Novel Lipid Nanoparticles Stable and Efficient for mRNA Transfection to Antigen-Presenting Cells
Abstract
1. Introduction
2. Results and Discussion
2.1. Preparation and Characterization of DMKD-PS
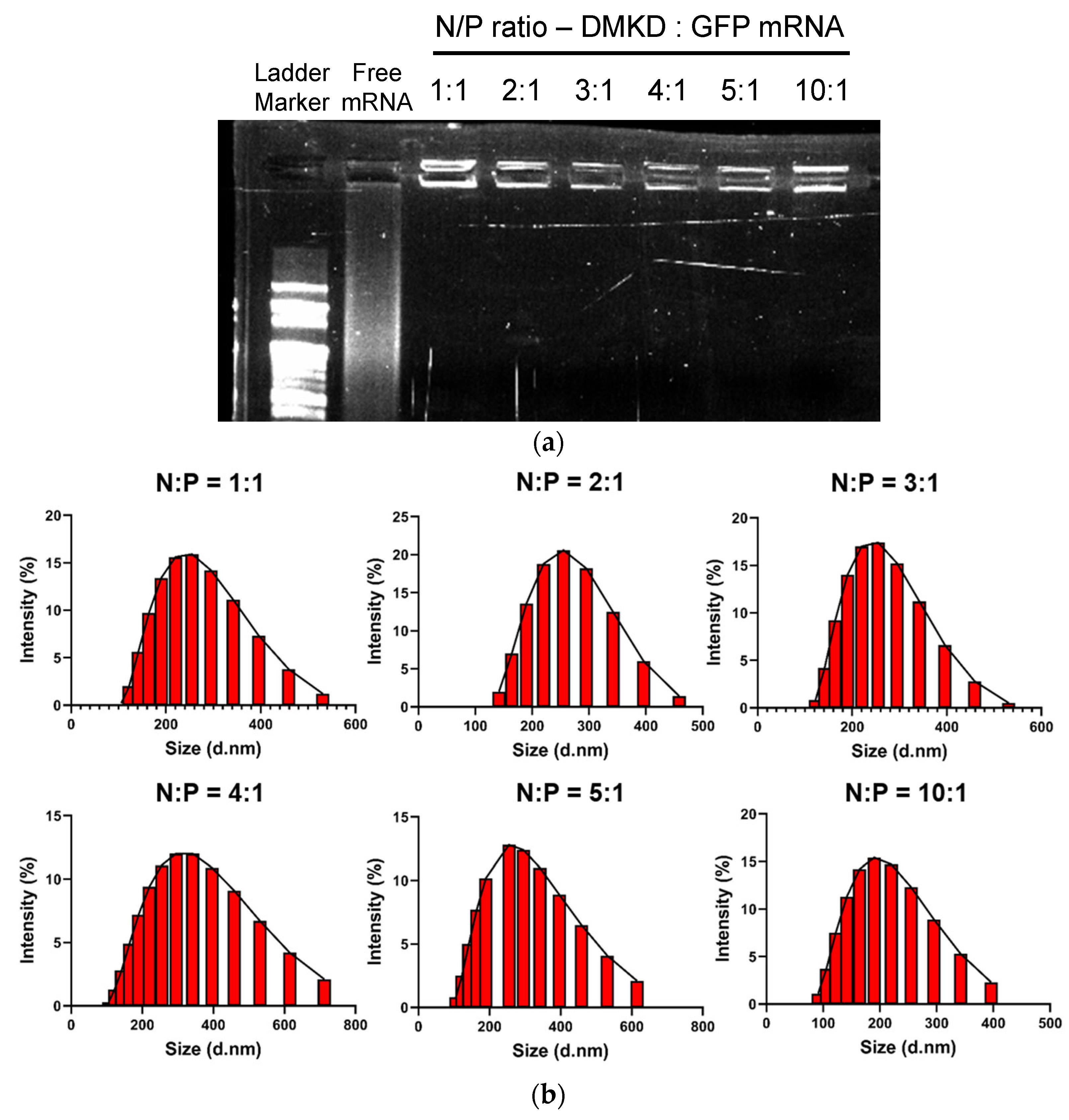
2.2. Optimization and Analyses of DMKD-PS Complexed with mRNA
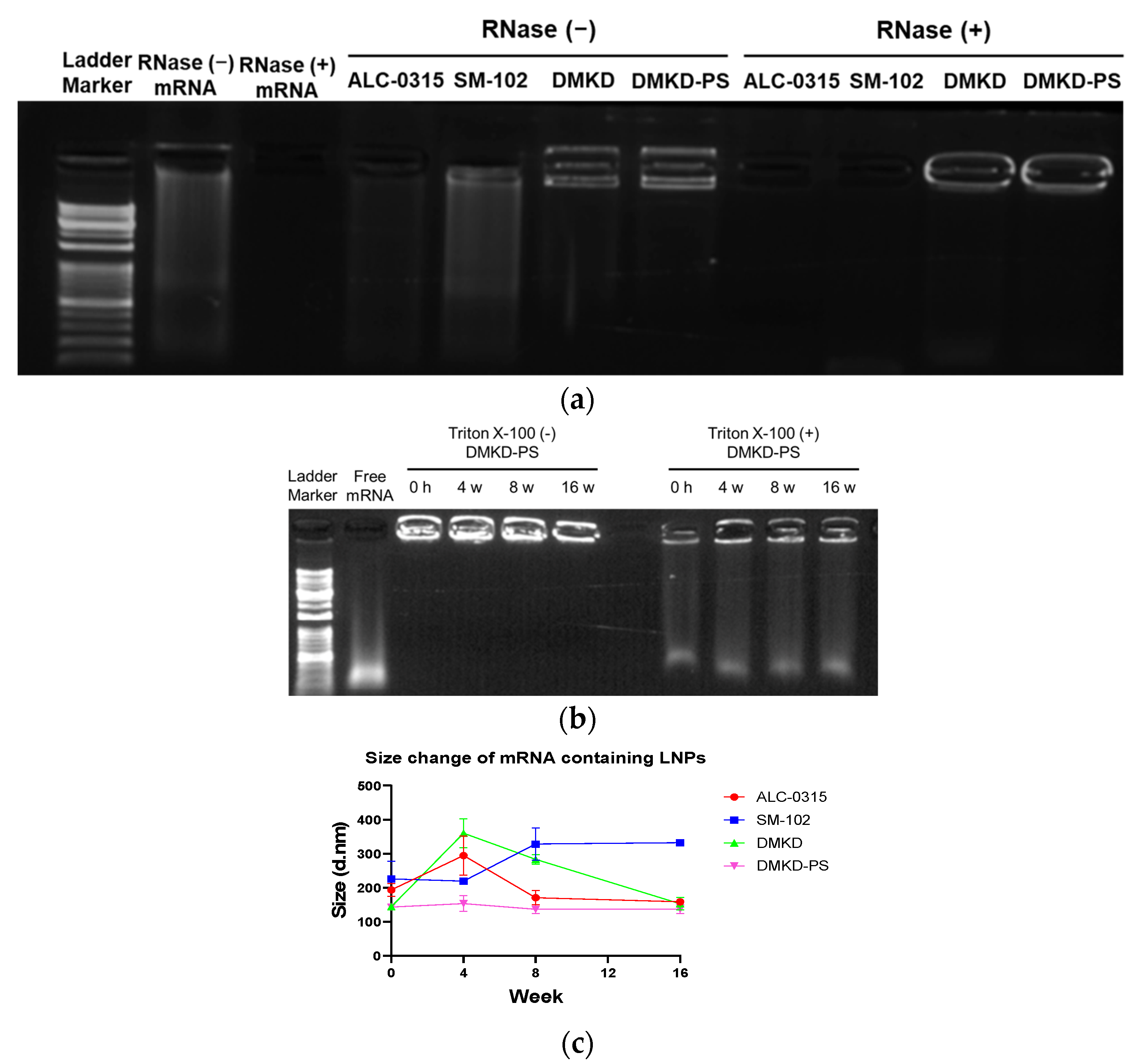
2.3. Physicochemical Properties of DMKD-PS
2.4. mRNA Protection Analyses
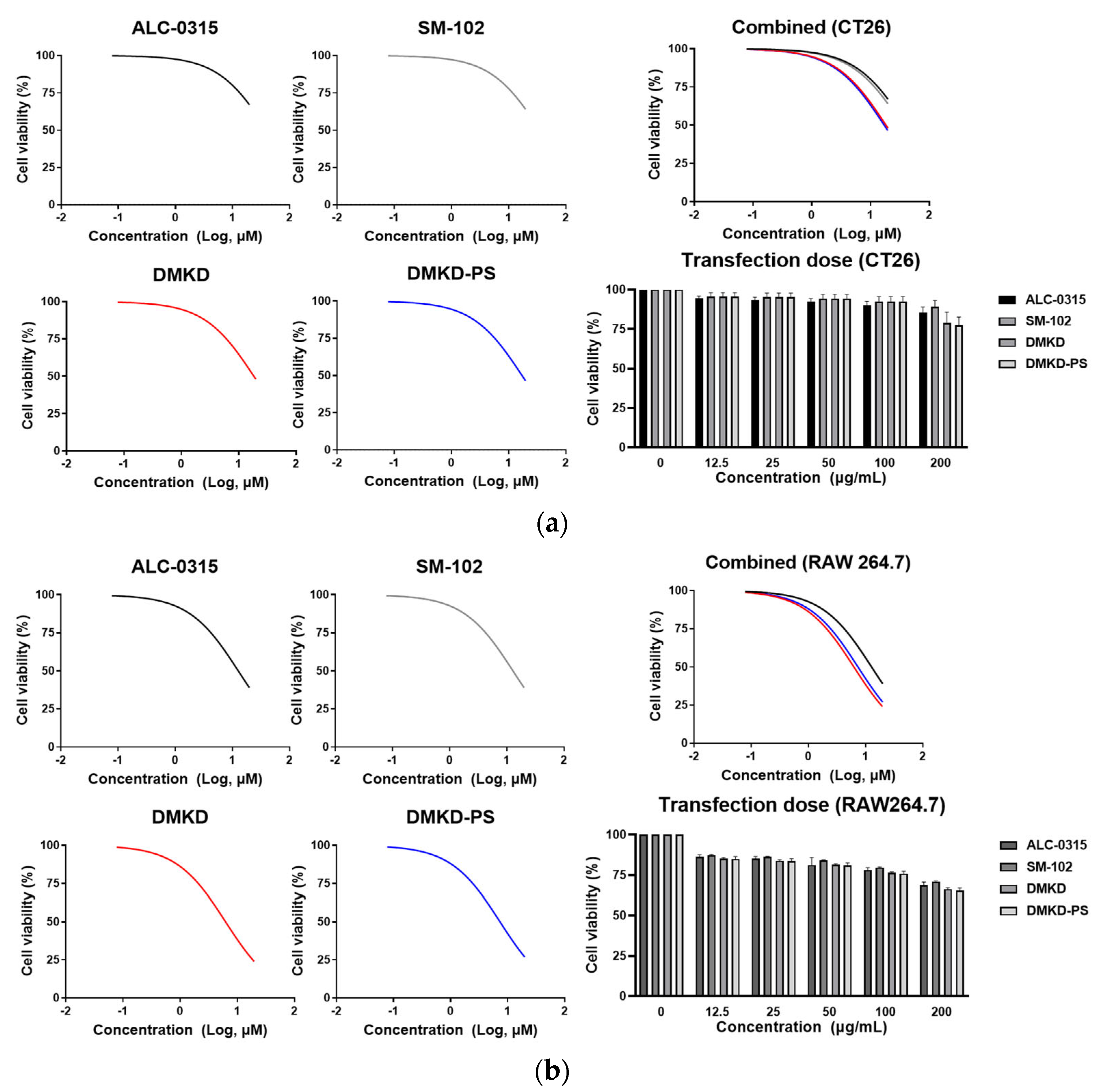
2.5. Inherent Cytotoxicity of DMKD-PS
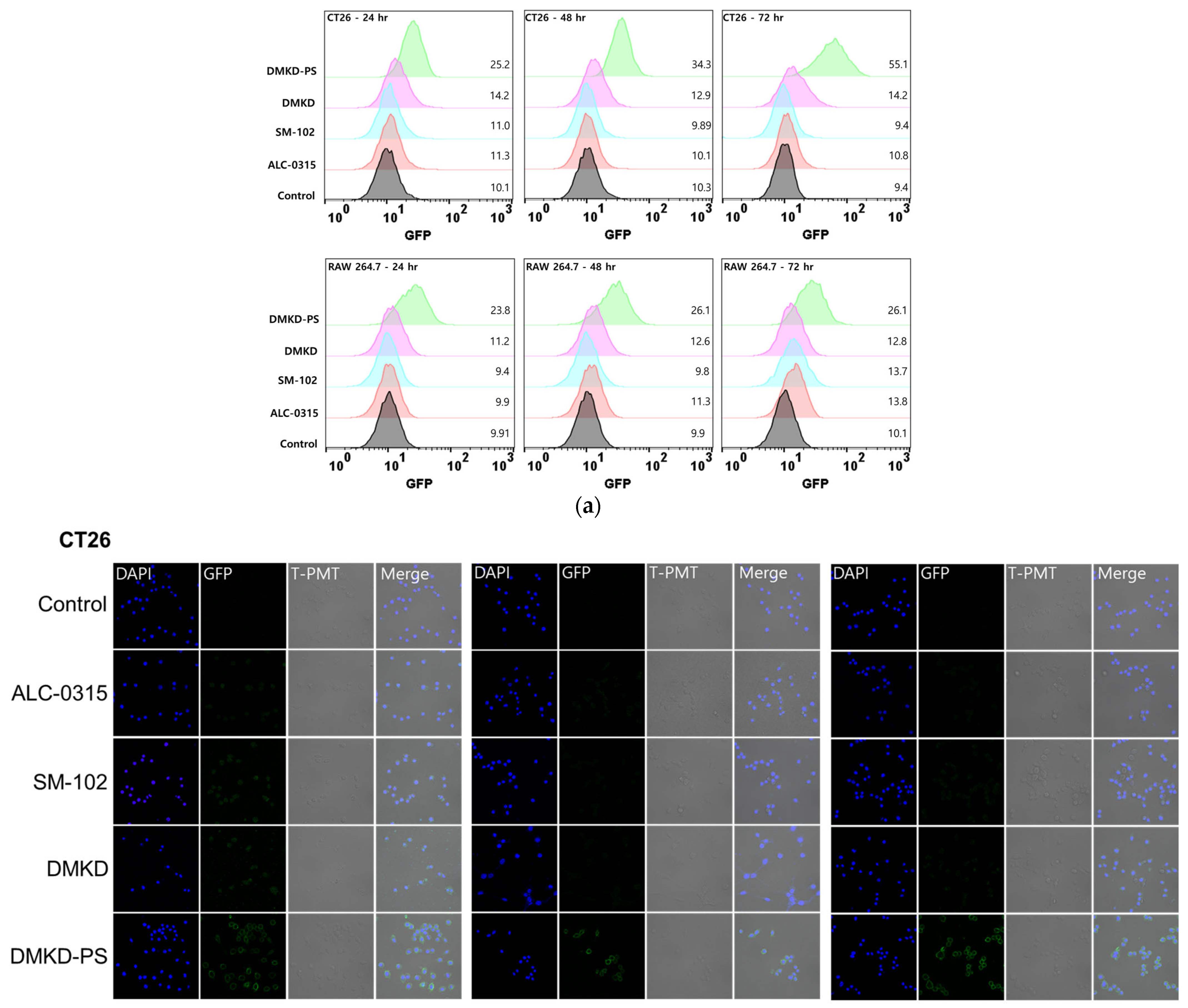
2.6. In-Vitro GFP mRNA Transfection of DMKD-PS
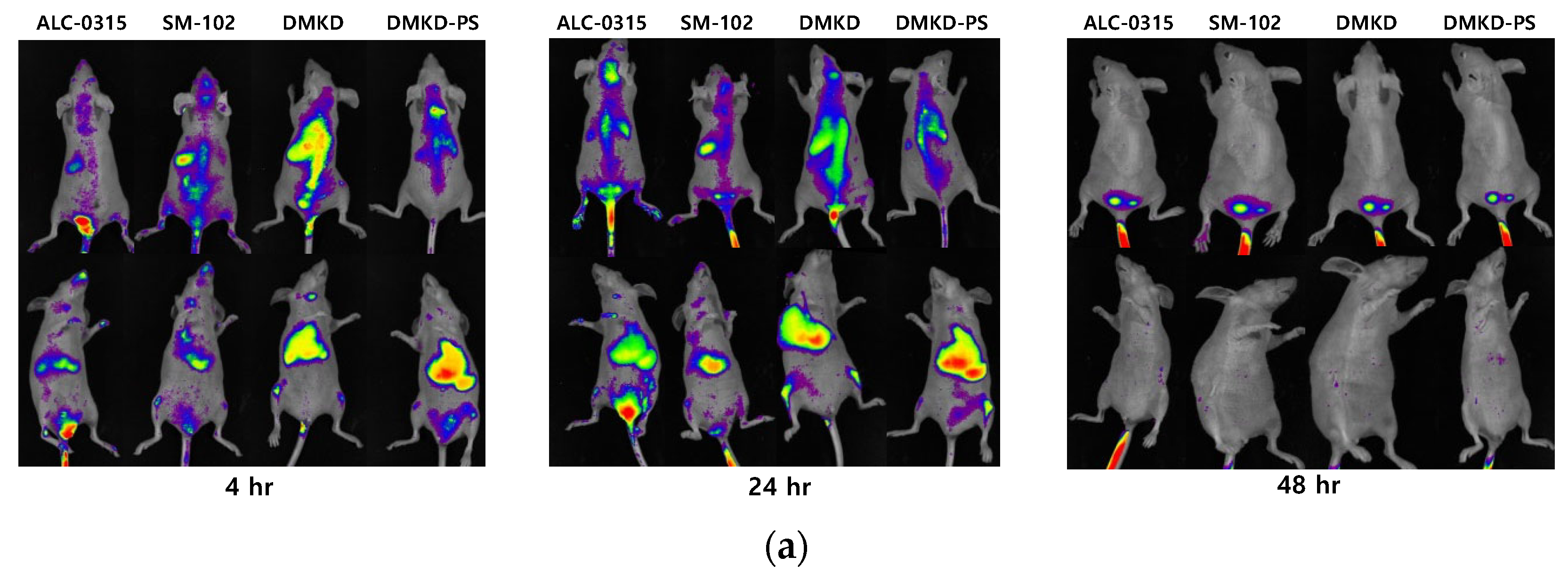
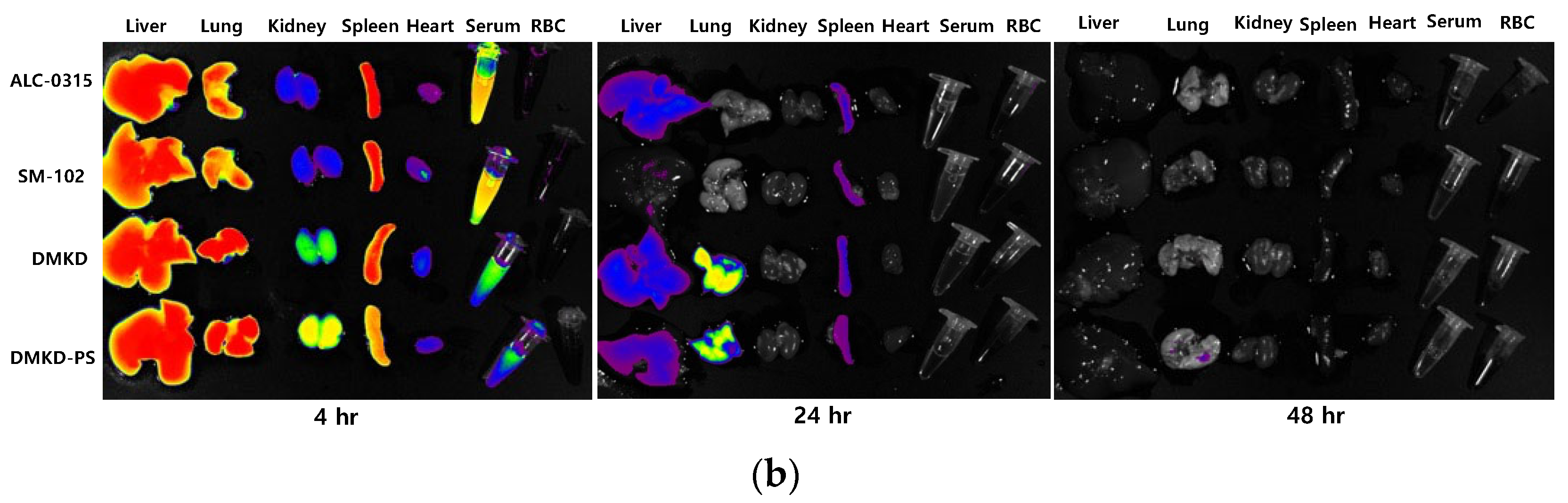
2.7. In-Vivo Biodistribution
3. Materials and Methods
3.1. Materials
3.2. Cell Lines and Cell Culture
3.3. Synthesis of Enhanced Green Fluorescence Protein mRNA
3.4. Preparation of DMKD-PS
3.5. Transmission Electron Microscopy Analysis
3.6. mRNA Protection Analysis
3.7. Cytotoxicity Assay
3.8. In Vitro GFP mRNA Transfection
3.8.1. Flow Cytometry Analysis
3.8.2. Confocal Microscopy Analysis
3.9. In Vivo Biodistribution Analysis
4. Conclusions
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karam, M.; Daoud, G. mRNA vaccines: Past, present, future. Asian J. Pharm. Sci. 2022, 17, 491–522. [Google Scholar] [CrossRef]
- Mascola, J.R.; Fauci, A.S. Novel vaccine technologies for the 21st century. Nat. Rev. Immunol. 2020, 20, 87–88. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.D.; Reidenbach, D.; Salomon, N.; Sahin, U.; Türeci, Ö.; Vormehr, M.; Kranz, L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer 2021, 20, 69. [Google Scholar] [CrossRef] [PubMed]
- Whitley, J.; Zwolinski, C.; Denis, C.; Maughan, M.; Hayles, L.; Clarke, D.; Snare, M.; Liao, H.; Chiou, S.; Marmura, T.; et al. Development of mRNA manufacturing for vaccines and therapeutics: mRNA platform requirements and development of a scalable production process to support early phase clinical trials. Transl. Res. 2022, 242, 38–55. [Google Scholar] [CrossRef] [PubMed]
- Hengelbrock, A.; Schmidt, A.; Helgers, H.; Vetter, F.L.; Strube, J. Scalable mRNA Machine for Regulatory Approval of Variable Scale between 1000 Clinical Doses to 10 Million Manufacturing Scale Doses. Processes 2023, 11, 745. [Google Scholar] [CrossRef]
- Kis, Z.; Kontoravdi, C.; Shattock, R.; Shah, N. Resources, Production Scales and Time Required for Producing RNA Vaccines for the Global Pandemic Demand. Vaccines 2021, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Conry, R.M.; LoBuglio, A.F.; Wright, M.; Sumerel, L.; Pike, M.J.; Johanning, F.; Benjamin, R.; Lu, D.; Curiel, D.T. Characterization of a messenger RNA polynucleotide vaccine vector. Cancer Res. 1995, 55, 1397–1400. [Google Scholar] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef]
- Weng, Y.; Li, C.; Yang, T.; Hu, B.; Zhang, M.; Guo, S.; Xiao, H.; Liang, X.-J.; Huang, Y. The challenge and prospect of mRNA therapeutics landscape. Biotechnol. Adv. 2020, 40, 107534. [Google Scholar] [CrossRef]
- Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [Google Scholar] [CrossRef]
- Cheng, F.; Wang, Y.; Bai, Y.; Liang, Z.; Mao, Q.; Liu, D.; Wu, X.; Xu, M. Research Advances on the Stability of mRNA Vaccines. Viruses 2023, 15, 668. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Roni, M.A. Challenges of Storage and Stability of mRNA-Based COVID-19 Vaccines. Vaccines 2021, 9, 1033. [Google Scholar] [CrossRef] [PubMed]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef] [PubMed]
- Kauffman, K.J.; Webber, M.J.; Anderson, D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release 2016, 240, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Rosenecker, J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017, 24, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Verma, J.; Warsame, C.; Seenivasagam, R.K.; Katiyar, N.K.; Aleem, E.; Goel, S. Nanoparticle-mediated cancer cell therapy: Basic science to clinical applications. Cancer Metastasis Rev. 2023, 42, 601–627. [Google Scholar] [CrossRef]
- Ganguly, S.; Neelam; Grinberg, I.; Margel, S. Layer by layer controlled synthesis at room temperature of tri-modal (MRI, fluorescence and CT) core/shell superparamagnetic IO/human serum albumin nanoparticles for diagnostic applications. Polym. Adv. Technol. 2021, 32, 3909–3921. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Han, X.; Zhang, H.; Butowska, K.; Swingle, K.L.; Alameh, M.-G.; Weissman, D.; Mitchell, M.J. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 2021, 12, 7233. [Google Scholar] [CrossRef]
- Maugeri, M.; Nawaz, M.; Papadimitriou, A.; Angerfors, A.; Camponeschi, A.; Na, M.; Hölttä, M.; Skantze, P.; Johansson, S.; Sundqvist, M.; et al. Linkage between endosomal escape of LNP-mRNA and loading into EVs for transport to other cells. Nat. Commun. 2019, 10, 4333. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, P.; Franke, C.; Stöter, M.; Höijer, A.; Bartesaghi, S.; Sabirsh, A.; Lindfors, L.; Arteta, M.Y.; Dahlén, A.; Bak, A.; et al. Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale. J. Cell Biol. 2022, 221, e202110137. [Google Scholar] [CrossRef]
- Schlich, M.; Palomba, R.; Costabile, G.; Mizrahy, S.; Pannuzzo, M.; Peer, D.; Decuzzi, P. Cytosolic delivery of nucleic acids: The case of ionizable lipid nanoparticles. Bioeng. Transl. Med. 2021, 6, e10213. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Lu, Z.-R. Structure and Function of Cationic and Ionizable Lipids for Nucleic Acid Delivery. Pharm. Res. 2023, 40, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, J.J.; Anchordoquy, T.J. Questioning the Use of PEGylation for Drug Delivery. Drug Deliv. Transl. Res. 2013, 3, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Lange, Y.; Tabei, S.M.; Ye, J.; Steck, T.L. Stability and stoichiometry of bilayer phospholipid-cholesterol complexes: Relationship to cellular sterol distribution and homeostasis. Biochemistry 2013, 52, 6950–6959. [Google Scholar] [CrossRef]
- Crommelin, D.J.A.; Anchordoquy, T.J.; Volkin, D.B.; Jiskoot, W.; Mastrobattista, E. Addressing the Cold Reality of mRNA Vaccine Stability. J. Pharm. Sci. 2021, 110, 997–1001. [Google Scholar] [CrossRef]
- McMahon, M.E.; Abbott, A.; Babayan, Y.; Carhart, J.; Chen, C.-w.; Debie, E.; Fu, M.; Hoaglund-Hyzer, C.; Lennard, A.; Li, H.; et al. Considerations for Updates to ICH Q1 and Q5C Stability Guidelines: Embracing Current Technology and Risk Assessment Strategies. AAPS J. 2021, 23, 107. [Google Scholar] [CrossRef]
- Segawa, K.; Nagata, S. An Apoptotic ‘Eat Me’ Signal: Phosphatidylserine Exposure. Trends Cell Biol. 2015, 25, 639–650. [Google Scholar] [CrossRef]
- Kim, H.S.; Song, I.H.; Kim, J.C.; Kim, E.J.; Jang, D.O.; Park, Y.S. In vitro and in vivo gene-transferring characteristics of novel cationic lipids, DMKD (O,O′-dimyristyl-N-lysyl aspartate) and DMKE (O,O′-dimyristyl-N-lysyl glutamate). J. Control. Release 2006, 115, 234–241. [Google Scholar] [CrossRef]
- Kim, H.S.; Moon, J.; Kim, K.S.; Choi, M.M.; Lee, J.E.; Heo, Y.; Cho, D.H.; Jang, D.O.; Park, Y.S. Gene-Transferring Efficiencies of Novel Diamino Cationic Lipids with Varied Hydrocarbon Chains. Bioconjug. Chem. 2004, 15, 1095–1101. [Google Scholar] [CrossRef] [PubMed]
- Hope, M.J.; Bally, M.B.; Webb, G.; Cullis, P.R. Production of large unilamellar vesicles by a rapid extrusion procedure. Characterization of size distribution, trapped volume and ability to maintain a membrane potential. Biochim. Biophys. Acta (BBA)-Biomembr. 1985, 812, 55–65. [Google Scholar] [CrossRef]
- Trollmann, M.F.W.; Böckmann, R.A. mRNA lipid nanoparticle phase transition. Biophys. J. 2022, 121, 3927–3939. [Google Scholar] [CrossRef] [PubMed]
- Hald Albertsen, C.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Lee, R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016, 99, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Lechanteur, A.; Sanna, V.; Duchemin, A.; Evrard, B.; Mottet, D.; Piel, G. Cationic Liposomes Carrying siRNA: Impact of Lipid Composition on Physicochemical Properties, Cytotoxicity and Endosomal Escape. Nanomaterials 2018, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, K.; Jääskeläinen, I.; Syrjänen, K.; Urtti, A.; Syrjänen, S. Comparison of cell proliferation and toxicity assays using two cationic liposomes. Pharm. Res. 1994, 11, 1127–1131. [Google Scholar] [CrossRef]
- Cui, S.; Wang, Y.; Gong, Y.; Lin, X.; Zhao, Y.; Zhi, D.; Zhou, Q.; Zhang, S. Correlation of the cytotoxic effects of cationic lipids with their headgroups. Toxicol. Res. 2018, 7, 473–479. [Google Scholar] [CrossRef]
- Moradian, H.; Roch, T.; Lendlein, A.; Gossen, M. mRNA Transfection-Induced Activation of Primary Human Monocytes and Macrophages: Dependence on Carrier System and Nucleotide Modification. Sci. Rep. 2020, 10, 4181. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, L.; Zhu, W.; Guo, R.; Sun, H.; Chen, X.; Deng, N. Barriers and Strategies of Cationic Liposomes for Cancer Gene Therapy. Mol. Ther. Methods Clin. Dev. 2020, 18, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Li, S.D.; Huang, L. Nanoparticles evading the reticuloendothelial system: Role of the supported bilayer. Biochim. Biophys. Acta 2009, 1788, 2259–2266. [Google Scholar] [CrossRef] [PubMed]
- Pattipeiluhu, R.; Arias-Alpizar, G.; Basha, G.; Chan, K.Y.T.; Bussmann, J.; Sharp, T.H.; Moradi, M.-A.; Sommerdijk, N.; Harris, E.N.; Cullis, P.R.; et al. Anionic Lipid Nanoparticles Preferentially Deliver mRNA to the Hepatic Reticuloendothelial System. Adv. Mater. 2022, 34, 2201095. [Google Scholar] [CrossRef] [PubMed]


| mRNA | Size (a,b) (nm) | Polydispersity Index (PDI) | ζ-Potential (mV) | |
|---|---|---|---|---|
| ALC-0315 | - | 193.4 ± 15.6 | 0.424 ± 0.054 | −1.2 ± 1.3 |
| + | 143.7 ± 3.5 (▼) | 0.290 ± 0.025 (▼) | −4.3 ± 0.5 (▼) | |
| SM-102 | - | 226.0 ± 41.9 | 0.497 ± 0.011 | 5.6 ± 0.2 |
| + | 158.6 ± 5.4 (▼) | 0.415 ± 0.021 (▼) | −8.1 ± 0.7 (▼) | |
| DMKD | - | 144.5 ± 3.0 | 0.325 ± 0.010 | 29.9 ± 2.3 |
| + | 206.7 ± 3.5 (▲) | 0.367 ± 0.012(▲) | 27.7 ± 1.3 (▼) | |
| DMKD-PS | - | 134.1 ± 1.0 | 0.316 ± 0.026 | 42.8 ± 1.0 |
| + | 150.7 ± 8.6 (▲) | 0.223 ± 0.006 (▼) | 26.1 ± 0.8 (▼) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choi, K.C.; Lee, D.H.; Lee, J.W.; Lee, J.S.; Lee, Y.K.; Choi, M.J.; Jeong, H.Y.; Kim, M.W.; Lee, C.-G.; Park, Y.S. Novel Lipid Nanoparticles Stable and Efficient for mRNA Transfection to Antigen-Presenting Cells. Int. J. Mol. Sci. 2024, 25, 1388. https://doi.org/10.3390/ijms25031388
Choi KC, Lee DH, Lee JW, Lee JS, Lee YK, Choi MJ, Jeong HY, Kim MW, Lee C-G, Park YS. Novel Lipid Nanoparticles Stable and Efficient for mRNA Transfection to Antigen-Presenting Cells. International Journal of Molecular Sciences. 2024; 25(3):1388. https://doi.org/10.3390/ijms25031388
Chicago/Turabian StyleChoi, Kang Chan, Do Hyun Lee, Ji Won Lee, Jin Suk Lee, Yeon Kyung Lee, Moon Jung Choi, Hwa Yeon Jeong, Min Woo Kim, Chang-Gun Lee, and Yong Serk Park. 2024. "Novel Lipid Nanoparticles Stable and Efficient for mRNA Transfection to Antigen-Presenting Cells" International Journal of Molecular Sciences 25, no. 3: 1388. https://doi.org/10.3390/ijms25031388
APA StyleChoi, K. C., Lee, D. H., Lee, J. W., Lee, J. S., Lee, Y. K., Choi, M. J., Jeong, H. Y., Kim, M. W., Lee, C.-G., & Park, Y. S. (2024). Novel Lipid Nanoparticles Stable and Efficient for mRNA Transfection to Antigen-Presenting Cells. International Journal of Molecular Sciences, 25(3), 1388. https://doi.org/10.3390/ijms25031388







