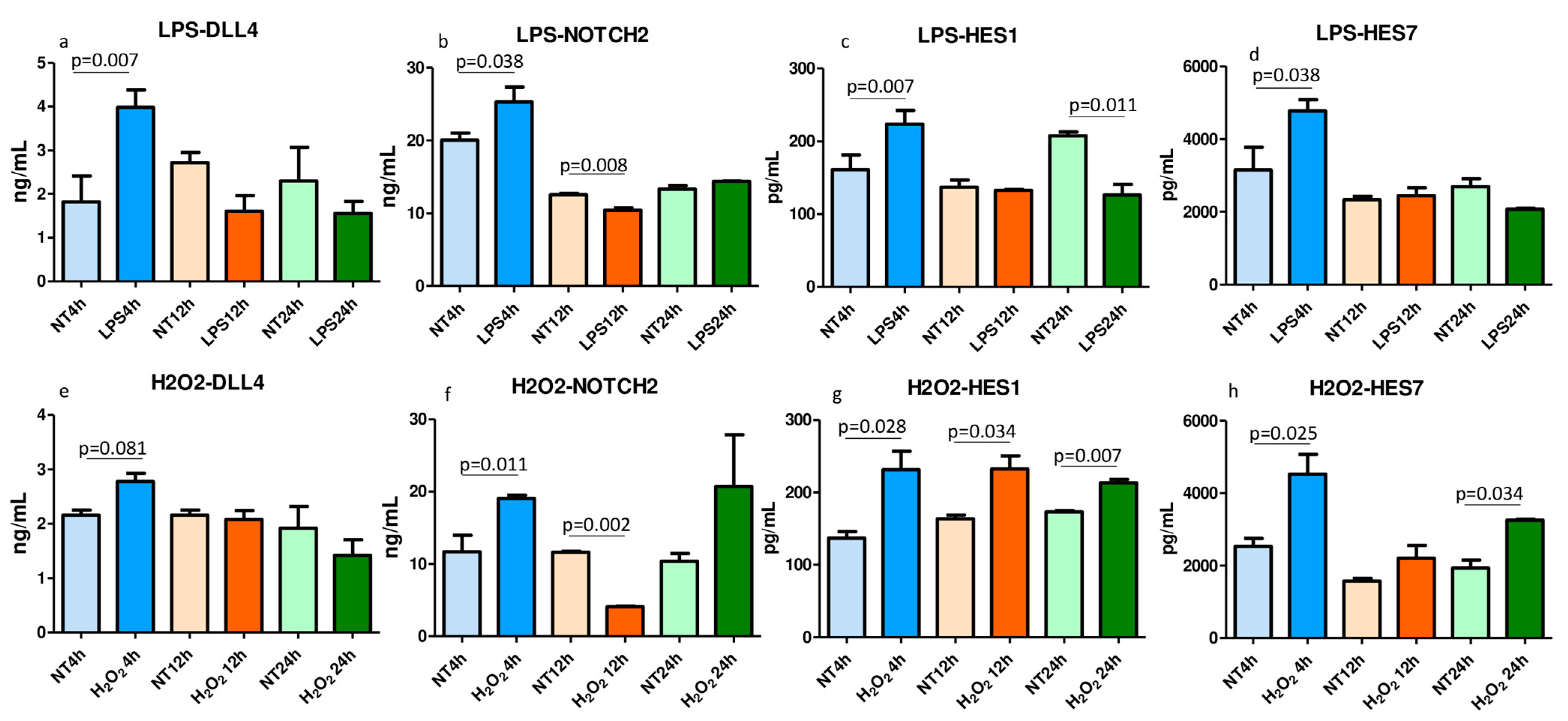
2.7. ELISA Tests for Notch Signaling Molecules in the LPS- and H2O2-Treated and Nontreated Lysed 16HBE Cells
3. Discussion
4. Materials and Methods
4.1. Subjects
4.2. Lung Function Tests and Volumes
4.3. Fiberoptic Bronchoscopy, Collection and Processing of Bronchial Biopsies
4.4. Collection and Processing of the Peripheral Lung Tissue
4.5. Immunohistochemistry on OCT-Embedded Bronchial Biopsies
4.6. Immunohistochemistry in Human Peripheral Lung Tissue
4.7. Scoring System for Immunohistochemistry in the Bronchial Biopsies
4.8. Scoring System for Immunohistochemistry in the Peripheral Lung Tissue
4.9. ELISA Tests in the Peripheral Lung Tissue Homogenates
4.10. RNA Extraction and Sequencing from Bronchial Rings and Lung Specimens
4.11. Data Analysis of RNA-Seq Data
4.12. Cell Culture and Treatments
4.13. ELISA Tests in the Cell Lysates of LPS- and H2O2-Treated and Untreated 16HBE Cells
4.14. Statistical Analysis Applied to Functional and Morphological Data
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| FEV1 | Forced expiratory volume in one second |
| FVC | Forced vital capacity |
| MCOPD | Mild/moderate chronic obstructive pulmonary disease |
| SCOPD | Severe/very severe chronic obstructive pulmonary disease |
| CS | Control smokers with normal lung function |
| CNS | Control nonsmokers |
| Notch | Notch receptor |
| NICD | Notch-intracellular domain |
| DLL | Delta-like ligand |
| Jagged | Jag ligand |
| RBP-JK | Notch intracellular domain |
| HES | Hairy and enhancer of split |
| HEY2 | Hairy/enhancer-of-split related with YRPW motif protein 2 |
| HEYL | Hairy/enhancer-of-split related with YRPW motif-like protein |
References
- Bi, P.; Kuang, S. Notch signaling as a novel regulator of metabolism. Trends Endocrinol. Metab. 2015, 26, 248–255. [Google Scholar] [CrossRef]
- Collins, B.J.; Kleeberger, W.; Ball, D.W. Notch in lung development and lung cancer. Semin. Cancer Biol. 2004, 14, 357–364. [Google Scholar] [CrossRef]
- Hansson, E.M.; Lendahl, U.; Chapman, G. Notch signaling in development and disease. Semin. Cancer Biol. 2004, 14, 320–328. [Google Scholar] [CrossRef]
- Li, X.; Shu, R.; Filippatos, G.; Uhal, B.D. Apoptosis in lung injury and remodeling. J. Appl. Physiol. (1985) 2004, 97, 1535–1542. [Google Scholar] [CrossRef]
- Zong, D.; Ouyang, R.; Li, J.; Chen, Y.; Chen, P. Notch signaling in lung diseases: Focus on Notch1 and Notch3. Ther. Adv. Respir. Dis. 2016, 10, 468–484. [Google Scholar] [CrossRef]
- Hogg, J.C.; McDonough, J.E.; Suzuki, M. Small airway obstruction in COPD: New insights based on micro-CT imaging and MRI imaging. Chest 2013, 143, 1436–1443. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.C.; McDonough, J.E.; Sanchez, P.G.; Cooper, J.D.; Coxson, H.O.; Elliott, W.M.; Naiman, D.; Pochettino, M.; Horng, D.; Gefter, W.B.; et al. Micro-computed tomography measurements of peripheral lung pathology in chronic obstructive pulmonary disease. Proc. Am. Thorac. Soc. 2009, 6, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, D.M.; Martinez, F.J.; Marchetti, N.; Galbán, C.J.; Hatt, C.; Meldrum, C.A.; Dass, C.; Tanabe, N.; Reddy, R.M.; Lagstein, A.; et al. Noninvasive Imaging Biomarker Identifies Small Airway Damage in Severe Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2019, 200, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Vasilescu, D.M.; Hackett, T.L.; Martinez, F.J.; Curtis, J.L.; Hogg, J.C.; Han, M.K. Reply to Janssen and Wouters: Loss of Alveolar Attachments as a Pathomechanistic Link between Small Airway Disease and Emphysema. Am. J. Respir. Crit. Care Med. 2020, 201, 879–880. [Google Scholar] [CrossRef] [PubMed]
- Tilley, A.E.; Harvey, B.G.; Heguy, A.; Hackett, N.R.; Wang, R.; O’Connor, T.P.; Crystal, R.G. Down-regulation of the notch pathway in human airway epithelium in association with smoking and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2009, 179, 457–466. [Google Scholar] [CrossRef]
- Zong, D.; Li, J.; Cai, S.; He, S.; Liu, Q.; Jiang, J.; Chen, S.; Long, Y.; Chen, Y.; Chen, P.; et al. Notch1 regulates endothelial apoptosis via the ERK pathway in chronic obstructive pulmonary disease. Am. J. Physiol. Cell Physiol. 2018, 315, C330–C340. [Google Scholar] [CrossRef]
- Boucherat, O.; Chakir, J.; Jeannotte, L. The loss of Hoxa5 function promotes Notch-dependent goblet cell metaplasia in lung airways. Biol. Open 2012, 1, 677–691. [Google Scholar] [CrossRef] [PubMed]
- Guseh, J.S.; Bores, S.A.; Stanger, B.Z.; Zhou, Q.; Anderson, W.J.; Melton, D.A.; Rajagopal, J. Notch signaling promotes airway mucous metaplasia and inhibits alveolar development. Development 2009, 136, 1751–1759. [Google Scholar] [CrossRef] [PubMed]
- Tsao, P.N.; Wei, S.C.; Wu, M.F.; Huang, M.T.; Lin, H.Y.; Lee, M.C.; Lin, K.M.; Wang, I.J.; Kaartinen, V.; Yang, L.T.; et al. Notch signaling prevents mucous metaplasia in mouse conducting airways during postnatal development. Development 2011, 138, 3533–3543. [Google Scholar] [CrossRef]
- Hellström, M.; Phng, L.K.; Hofmann, J.J.; Wallgard, E.; Coultas, L.; Lindblom, P.; Alva, J.; Nilsson, A.K.; Karlsson, L.; Gaiano, N.; et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 2007, 445, 776–780. [Google Scholar] [CrossRef]
- Siekmann, A.F.; Lawson, N.D. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature 2007, 445, 781–784. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.S.; Tsaryk, R.; Lange, M.; Wisniewski, L.; Moore, J.C.; Lawson, N.D.; Wojciechowska, K.; Schnittler, H.; Siekmann, A.F. Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat. Cell Biol. 2017, 19, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Osborne, B.A.; Minter, L.M. Notch signalling during peripheral T-cell activation and differentiation. Nat. Rev. Immunol. 2007, 7, 64–75. [Google Scholar] [CrossRef]
- Ito, T.; Schaller, M.; Hogaboam, C.M.; Standiford, T.J.; Sandor, M.; Lukacs, N.W.; Chensue, S.W.; Kunkel, S.L. TLR9 regulates the mycobacteria-elicited pulmonary granulomatous immune response in mice through DC-derived Notch ligand delta-like 4. J. Clin. Investig. 2009, 119, 33–46. [Google Scholar] [CrossRef]
- Ito, T.; Allen, R.M.; Carson WF 4th Schaller, M.; Cavassani, K.A.; Hogaboam, C.M.; Lukacs, N.W.; Matsukawa, A.; Kunkel, S.L. The critical role of Notch ligand Delta-like 1 in the pathogenesis of influenza A virus (H1N1) infection. PLoS Pathog. 2011, 7, e1002341. [Google Scholar] [CrossRef]
- Ito, T.; Connett, J.M.; Kunkel, S.L.; Matsukawa, A. Notch system in the linkage of innate and adaptive immunity. J. Leukoc. Biol. 2012, 92, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.; Rosani, U.; Levra, S.; Gnemmi, I.; Brun, P.; Maniscalco, M.; D’Anna, S.E.; Carriero, V.; Bertolini, F.; Ricciardolo, F.L.M. Bone Morphogenic Proteins and Their Antagonists in the Lower Airways of Stable COPD Patients. Biology 2023, 12, 1304. [Google Scholar] [CrossRef]
- Di Stefano, A.; Sangiorgi, C.; Gnemmi, I.; Casolari, P.; Brun, P.; Ricciardolo, F.L.M.; Contoli, M.; Papi, A.; Maniscalco, P.; Ruggeri, P.; et al. TGF-β Signaling Pathways in Different Compartments of the Lower Airways of Patients With Stable COPD. Chest 2018, 153, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Auderset, F.; Schuster, S.; Fasnacht, N.; Coutaz, M.; Charmoy, M.; Koch, U.; Favre, S.; Wilson, A.; Trottein, F.; Alexander, J.; et al. Notch signaling regulates follicular helper T cell differentiation. J. Immunol. 2013, 191, 2344–2350. [Google Scholar] [CrossRef] [PubMed]
- Radtke, F.; Fasnacht, N.; Macdonald, H.R. Notch signaling in the immune system. Immunity 2010, 32, 14–27. [Google Scholar] [CrossRef]
- Vallese, D.; Ricciardolo, F.L.; Gnemmi, I.; Casolari, P.; Brun, P.; Sorbello, V.; Capelli, A.; Cappello, F.; Cavallesco, G.N.; Papi, A.; et al. Phospho-p38 MAPK expression in COPD patients and asthmatics and in challenged bronchial epithelium. Respiration 2015, 89, 329–342. [Google Scholar] [CrossRef]
- Di Stefano, A.; Ricciardolo, F.L.M.; Caramori, G.; Adcock, I.M.; Chung, K.F.; Barnes, P.J.; Brun, P.; Leonardi, A.; Andò, F.; Vallese, D.; et al. Bronchial inflammation and bacterial load in stable COPD is associated with TLR4 overexpression. Eur. Respir. J. 2017, 49, 1602006. [Google Scholar] [CrossRef]
- Skokos, D.; Nussenzweig, M.C. CD8- DCs induce IL-12-independent Th1 differentiation through Delta 4 Notch-like ligand in response to bacterial LPS. J. Exp. Med. 2007, 204, 1525–1531. [Google Scholar] [CrossRef]
- Schaller, M.A.; Neupane, R.; Rudd, B.D.; Kunkel, S.L.; Kallal, L.E.; Lincoln, P.; Lowe, J.B.; Man, Y.; Lukacs, N.W. Notch ligand Delta-like 4 regulates disease pathogenesis during respiratory viral infections by modulating Th2 cytokines. J. Exp. Med. 2007, 204, 2925–2934. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Cozens, A.L.; Yezzi, M.J.; Kunzelmann, K.; Ohrui, T.; Chin, L.; Eng, K.; Finkbeiner, W.E.; Widdicombe, J.H.; Gruenert, D.C. CFTR expression and chloride secretion in polarized immortal human bronchial epithelial cells. Am. J. Respir. Cell Mol. Biol. 1994, 10, 38–47. [Google Scholar] [CrossRef] [PubMed]

| Groups | n | Age (years) | M/F | Pack Years | Ex/Current Smokers | FEV1 (% pred) pre-β2 | FEV1 (% pred) post-β2 | FEV1/FVC (%) |
|---|---|---|---|---|---|---|---|---|
| Control nonsmokers | 11 | 64 ± 11 | 10/1 | 0 | 0 | 111 ± 16 | ND | 84 ± 9 |
| Control smokers with normal lung function | 13 | 60 ± 8 | 9/4 | 40 ± 9 | 2/11 | 101 ± 14 | ND | 81 ± 5 |
| COPD grades I and II (mild/moderate) | 18 | 68 ± 9 | 15/3 | 46 ± 6.3 | 6/12 | 66 ± 13 # | 70 ± 12 | 58 ± 9 # |
| COPD grades III and IV (severe/very severe) | 16 | 66 ± 12 | 11/5 | 61 ± 11 | 13/3 | 36 ± 7 #& | 41 ± 6 | 44 ± 9 #& |
| Groups | N° | Age (year) | M/F | Ex/Current Smokers | Pack Years | FEV1 (% pred) pre-β2 | FEV1 (% pred) post-β2 | FEV1/FVC (%) |
|---|---|---|---|---|---|---|---|---|
| Control nonsmokers | 10 | 71 ± 3 | 5/5 | --- | --- | 116 ± 5.1 | ND | 80 ± 1.6 |
| Control smokers | 10 | 68 ± 1.7 | 7/3 | 7/3 | 38 ± 4.3 | 96 ± 2.6 | ND | 74 ± 1.4 |
| Patients with COPD | 13 | 70 ± 1.3 | 11/2 | 11/2 | 61 ± 8.8 | 74 ± 3.3 # | 81 ± 3.7 | 62 ± 1.3 # |
| Healthy Nonsmokers | Healthy Smokers | Mild/Moderate COPD | Severe COPD | Kruskal–Wallis p-Value | |
|---|---|---|---|---|---|
| Bronchial Epithelium (score 0–3) | Median (Range) | Median (Range) | Median (Range) | Median (Range) | |
| JAGGED 1 | 0 (0–0.25) | 0 (0–0) | 0 (0–0) | 0 (0–0) | 0.947 |
| JAGGED 2 | 1.25 (0.5–2.75) | 2 (0.5–3) | 1.13 (0.5–3) | 1.75 (0.5–2.75) | 0.244 |
| DELTA 1 | 0 (0–0) | 0 (0–1) | 0 (0–0) | 0 (0–0) | 0.987 |
| DELTA 4 | 0 (0–0.5) | 0 (0–0.25) | 0 (0–0.5) | 0 (0–0.5) | 0.869 |
| NOTCH 1 | 0 (0–0) | 0 (0–0.25) | 0 (0–0) | 0 (0–0.5) | 0.978 |
| NOTCH 2 | 0 (0–1) | 0 (0–1) | 0 (0–2) | 0 (0–1) | 0.884 |
| NOTCH 3 | 0 (0–0.25) | 0 (0–1) | 0 (0–0.25) | 0 (0–0.5) | 0.980 |
| NOTCH 4 | 1.25 (0–2.5) | 2 (1–5) | 1.88 (0.5–2.5) | 2 (1–3) | 0.360 |
| RBP-JK | 0.13 (0–0.75) | 0 (0–0.25) | 0 (0–0.5) | 0 (0–0.5) | 0.600 |
| HES-1 | 2 (1–2.5) | 1.5 (0.25–3) | 1.75 (0.75–2.5) | 1.75 (0.5–2.5) | 0.695 |
| HES-3 | 2.37 (1.5–2.75) | 2.25 (0.75–3) | 2 (1.5–2.5) | 2.5 (1.25–3) | 0.547 |
| HES-5 | 3 (2.5–3) | 2.5 (1.75–3) | 2.5 (2–3) | 2.87 (1.75–3) | 0.187 |
| HES-6 | 0.25 (0–0.75) | 0.25 (0–0.75) | 0 (0–1.25) | 0.25 (0–1.5) | 0.783 |
| HES-7 | 0.5 (0.25–1.5) | 0.5 (0.25–1.25) | 0.5 (0–1.75) | 1 (0.25–1.75) | 0.179 |
| HEY2 | 1.5 (1–2) | 1 (0.75–2) | 1.25 (0.75–2) | 1.5 (0.5–2.25) | 0.404 |
| HEYL | 0.62 (0–1) | 0.5 (0–1) | 0.5 (0–1) | 0.5 (0–0.75) | 0.815 |
| Bronchial Lamina propria (cells/mm2) | Median (Range) | Median (Range) | Median (Range) | Median (Range) | |
| JAGGED 1 | 0 (0–8) | 2.5 (0–12) | 4 (0–25) | 0 (0–16) | 0.547 |
| JAGGED 2 | 21 (8–206) | 64 (8–166) | 37 (5–202) | 62 (5–223) | 0.470 |
| DELTA 1 | 0 (0–6) | 0 (0–12) | 0 (0–7) | 3 (0–19) | 0.584 |
| DELTA 4 | 0 (0–11) | 0 (0–11) | 6 (0–113)& | 1.5 (0–38) | 0.207 |
| NOTCH 1 | 0 (0–16) | 0 (0–6) | 0 (0–9) | 0 (0–51) | 0.577 |
| NOTCH 2 | 11 (0–116) | 9 (0–113) | 9 (0–172) | 5.5 (0–123) | 0.795 |
| NOTCH 3 | 1 (0–9) | 2.5 (0–11) | 3.5 (0–55) | 7 (0–48) | 0.494 |
| NOTCH 4 | 129 (68–279) | 140 (71–330) | 161 (48–369) | 218 (107–419) &£ | 0.035 |
| RBP-JK | 12 (0–89) | 4 (0–72) | 10 (0–165) & | 4 (0–124) | 0.266 |
| HES-1 | 89 (43–113) | 69 (8–161) | 79 (28–206) | 131 (12–306) | 0.176 |
| HES-3 | 0 (0–7) | 0 (0–6) | 0 (0–8) | 0 (0–5) | 0.955 |
| HES-5 | 281 (226–363) | 267 (173–393) | 293 (193–403) | 317 (225–363) & | 0.214 |
| HES-6 | 0 (0–74) | 0 (0–204) | 0 (0–358) | 2 (0–38) | 0.662 |
| HES-7 | 84 (21–184) | 66 (0–229) | 97 (0–181) | 149 (97–226) *&£ | 0.003 |
| HEY2 | 218 (153–252) | 193 (97–306) | 221 (116–339) | 213 (2–336) | 0.890 |
| HEYL | 24 (0–97) | 43 (16–134) | 51 (11–139) | 50 (11–107) | 0.773 |
| Localization | Control Nonsmokers | Control Smokers | COPD Patients | Kruskal–Wallis p-Value |
|---|---|---|---|---|
| Bronchiolar Epithelium (score 0–3) | ||||
| JAGGED 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.928 |
| JAGGED 2 | 0.5 (0.62) | 0.81 (0.92) | 0.5 (0.65) | 0.528 |
| DELTA 1 | 0.0 (0.19) | 0.02 (0.25) | 0.0 (0.0) | 0.374 |
| DELTA 4 | 0.17 (0.37) | 0.37 (0.49) | 0.15 (0.38) | 0.425 |
| NOTCH 1 | 0.0 (0.0) | 0.26 (0.20) * | 0.10 (0.25) * | 0.0075 |
| NOTCH 2 | 0.0 (0.15) | 0.0 (0.20) | 0.0 (0.0) | 0.585 |
| NOTCH 3 | 0.37 (0.36) | 0.27 (0.35) | 0.0 (0.41) | 0.224 |
| NOTCH 4 | 1.6 (1.0) | 1.58 (0.75) | 1.5 (0.67) | 0.909 |
| RBP-JK | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.654 |
| HES-1 | 0.5 (1.31) | 1.37 (1.12) | 1.5 (1.40) | 0.229 |
| HES-3 | 1.0 (1.25) | 0.68 (1.34) | 0.54 (1.0) | 0.626 |
| HES-5 | 2.0 (0.55) | 1.75 (1.07) | 2.0 (0.35) | 0.649 |
| HES-6 | 0.0 (0.14) | 0.0 (0.0) | 0.0 (0.0) | 0.658 |
| HES-7 | 0.62 (0.77) | 0.93 (1.32) | 0.91 (0.59) | 0.797 |
| HEY2 | 0.60 (1.02) | 0.25 (0.90) | 0.50 (1.13) | 0.992 |
| HEYL | 0.33 (0.40) | 0.57 (0.63) | 0.25 (0.35) | 0.579 |
| Bronchiolar Lamina Propria (score 0–3) | ||||
| JAGGED 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| JAGGED 2 | 0.0 (0.12) | 0.0 (0.10) | 0.0 (0.0) | 0.770 |
| DELTA 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| DELTA 4 | 0.0 (0.0) | 0.0 (0.25) | 0.0 (0.0) | 0.210 |
| NOTCH 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.882 |
| NOTCH 2 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| NOTCH 3 | 0.0 (0.0) | 0.0 (0.0) | 0. (0.0) | 0.919 |
| NOTCH 4 | 0.85 (0.50) | 0.87 (0.48) | 1.0 (0.2) | 0.290 |
| RBP-JK | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-1 | 0.05 (0.70) | 0.75 (0.97) | 0.5 (0.95) | 0.324 |
| HES-3 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.744 |
| HES-5 | 1.0 (0.25) | 0.5 (0.50) | 1.0 (0.50) | 0.708 |
| HES-6 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-7 | 0.5 (0.25) | 0.5 (0.72) | 0.5 (0.21) | 0.889 |
| HEY2 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HEYL | 0.0 (0.10) | 0.20 (0.40) | 0.0 (0.12) | 0.385 |
| Alveolar Macrophages (score 0–3) | ||||
| JAGGED 1 | 0.0 (0.0) | 0.10 (0.24) | 0.0 (0.11) | 0.103 |
| JAGGED 2 | 0.50 (0.88) | 1.25 (1.41) | 0.50 (0.55) | 0.298 |
| DELTA 1 | 0.0 (0.25) | 0.87 (1.5) | 0.05 (0.93) | 0.148 |
| DELTA 4 | 0.16 (0.5) | 0.81 (0.70) * | 0.62 (1.0) | 0.028 |
| NOTCH 1 | 0.37 (0.83) | 0.72 (0.70) | 0.50 (0.95) | 0.553 |
| NOTCH 2 | 0.0 (0.10) | 0.37 (0.40) * | 0.0 (0.11) & | 0.025 |
| NOTCH 3 | 0.10 (0.20) | 0.33 (1.25) | 0.17 (0.45) | 0.107 |
| NOTCH 4 | 1.25 (0.90) | 1.5 (0.29) | 1.5 (0.50) | 0.817 |
| RBP-JK | 0.0 (0.0) | 0.25 (0.50) | 0.0 (0.50) | 0.481 |
| HES-1 | 0.75 (0.50) | 1.0 (0.45) | 0.85 (0.65) | 0.283 |
| HES-3 | 0.0 (0.09) | 0.41 (0.88) * | 0.0 (0.30) | 0.051 |
| HES-5 | 2.0 (0.62) | 2.0 (1.12) | 1.75 (0.50) | 0.717 |
| HES-6 | 0.0 (0.37) | 0.25 (0.32) | 0.0 (0.54) | 0.606 |
| HES-7 | 1.31 (1.05) | 1.60 (1.20) | 1.75 (0.79) | 0.568 |
| HEY2 | 0.55 (0.87) | 0.74 (0.66) | 0.50 (0.87) | 0.621 |
| HEYL | 0.25 (0.40) | 0.75 (0.56) | 0.66 (0.62) | 0.073 |
| Alveolar Septa (score 0–3) | ||||
| JAGGED 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| JAGGED 2 | 0.0 (0.50) | 0.07 (0.50) | 0.0 (0.22) | 0.886 |
| DELTA 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| DELTA 4 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.991 |
| NOTCH 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.810 |
| NOTCH 2 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.903 |
| NOTCH 3 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.927 |
| NOTCH 4 | 1.0 (0.46) | 1.2 (0.50) | 1.37 (0.50) | 0.550 |
| RBP-JK | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-1 | 0.5 (0.62) | 0.55 (0.40) | 0.50 (0.58) | 0.973 |
| HES-3 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-5 | 1.0 (0.50) | 1.0 (0.81) | 0.75 (0.50) | 0.532 |
| HES-6 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-7 | 0.50 (0.33) | 0.55 (0.62) | 0.75 (0.33) | 0.813 |
| HEY2 | 0.0 (0.25) | 0.0 (0.10) | 0.0 (0.0) | 0.663 |
| HEYL | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.878 |
| Lung vessels-(score 0–3) | ||||
| JAGGED 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| JAGGED 2 | 0.5 (0.81) | 0.75 (0.75) | 0.25 (0.50) | 0.182 |
| DELTA 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| DELTA 4 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.911 |
| NOTCH 1 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| NOTCH 2 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.903 |
| NOTCH 3 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.918 |
| NOTCH 4 | 1.5 (0.62) | 1.5 (0.62) | 1.5 (0.50) | 0.712 |
| RBP-JK | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-1 | 0.62 (0.75) | 0.79 (0.87) | 0.50 (1.17) | 0.997 |
| HES-3 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | 0.907 |
| HES-5 | 1.0 (0.50) | 1.0 (0.62) | 1.0 (0.50) | 0.555 |
| HES-6 | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
| HES-7 | 0.50 (0.43) | 0.35 (1.00) | 0.75 (0.60) | 0.936 |
| HEY2 | 0.0 (0.50) | 0.0 (0.5) | 0.0 (0.0) | 0.665 |
| HEYL | 0.0 (0.0) | 0.0 (0.0) | 0.0 (0.0) | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Stefano, A.; Gnemmi, I.; Rosani, U.; Maniscalco, M.; D’Anna, S.E.; Brun, P.; Carriero, V.; Bertolini, F.; Balbi, B.; Ricciardolo, F.L.M. Upregulation of Notch Signaling and Cell-Differentiation Inhibitory Transcription Factors in Stable Chronic Obstructive Pulmonary Disease Patients. Int. J. Mol. Sci. 2024, 25, 3287. https://doi.org/10.3390/ijms25063287
Di Stefano A, Gnemmi I, Rosani U, Maniscalco M, D’Anna SE, Brun P, Carriero V, Bertolini F, Balbi B, Ricciardolo FLM. Upregulation of Notch Signaling and Cell-Differentiation Inhibitory Transcription Factors in Stable Chronic Obstructive Pulmonary Disease Patients. International Journal of Molecular Sciences. 2024; 25(6):3287. https://doi.org/10.3390/ijms25063287
Chicago/Turabian StyleDi Stefano, Antonino, Isabella Gnemmi, Umberto Rosani, Mauro Maniscalco, Silvestro Ennio D’Anna, Paola Brun, Vitina Carriero, Francesca Bertolini, Bruno Balbi, and Fabio Luigi Massimo Ricciardolo. 2024. "Upregulation of Notch Signaling and Cell-Differentiation Inhibitory Transcription Factors in Stable Chronic Obstructive Pulmonary Disease Patients" International Journal of Molecular Sciences 25, no. 6: 3287. https://doi.org/10.3390/ijms25063287
APA StyleDi Stefano, A., Gnemmi, I., Rosani, U., Maniscalco, M., D’Anna, S. E., Brun, P., Carriero, V., Bertolini, F., Balbi, B., & Ricciardolo, F. L. M. (2024). Upregulation of Notch Signaling and Cell-Differentiation Inhibitory Transcription Factors in Stable Chronic Obstructive Pulmonary Disease Patients. International Journal of Molecular Sciences, 25(6), 3287. https://doi.org/10.3390/ijms25063287







