Absolute Quantification of Selected microRNAs Expression in Endometrial Cancer by Digital PCR
Abstract
1. Introduction
1.1. Endometrial Cancer
- -
- POLE mutation (POLEmut group) (Polymerase Epsilon) (7%);
- -
- Microsatellite instability (MSI group) which results from mismatch repair deficiency (MMRd) (28%);
- -
- High somatic copy number changes (driven by the TP53 mutation, also called the p53abn group) (26%);
- -
- Low copy number without defined molecular profile, no specific molecular profile (NSMP group) (39%) [5].
1.2. MicroRNA in Control of Gene Expression
1.3. The Aim of Study
2. Results
- -
- G1—highly differentiated cancer (<5% of solid tissue)—30 patients;
- -
- G2—moderately differentiated cancer (6–50% of the solid part)—47 patients;
- -
- G3—poorly differentiated cancer (>50% of the solid tissue)—12 patients.
- -
- 18.5–24.9—normal weight (14 patients);
- -
- 25–29.9—overweight (43 patients);
- -
- 30 and above means obesity (54 patients).
3. Discussion
4. Materials and Methods
4.1. Tissues Samples
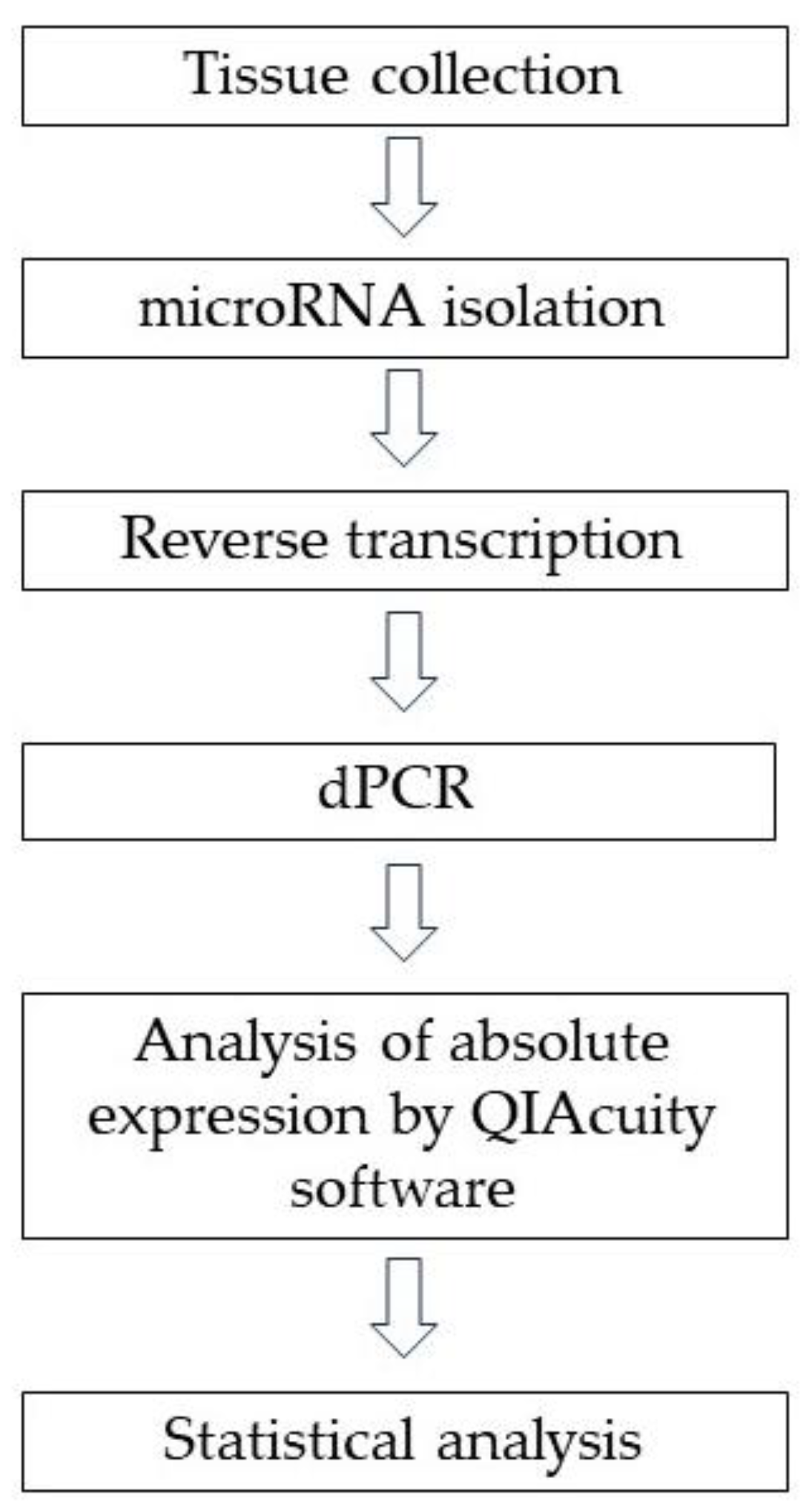
4.2. MiRNA Isolation from Tissue Samples
4.3. Reverse Transcriptase Reaction and dPCR Method
4.4. Statistical Analysis
5. Conclusions
Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Makker, V.; MacKay, H.; Ray-Coquard, I.; Levine, D.A.; Westin, S.N.; Aoki, D.; Oaknin, A. Endometrial cancer. Nat. Rev. Dis. Primers 2021, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Yen, T.T.; Wang, T.L.; Fader, A.N.; Shih, I.M.; Gaillard, S. Molecular Classification and Emerging Targeted Therapy in Endometrial Cancer. Int. J. Gynecol. Pathol. 2020, 39, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; Benz, C.C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Stavropoulos, A.; Varras, M.; Vasilakaki, T.; Varra, V.K.; Tsavari, A.; Varra, F.N.; Nonni, A.; Kavantzas, N.; Lazaris, A.C. Expression of p53 and PTEN in human primary endometrial carcinomas: Clinicopathological and immunohistochemical analysis and study of their concomitant expression. Oncol. Lett. 2019, 17, 4575–4589. [Google Scholar] [CrossRef]
- Konopka, B.; Paszko, Z.; Janiec-Jankowska, A.; Goluda, M. Assessment of the quality and frequency of mutations occurrence in PTEN gene in endometrial carcinomas and hyperplasias. Cancer Lett. 2002, 178, 43–51. [Google Scholar] [CrossRef]
- Gbelcová, H.; Gergely, L.; Šišovský, V.; Straka, Ľ.; Böhmer, D.; Pastoráková, A.; Sušienková, K.; Repiská, V.; Korbeľ, M.; Danihel, Ľ.; et al. PTEN mutations as predictive marker for the high-grade endometrial cancer development in slovak women. Physiol. Res. 2022, 71, S125–S135. [Google Scholar] [CrossRef]
- Khatami, F.; Shahriari, S.; Aminimoghaddam, S.; Klashami, Z.N.; Farahani, M.S.; Teimoori-Toolabi, L.; Amoli, M.M.; Asadi, M.; Rashidi, B.H. PTEN promoter methylation and expression in endometrial cancer tissues. Epigenomics 2023, 15, 507–516. [Google Scholar] [CrossRef]
- Wang, C.; Li, Q.; He, Y. MicroRNA-21-5p promotes epithelial to mesenchymal transition by targeting SRY-box 17 in endometrial cancer. Oncol. Rep. 2020, 43, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Stampoliou, A.; Arapantoni-Dadioti, P.; Pavlakis, K. Epigenetic mechanisms in endometrial cancer. J. BUON 2016, 21, 301–306. [Google Scholar] [PubMed]
- Favier, A.; Rocher, G.; Larsen, A.K.; Delangle, R.; Uzan, C.; Sabbah, M.; Castela, M.; Duval, A.; Mehats, C.; Canlorbe, G. MicroRNA as Epigenetic Modifiers in Endometrial Cancer: A Systematic Review. Cancers 2021, 13, 1137. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009, 4, 199–227. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yan, L.; Zhao, X.; Li, C.; Fu, Y. microRNA-21 overexpression contributes to cell proliferation by targeting PTEN in endometrioid endometrial cancer. Oncol. Lett. 2012, 4, 1290–1296. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Su, J.; Ma, Z.; Wu, Y.; Ma, H. lncRNA NBAT1 Inhibits Cell Metastasis and Promotes Apoptosis in Endometrial Cancer by Sponging miR-21-5p to Regulate PTEN. Comput. Math. Methods Med. 2022, 2022, 9304392. [Google Scholar] [CrossRef]
- Li, Q.; Li, B.; Wei, S.; He, Z.; Huang, X.; Wang, L.; Xia, Y.; Xu, Z.; Li, Z.; Wang, W.; et al. Exosomal miR-21-5p derived from gastric cancer promotes peritoneal metastasis via mesothelial-to-mesenchymal transition. Cell Death Dis. 2018, 9, 854. [Google Scholar] [CrossRef]
- Tang, J.; Li, X.; Cheng, T.; Wu, J. miR-21-5p/SMAD7 axis promotes the progress of lung cancer. Thorac. Cancer 2021, 12, 2307–2313. [Google Scholar] [CrossRef]
- He, Q.; Ye, A.; Ye, W.; Liao, X.; Qin, G.; Xu, Y.; Yin, Y.; Luo, H.; Yi, M.; Xian, L.; et al. Cancer-secreted exosomal miR-21-5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis. 2021, 12, 576. [Google Scholar] [CrossRef]
- Liu, M.; Mo, F.; Song, X.; He, Y.; Yuan, Y.; Yan, J.; Yang, Y.; Huang, J.; Zhang, S. Exosomal hsa-miR-21-5p is a biomarker for breast cancer diagnosis. PeerJ 2021, 9, e12147. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhang, Y.; Mu, J.; Yang, D.; Gu, X.; Zhang, J. Exosomal miR-21-5p contributes to ovarian cancer progression by regulating CDK6. Hum. Cell 2021, 34, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Greene, S.B.; Gunaratne, P.H.; Hammond, S.M.; Rosen, J.M. A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci. 2010, 123, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Lee, E.J.; Gusev, Y.; Schmittgen, T.D. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005, 33, 5394–5403. [Google Scholar] [CrossRef]
- Wang, X.; Tang, S.; Le, S.Y.; Lu, R.; Rader, J.S.; Meyers, C.; Zheng, Z.M. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 2008, 3, e2557. [Google Scholar] [CrossRef] [PubMed]
- Gottardo, F.; Liu, C.G.; Ferracin, M.; Calin, G.A.; Fassan, M.; Bassi, P.; Sevignani, C.; Byrne, D.; Negrini, M.; Pagano, F.; et al. Micro-RNA profiling in kidney and bladder cancers. Urol. Oncol. 2007, 25, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Markou, A.; Tsaroucha, E.G.; Kaklamanis, L.; Fotinou, M.; Georgoulias, V.; Lianidou, E.S. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin. Chem. 2008, 54, 1696–1704. [Google Scholar] [CrossRef] [PubMed]
- Karaayvaz, M.; Zhang, C.; Liang, S.; Shroyer, K.R.; Ju, J. Prognostic significance of miR-205 in endometrial cancer. PLoS ONE 2012, 7, e35158. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, H. miR-205 inhibits cell growth by targeting AKT-mTOR signaling in progesterone-resistant endometrial cancer Ishikawa cells. Oncotarget 2017, 8, 28042–28051. [Google Scholar] [CrossRef]
- Mutter, G.L. PTEN, a protean tumor suppressor. Am. J. Pathol. 2001, 158, 1895–1898. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, L.; Geng, J.; Chen, Z.; Cui, X. MiR-205-5p Functions as a Tumor Suppressor in Gastric Cancer Cells through Downregulating FAM84B. J. Oncol. 2022, 2022, 8267891. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Xu, Y.; Yao, Y.; Jiang, S. miR-205-5p contributes to paclitaxel resistance and progression of endometrial cancer by downregulating FOXO1. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 5, 43–57. [Google Scholar] [CrossRef]
- Xin, W.; Zhao, S.; Han, X.; Zhao, P.; Yu, H.; Gao, X.; Li, P.; Wu, Q.; Ding, J.; Hua, K. lncRNA LA16c-313D11.11 modulates the development of endometrial cancer by binding to and inhibiting microRNA-205-5p function and indirectly increasing PTEN activity. Int. J. Oncol. 2020, 57, 355–363. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Liu, X.; Ding, J.; Zhao, J.; Zhou, Y.; Wu, Q.; Hua, K. Long non-coding RNA derived miR-205-5p modulates human endometrial cancer by targeting PTEN. Am. J. Transl. Res. 2015, 7, 2433–2441. [Google Scholar] [PubMed]
- Liu, B.; Che, Q.; Qiu, H.; Bao, W.; Chen, X.; Lu, W.; Li, B.; Wan, X. Elevated MiR-222-3p promotes proliferation and invasion of endometrial carcinoma via targeting ERα. PLoS ONE 2014, 9, e87563. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Li, Y.; Alvero, A.; Li, J.; Wu, Q.; Xiao, Q.; Peng, Y.; Hu, Y.; Li, X.; Yan, W.; et al. MicroRNA-222-3p/GNAI2/AKT axis inhibits epithelial ovarian cancer cell growth and associates with good overall survival. Oncotarget 2016, 7, 80633–80654. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, X. MiR-222-3p Promotes Cell Proliferation and Inhibits Apoptosis by Targeting PUMA (BBC3) in Non-Small Cell Lung Cancer. Technol. Cancer Res. Treat. 2020, 19, 1533033820922558. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Wdowiak, P.; Paszkowski, T.; Maciejewski, R. Selection and validation of endogenous controls for microRNA expression studies in endometrioid endometrial cancer tissues. Gynecol. Oncol. 2013, 130, 588–594. [Google Scholar] [CrossRef]
- Bignotti, E.; Calza, S.; Tassi, R.A.; Zanotti, L.; Bandiera, E.; Sartori, E.; Odicino, F.E.; Ravaggi, A.; Todeschini, P.; Romani, C. Identification of stably expressed reference small non-coding RNAs for microRNA quantification in high-grade serous ovarian carcinoma tissues. J. Cell. Mol. Med. 2016, 20, 2341–2348. [Google Scholar] [CrossRef]
- Egidi, M.G.; Cochetti, G.; Guelfi, G.; Zampini, D.; Diverio, S.; Poli, G.; Mearini, E. Stability Assessment of Candidate Reference Genes in Urine Sediment of Prostate Cancer Patients for miRNA Applications. Dis. Markers 2015, 2015, 973597. [Google Scholar] [CrossRef]
- Lawlor, H.; Meunier, A.; McDermott, N.; Lynch, T.H.; Marignol, L. Identification of suitable endogenous controls for gene and miRNA expression studies in irradiated prostate cancer cells. Tumour Biol. 2015, 36, 6019–6028. [Google Scholar] [CrossRef] [PubMed]
- Jurcevic, S.; Olsson, B.; Klinga-Levan, K. Validation of suitable endogenous control genes for quantitative PCR analysis of microRNA gene expression in a rat model of endometrial cancer. Cancer Cell Int. 2013, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Lou, G.; Ma, N.; Xu, Y.; Jiang, L.; Yang, J.; Wang, C.; Jiao, Y.; Gao, X. Differential distribution of U6 (RNU6-1) expression in human carcinoma tissues demonstrates the requirement for caution in the internal control gene selection for microRNA quantification. Int. J. Mol. Med. 2015, 36, 1400–1408. [Google Scholar] [CrossRef] [PubMed]
- Kulinczak, M.; Sromek, M.; Panek, G.; Zakrzewska, K.; Lotocka, R.; Szafron, L.M.; Chechlinska, M.; Siwicki, J.K. Endometrial Cancer-Adjacent Tissues Express Higher Levels of Cancer-Promoting Genes than the Matched Tumors. Genes 2022, 13, 1611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, J.; Hao, J.; Shi, Z.; Wang, Y.; Han, L.; Yu, S.; You, Y.; Jiang, T.; Wang, J.; et al. High level of miR-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Stinson, S.; Lackner, M.R.; Adai, A.T.; Yu, N.; Kim, H.J.; O’Brien, C.; Spoerke, J.; Jhunjhunwala, S.; Boyd, Z.; Januario, T.; et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci. Signal. 2011, 4, ra41. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Zeng, J.J.; Wang, W.; Wu, C.T.; Lei, S.T.; Li, G.X. MicroRNA-221 controls CDKN1C/P57 expression in human colorectal carcinoma. Chin. J. Gastrointest. Surg. 2011, 14, 279–283. [Google Scholar]
- Sato, K.; Miyamoto, M.; Takano, M.; Tsuda, H. MicroRNA-21 expression in cancer cells is an independent biomarker of progression-free survival of endometrioid endometrial carcinoma. Virchows Arch. 2021, 479, 883–891. [Google Scholar] [CrossRef]
- Bouziyane, A.; Lamsisi, M.; Benaguida, H.; Benhessou, M.; El Kerroumi, M.; Ennaji, M.M. Diagnostic Value of MicroRNA 21 in Endometrial Cancer and Benign Lesions and its Differential Expression with Clinicopathological Parameters. Microrna 2021, 10, 146–152. [Google Scholar] [CrossRef]
- Braun, M.M.; Overbeek-Wager, E.A.; Grumbo, R.J. Diagnosis and Management of Endometrial Cancer. Am. Fam. Phys. 2016, 93, 468–474. [Google Scholar]
- Kumar, S.; Vijayan, M.; Bhatti, J.S.; Reddy, P.H. MicroRNAs as Peripheral Biomarkers in Aging and Age-Related Diseases. Prog. Mol. Biol. Transl. Sci. 2017, 146, 47–94. [Google Scholar] [CrossRef] [PubMed]
- Van Arsdale, A.; Miller, D.T.; Kuo, D.Y.; Isani, S.; Sanchez, L.; Nevadunsky, N.S. Association of obesity with survival in patients with endometrial cancer. Gynecol. Oncol. 2019, 154, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Purdie, D.M.; Green, A.C. Epidemiology of endometrial cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2001, 15, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Donkers, H.; Hirschfeld, M.; Weiß, D.; Erbes, T.; Jaeger, M.; Pijnenborg, J.M.A.; Bekkers, R.; Galaal, K.; ENITEC-Consortium. Usefulness of microRNA detection in the diagnostics of endometrial cancer. Acta Obstet. Gynecol. Scand. 2021, 100, 1148–1154. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, F.; Catellani, C.; Sartori, C.; Lazzeroni, P.; Amarri, S.; Street, M.E. Obesity, Insulin Resistance, and Colorectal Cancer: Could miRNA Dysregulation Play A Role? Int. J. Mol. Sci. 2019, 20, 2922. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.M.; Afonso, M.B.; Simão, A.L.; Islam, T.; Gaspar, M.M.; O’Rourke, C.J.; Lewinska, M.; Andersen, J.B.; Arretxe, E.; Alonso, C.; et al. miR-21-5p promotes NASH-related hepatocarcinogenesis. Liver Int. 2023, 43, 2256–2274. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Lu, W.; Huang, Y.; He, J.; Wang, Q.; Zheng, X.; Wang, Z. SNORD15B and SNORA5C: Novel Diagnostic and Prognostic Biomarkers for Colorectal Cancer. BioMed Res. Int. 2022, 2022, 8260800. [Google Scholar] [CrossRef] [PubMed]
- Rapti, S.M.; Kontos, C.K.; Papadopoulos, I.N.; Scorilas, A. High miR-96 levels in colorectal adenocarcinoma predict poor prognosis, particularly in patients without distant metastasis at the time of initial diagnosis. Tumour Biol. 2016, 37, 11815–11824. [Google Scholar] [CrossRef]
- Masè, M.; Grasso, M.; Avogaro, L.; D’Amato, E.; Tessarolo, F.; Graffigna, A.; Denti, M.A.; Ravelli, F. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci. Rep. 2017, 7, 41127. [Google Scholar] [CrossRef]
| Study Group, N = 111 1 | Control Group, N = 19 1 | p-Value 2 | |
|---|---|---|---|
| Age | 61 (39–88) | 48 (42–92) | <0.001 |
| First period | 13 (10–18) | 14 (11–17) | 0.4 |
| Last menstrual period | 51 (37–60) | 48 (42–61) | 0.043 |
| Birth | 2 (0–7) | 2 (0–3) | 0.2 |
| Cesarean section | 0 (0–3) | 0 (0–3) | 0.5 |
| Miscarriages | 0 (0–3) | 0 (0–4) | 0.2 |
| BMI | 29.50 (21.30–50.20) | 25.39 (20.08–34.19) | <0.001 |
| Hypertension | 0.002 | ||
| No | 48 (43%) | 16 (84%) | |
| Yes | 63 (57%) | 3 (16%) | |
| DM | 0.042 | ||
| No | 90 (81%) | 19 (100%) | |
| Yes | 21 (19%) | 0 (0%) | |
| Hypothyroidism | 0.024 | ||
| No | 88 (79%) | 19 (100%) | |
| Yes | 23 (21%) | 0 (0%) | |
| Hyperthyroidism | >0.9 | ||
| No | 108 (97%) | 19 (100%) | |
| Yes | 3 (2.7%) | 0 (0%) |
| Absolute Expression (Copies/µL) | Study Group N = 111 1 | Control Group N = 19 1 | p-Value 2 |
|---|---|---|---|
| miR-21-5p | 698,576 (56.96–2,858,160) | 938,736 (118,688–3,424,496) | 0.2 |
| miR-205-5p | 4009.2 (64.08–611,824) | 181.52 (32.5–7336.80) | <0.001 |
| miR-222-3p | 69,752 (819.2–315 144) | 112,168 (35,104–421,912) | 0.013 |
| U6 | 19,390.4 (1034.72–3,321,321) | 47,264 (9926.4–119,536) | 0.074 |
| SNORD48 | 79 760 (665.6–581,552) | 214,400 (37,168–394,592) | <0.001 |
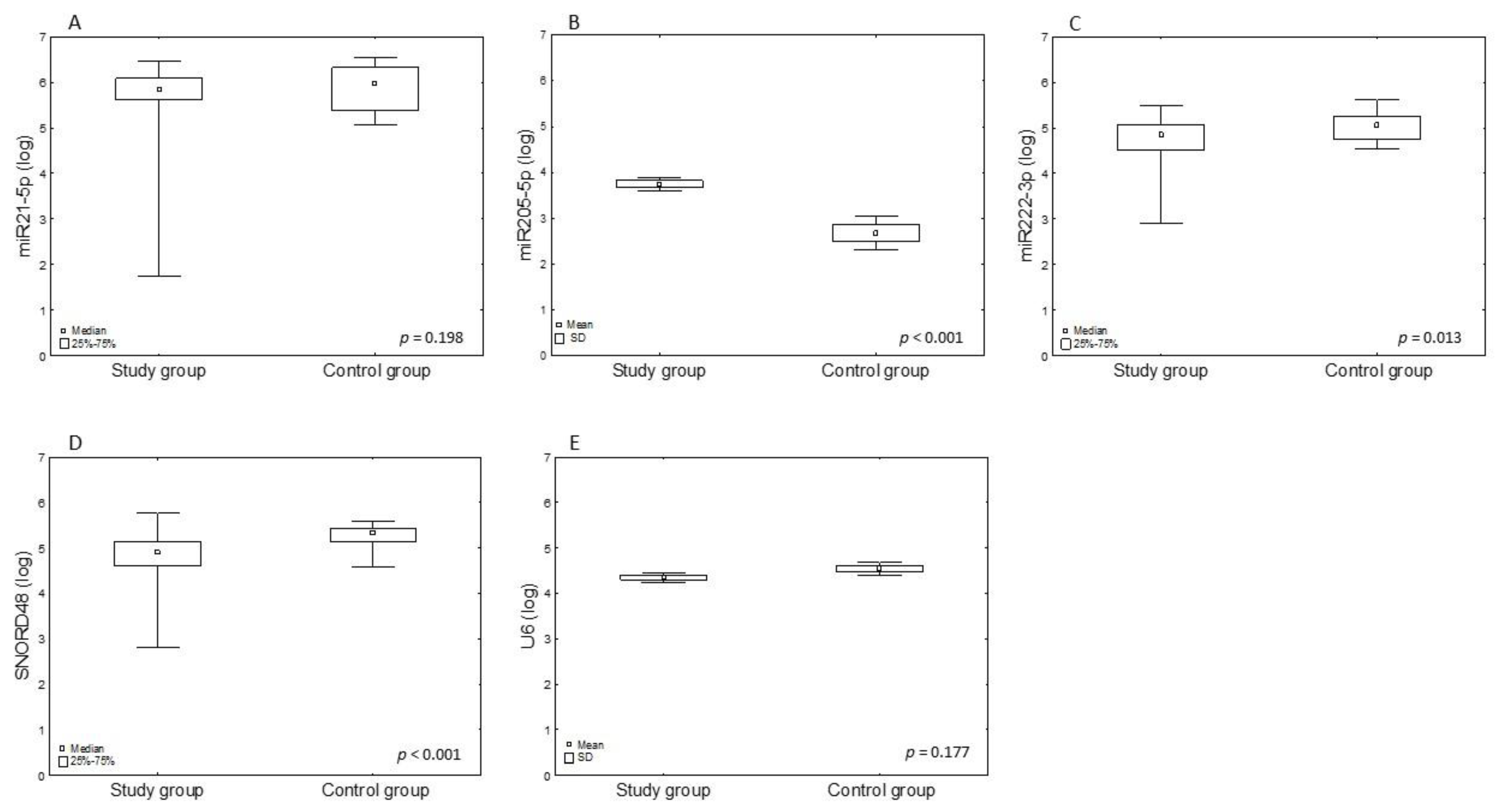
| miRNA | Study Group Mean ± SD | Control Group Mean ± SD | Fold Change | log2FC | p-Value 1 |
|---|---|---|---|---|---|
| miR-21-5 | 5.73 ±0.72 | 5.92 ± 0.49 | 0.97 | −0.05 | 0.198 |
| miR-205-5p | 3.74 ± 0.78 | 2.68 ± 0.71 | 1.4 | 0.48 | <0.001 |
| miR-222-3p | 4.74 ± 0.45 | 5.03 ± 0.32 | 0.94 | −0.09 | 0.022 |
| U6 | 4.35 ± 0.59 | 4.54 ± 0.33 | 0.96 | −0.06 | 0.177 |
| SNORD48 | 4.85 ± 0.44 | 5.3 ± 0.25 | 0.92 | −0.12 | <0.001 |
| OR (Odds Ratio) | 95% CI Lower | 95% CI Upper | p-Value | |
|---|---|---|---|---|
| Age | 1.12 | 1.06 | 1.20 | <0.001 |
| First period | 0.88 | 0.65 | 1.19 | 0.405 |
| Last menstrual period | 1.12 | 0.995 | 1.26 | 0.061 |
| Number of Births | 1.36 | 0.92 | 2.09 | 0.140 |
| At least one birth | 1.37 | 0.41 | 3.996 | 0.585 |
| More than one birth | 1.598 | 0.59 | 4.29 | 0.349 |
| More than two births | 3.29 | 0.87 | 21.54 | 0.125 |
| Number of Cesarean sections | 0.76 | 0.41 | 1.59 | 0.41 |
| At least one CS | 0.68 | 0.21 | 2.596 | 0.532 |
| More than one CS | 0.57 | 0.13 | 4.05 | 0.508 |
| Number of miscarriages | 0.61 | 0.31 | 1.24 | 0.14 |
| At least one miscarriage | 0.51 | 0.17 | 1.73 | 0.244 |
| BMI | 1.23 | 1.096 | 1.42 | 0.001 |
| Normal BMI (20–25) | 0.16 | 0.05 | 0.47 | <0.001 |
| Obesity (BMI >= 30) | 5.05 | 1.57 | 22.597 | 0.0137 |
| Hypertension | 7 | 2.18 | 31.33 | 0.003 |
| Smoking | 0.24 | 0.04 | 1.89 | 0.128 |
| miR-21-5p | 0.48 | 0.11 | 1.28 | 0.263 |
| miR-205-5p | 7.27 | 2.96 | 22.4 | <0.001 |
| miR-222-3p | 0.12 | 0.02 | 0.53 | 0.011 |
| U6 | 0.56 | 0.24 | 1.31 | 0.178 |
| SNORD48 | 0.04 | 0.008 | 0.24 | <0.001 |
| OR (Odds Ratio) | 95% CI Lower | 95% CI Upper | p-Value | |
|---|---|---|---|---|
| Age | 1.01 | 0.93 | 1.1 | 0.739 |
| Last menstrual period | 0.98 | 0.78 | 1.22 | 0.836 |
| BMI | 1.16 | 0.97 | 1.4 | 0.11 |
| Hypertension | 2.76 | 0.38 | 20.05 | 0.314 |
| miR-205-5 (log) | 6.05 | 1.87 | 19.59 | 0.003 |
| miR-222-3 (log) | 2.76 | 0.38 | 20.05 | 0.264 |
| SNORD48 (log) | 0.01 | 0.0003 | 0.47 | 0.018 |
| Absolute Expression (Copies/µL) | Control Group N = 19 1 | EIN N = 22 1 | G1 N = 30 1 | G2 N = 47 1 | G3 N = 12 1 | p-Value 2 |
|---|---|---|---|---|---|---|
| miR-21-5p | 938,736 (118,688–3,424,496) | 559,560 (177,664–2,858,160) | 630,336 (39,392–2,333,184) | 711,920 (115.04–2,591,056) | 860,536 (56.96–2,264,720) | 0.522 |
| miR-205-5p | 399.8 (32.5–7337) | 1590 (155.5–45,008) | 3367.2 (246.72–99,696) | 8067 (64.1–113,472) | 20,360 (1858.6–94,824) | <0.001 |
| miR-222-3p | 112,168 (35,104–421,912) | 74,548 (7464–224,816) | 83,720 (819.2–261,680) | 63,376 (4096.8–315,144) | 43,648 (13,256–117,560) | 0.058 |
| U6 | 47,264 (9926.4–119,536) | 19,032 (3761.6–113,680) | 16,482.4 (1283.84–134,320) | 16,656 (1034.72–3,321,312) | 133,008 (14,384–590,000) | <0.001 |
| SNORD48 | 214,400 (37,168–394,592) | 97,088 (18,800–304,272) | 59,520 (665.6–493,216) | 49,520 (11,449.6–459,344) | 227,200 (9840–581,552) | <0.001 |
| miRNA | OR | 95% CI Lower | 95% CI Upper | p-Value |
|---|---|---|---|---|
| miR-21-5 | 0.48 | 0.11 | 1.28 | 0.263 |
| miR-205-5p | 0.48 | 0.25 | 0.86 | 0.018 |
| miR-222-3p | 1.74 | 0.59 | 6.12 | 0.35 |
| U6 | 0.8 | 0.34 | 1.8 | 0.594 |
| SNORD48 | 1.48 | 0.51 | 4.76 | 0.492 |
| miRNA | OR | 95% CI Lower | 95% CI Upper | p-Value |
|---|---|---|---|---|
| miR-21-5p | 0.49 | 0.26 | 0.91 | 0.019 |
| miR-205-5p | 3.04 | 1.14 | 9.73 | 0.037 |
| miR-222-3p | 0.49 | 0.15 | 1.84 | 0.26 |
| U6 | 9.67 | 2.96 | 41.97 | <0.001 |
| SNORD48 | 5.69 | 1.09 | 38.01 | 0.053 |
| Absolute Expression (Copies/µL) | Age < 50, N = 17 1 | Age > 50, N = 94 1 | p-Value 2 |
|---|---|---|---|
| miR-21-5p | 764,944 (177,664–2,061,856) | 698,576 (56.96–2,858,160) | 0.509 |
| miR-205-5p | 1590 (155.5–113,472) | 5057.6 (64.08–611,824) | 0.041 |
| miR-222-3p | 42,850.7 (7464–223,040) | 78,764 (819.2–315,144) | 0.063 |
| U6 | 26,192 (3761.6–107,856) | 19,167.2 (1034.72–3,321,312) | >0.9 |
| SNORD48 | 77,744 (14,462.4–184,144) | 80,904 (665.6–581,552) | 0.6 |
| BMI | Study Group N = 111 1 | Control Group N = 19 1 | p-Value 2 |
|---|---|---|---|
| Normal | 14 (13%) | 9 (47%) | <0.001 |
| Overweight | 43 (39%) | 7 (37%) | |
| Obesity | 54 (49%) | 3 (16%) |
| BMI | EIN N = 22 1 | G1 N = 30 1 | G2 N = 47 1 | G3 N = 12 1 | p-Value 2 |
|---|---|---|---|---|---|
| Normal | 3 (14%) | 3 (10%) | 5 (10.6%) | 3 (18.2%) | 0.72 |
| Overweight | 10 (45%) | 11 (37%) | 16 (34%) | 6 (54.5%) | |
| Obesity | 9 (41%) | 16 (53%) | 26 (55.3%) | 3 (27.3%) |
| Absolute Expression (Copies/µL) | Normal N = 14 1 | Overweight N = 43 1 | Obesity N = 54 1 | p-Value 2 |
|---|---|---|---|---|
| miR-21-5p | 1,092,576 (197,296–2,264,720) | 694,496 (56.96–2,858,160) | 631,112 (114.08–2,591,056) | 0.088 |
| miR-205-5p | 6002 (259.5–96,128) | 4009.2 (155.52–113,472) | 4209.6 (64.08–611,824) | 0.869 |
| miR-222-3p | 69,112 (4460.8–315,144) | 69,752 (7088.8–261,680) | 66,988 (819.2–233,960) | 0.728 |
| U6 | 30,168 (5124.8–314,608) | 18,704 (1283.84–3,321,312) | 18,936 (1034.72–388,256) | 0.421 |
| SNORD48 | 94,232 (26,272–354,656) | 91,632 (665.6–581,552) | 64,048 (6481.6–519,488) | 0.473 |
| Absolute Expression (Copies/µL) | HA−, N = 48 1 | HA+, N = 63 1 | p-Value 2 |
|---|---|---|---|
| miR-21-5p | 747,056 (115.04–2,858,160) | 697 472 (56.96–2,591,056) | 0.875 |
| miR-205-5p | 3674.8 (64.08–113,472) | 4 379.6 (0–611,824) | 0.421 |
| miR-222-3p | 73,768 (4460.8–223,040) | 63,240 (819.2–315,144) | 0.554 |
| U6 | 19,104 (1283.84–590,000) | 19,390.4 (1034.72–3,321,312) | 0.52 |
| SNORD48 | 83,224 (665.6–354,656) | 77,488 (6481.6–581,552) | 0.882 |
| Absolute Expression (Copies/µL) | DM−, N = 90 1 | DM+, N = 21 1 | p-Value 2 |
|---|---|---|---|
| miR-21-5p | 631,728 (56.96–2,858,160) | 799,984 (161,648–2,210,368) | 0.579 |
| miR-205-5p | 3855.2 (64.08–611,824) | 8424 (692.24–92,712) | 0.164 |
| miR-222-3p | 71,460 (4096.8–315,144) | 59 248 (819.2–261,680) | 0.44 |
| U6 | 18,816 (1034.72–3,321,312) | 24,624 (3926.4–134,320) | 0.472 |
| SNORD48 | 86,920 (665.6–581,552) | 58,704 (6481.6–493,216) | 0.401 |
| Absolute Expression (Copies/µL) | Hypothyroidism−, N = 88 1 | Hypothyroidism+, N = 23 1 | p-Value 2 |
|---|---|---|---|
| miR-21-5p | 680,912 (56.96–2,858,160) | 740,480 (166,528–2,210,368) | 0.746 |
| miR-205-5p | 3894.8 (64.08–108,664) | 4549.6 (692.24–611,824) | 0.16 |
| miR-222-3p | 78,140 (4096.8–315,144) | 49,504 (819.2–224,816) | 0.192 |
| U6 | 20,520 (1034.72–3,321,312) | 18,704 (1964.8–134,320) | 0.859 |
| SNORD48 | 83,056 (665.6–581,552) | 62,112 (6481.6–493,216) | 0.178 |
| miRNA | GeNorm Stability | NormFinder Stability | BestKeeper Stability | Average Stability |
|---|---|---|---|---|
| U6 | 0.29 | 0.07 | 0.27 | 0.21 |
| SNORD48 | 0.29 | 0.05 | 0.17 | 0.17 |
| U6+SNORD48 | 0.17 | 0.04 | 0.11 | 0.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bogaczyk, A.; Potocka, N.; Paszek, S.; Skrzypa, M.; Zuchowska, A.; Kośny, M.; Kluz, M.; Zawlik, I.; Kluz, T. Absolute Quantification of Selected microRNAs Expression in Endometrial Cancer by Digital PCR. Int. J. Mol. Sci. 2024, 25, 3286. https://doi.org/10.3390/ijms25063286
Bogaczyk A, Potocka N, Paszek S, Skrzypa M, Zuchowska A, Kośny M, Kluz M, Zawlik I, Kluz T. Absolute Quantification of Selected microRNAs Expression in Endometrial Cancer by Digital PCR. International Journal of Molecular Sciences. 2024; 25(6):3286. https://doi.org/10.3390/ijms25063286
Chicago/Turabian StyleBogaczyk, Anna, Natalia Potocka, Sylwia Paszek, Marzena Skrzypa, Alina Zuchowska, Michał Kośny, Marta Kluz, Izabela Zawlik, and Tomasz Kluz. 2024. "Absolute Quantification of Selected microRNAs Expression in Endometrial Cancer by Digital PCR" International Journal of Molecular Sciences 25, no. 6: 3286. https://doi.org/10.3390/ijms25063286
APA StyleBogaczyk, A., Potocka, N., Paszek, S., Skrzypa, M., Zuchowska, A., Kośny, M., Kluz, M., Zawlik, I., & Kluz, T. (2024). Absolute Quantification of Selected microRNAs Expression in Endometrial Cancer by Digital PCR. International Journal of Molecular Sciences, 25(6), 3286. https://doi.org/10.3390/ijms25063286






